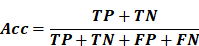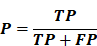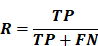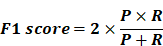Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(16):4236-4249. doi:10.7150/ijms.120138 This issue Cite
Research Paper
Real-Time Ultrasound Diagnosis of Developmental Dysplasia of the Hip Using an Attention-Enhanced YOLOv11 Model
1. Department of Medical Information, Chung Shan Medical University, Taichung 402201, Taiwan.
2. Informatics Office Technology, Chung Shan Medical University Hospital, Taichung 402201, Taiwan.
3. Program for Precision Health and Intelligent Medicine, Graduate School of Advanced Technology, National Taiwan University, Taipei 106319, Taiwan.
4. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung 402202, Taiwan.
5. Department of Golden-Ager Industry Management, Chaoyang University of Technology, Taichung 413310, Taiwan.
6. Department of Orthopedic Surgery, Changhua Christian Hospital, Changhua 500209, Taiwan.
7. Department of Medical Imaging and Radiology, Shu-Zen Junior College of Medicine and Management, Kaohsiung 821, Taiwan.
Received 2025-6-23; Accepted 2025-9-15; Published 2025-10-1
Abstract
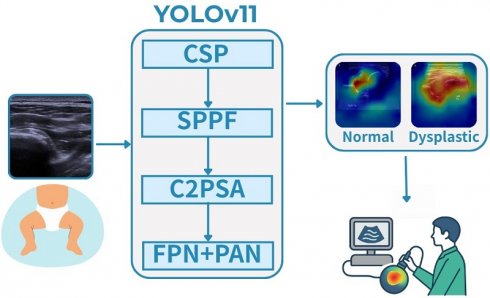
Developmental dysplasia of the hip (DDH) is a common pediatric orthopedic disorder that can lead to lifelong disability if undetected. Ultrasound is the primary diagnostic modality but is subject to operator dependence and inter-observer variability. To address this challenge, we propose an attention-enhanced YOLOv11 framework for automated DDH classification. A dataset of 6,075 hip ultrasound images was preprocessed with augmentation and dimensionality reduction via UMAP. The model integrates Cross-Stage Partial (CSP) modules and C2PSA spatial attention to improve feature extraction, and was trained using Focal Loss and IoU Loss. It achieved 95.05% accuracy with an inference speed of 11.5 ms per image, substantially outperforming MobileNetV3 and ShuffleNetV2. Grad-CAM visualizations confirmed that the model consistently attends to the acetabular roof and femoral head, landmarks central to Graf classification, thereby enhancing clinical interpretability. These findings demonstrate that the proposed framework combines technical robustness with clinical relevance. Future work will emphasize multi-center validation and multimodal integration to ensure generalizability and support widespread clinical adoption.
Keywords: Developmental Dysplasia of the Hip (DDH), Ultrasound Imaging, Deep Learning, YOLOv11, Medical Image Classification, Automated Diagnosis.
1. Introduction
Developmental Dysplasia of the Hip (DDH) is a prevalent pediatric orthopedic disorder that impairs hip joint development in infants [1]. If undiagnosed or untreated, DDH may progress to hip joint instability, impaired mobility, and early-onset osteoarthritis, ultimately diminishing quality of life. Timely and accurate diagnosis is therefore essential to enable early intervention and prevent long-term complications. Ultrasound, particularly the Graf classification system, remains the standard diagnostic modality, as it assesses hip joint alignment through key anatomical angles [2]. However, this approach requires substantial clinical expertise, rendering it highly susceptible to inter-observer variability [3]. Moreover, variations in image quality and infant positioning further complicate interpretation, often leading to diagnostic inconsistencies [4]. Given the high prevalence of DDH and the limitations of manual ultrasound interpretation, there is a pressing need for an automated, deep learning-based classification system to enhance diagnostic accuracy and efficiency.
Recent advances in deep learning have shown substantial promise in automating medical image analysis [5]. Convolutional Neural Networks (CNNs), particularly architectures such as U-Net and Fully Convolutional Networks (FCNs), have achieved high accuracy in segmentation tasks across diverse medical imaging modalities [6]. Nevertheless, most existing deep learning approaches for DDH diagnosis have focused on segmentation rather than direct classification, and their success often relies on large, expertly annotated datasets that are both costly and time-consuming to obtain [7]. Furthermore, CNN-based models must contend with challenges such as dataset imbalance, variability in image quality, and limited availability of labeled samples [8]. To address these challenges, this study introduces an automated DDH classification framework based on YOLOv11, a state-of-the-art real-time object detection model [9]. Unlike conventional CNN-based segmentation networks, YOLOv11 provides an efficient and unified approach to simultaneously detecting and classifying DDH-related hip structures in ultrasound images, thereby improving diagnostic accuracy and consistency [10]. In addition, the framework leverages data augmentation to mitigate class imbalance and employs UMAP for dataset visualization [11]. By integrating advanced attention mechanisms and optimized convolutional layers, the proposed method enhances both feature representation and classification reliability.
In summary, this study makes the following contributions. First, we propose an automated DDH classification framework based on YOLOv11, optimized for real-time ultrasound diagnosis. Second, we introduce a preprocessing pipeline that combines UMAP-based visualization with data augmentation to alleviate class imbalance and enhance model robustness. Third, we integrate advanced spatial attention mechanisms (C2PSA) into the YOLOv11 architecture to strengthen anatomical feature recognition. Finally, we validate the proposed model against lightweight benchmark networks, demonstrating superior accuracy and inference speed suitable for clinical application.
The remainder of this paper is organized as follows. Section 2 reviews related work on DDH diagnosis and deep learning in medical imaging. Section 3 describes the proposed methodology, including dataset acquisition, preprocessing, and model architecture. Section 4 presents experimental results and performance analyses. Section 5 discusses the findings and their clinical implications, and Section 6 concludes the study with future research directions.
2. Related Work
2.1 Ultrasound-Based DDH Diagnosis and Graf Classification
Due to the inherent challenges of accurately delineating the proximal femur and acetabular margin in neonatal hip X-ray imaging [12], ultrasound has become the preferred modality for diagnosing developmental dysplasia of the hip (DDH) [13]. Although its role in large-scale screening programs remains debated [14], ultrasound continues to be widely adopted across Europe [15]. Several diagnostic approaches have been developed, including the Graf, Harcke, Terjesen, and Suzuki methods, with the Graf technique being the most widely accepted for screening, diagnosis, and treatment monitoring of DDH [16].
The Graf method relies on predefined anatomical landmarks within the hip joint, identifying five critical points: the iliac outer edge, the lower limb of the ilium, the transition point where the bony acetabular roof curves toward the ilium, the center of the labrum, and the femoral head [17]. By constructing three intersecting lines—the baseline, the bony roof line, and the soft tissue covering line—two key angles can be measured: the α angle (bony roof angle) and the β angle (cartilage roof angle) [18, 19]. These parameters enable the classification of neonatal hips into distinct categories:
- Type I (Normal Hip): α > 60°
- Type IIa/IIb (Immature Hip): 50° ≤ α ≤ 59°
- Type IIc/D (Dysplastic Hip): α < 50°
- Type III/IV (Dislocated Hip): α < 43° [20-22].
Ultrasound imaging offers significant advantages, including ease of operation, reproducibility, and the absence of ionizing radiation [23]. However, the accuracy of the Graf method depends heavily on strict adherence to standardized imaging protocols [24]. Failure to capture the standard plane can result in measurement errors and subjective interpretation, thereby reducing the reliability of α and β angle assessments [25, 26]. These limitations underscore the importance of developing automated image analysis techniques to improve diagnostic precision and consistency.
2.2 Deep Learning for Medical Image Analysis
Deep learning has revolutionized medical image analysis, demonstrating remarkable performance in segmentation, classification, and anomaly detection tasks across diverse clinical domains [27-29]. While the concept of artificial neural networks (ANNs) originated in 1943 [30, 31], the advent of deep learning in 2006 enabled the development of multi-layer network architectures with enhanced representational capacity [32]. Among these, Convolutional Neural Networks (CNNs) have driven major breakthroughs in applications such as disease diagnosis, semantic segmentation, and object detection [33-35].
CNN-based approaches have become the dominant paradigm in medical imaging [36, 37], leveraging hierarchical feature extraction to achieve high precision in identifying and localizing pathological structures [38]. However, these methods often require large-scale, expert-labeled datasets, making data annotation labor-intensive and time-consuming [39-41]. This challenge highlights the need for scalable, automated, and real-time classification systems that can reduce reliance on manual labeling while maintaining diagnostic accuracy [42, 43].
2.3 Deep Learning for DDH Classification
Convolutional Neural Networks (CNNs) have shown encouraging results in ultrasound-based DDH classification, particularly for segmenting femoral head and acetabular structures [44]. Early studies primarily employed conventional machine learning techniques; however, the introduction of Fully Convolutional Networks (FCNs), U-Net, and transformer-based architectures has substantially improved segmentation accuracy in recent years [45, 46]. Despite these advances, most existing methods continue to emphasize segmentation rather than direct classification of DDH severity.
For reliable classification, access to sufficiently large and high-quality labeled datasets is essential [47]. Yet, training on imbalanced datasets frequently results in biased predictions, as underrepresented classes are inadequately learned [48]. Moreover, medical ultrasound images often suffer from noise, speckle artifacts, and incomplete anatomical visualization, all of which degrade CNN performance [49, 50]. Preprocessing techniques such as noise reduction and data augmentation can mitigate these limitations, but they inevitably increase computational complexity [48-50]. These challenges underscore the necessity for more robust, efficient, and clinically applicable classification frameworks tailored to DDH diagnosis.
2.4 YOLO-based Medical Image Classification
The You Only Look Once (YOLO) family of models has been widely adopted for real-time object detection and classification across diverse domains, including medical imaging [51]. Recent iterations such as YOLOv4, YOLOv5, and YOLOv8 have introduced optimized backbone networks, attention mechanisms, and enhanced feature fusion modules, leading to notable improvements in both accuracy and computational efficiency [52, 53]. Unlike conventional CNN-based classification approaches, YOLO simultaneously performs object localization and classification in a single forward pass, making it highly suitable for time-sensitive diagnostic applications.
The latest version, YOLOv11, incorporates Cross-Stage Partial (CSP) connections, C2PSA spatial attention mechanisms, and efficient convolutional blocks, enabling superior feature extraction while maintaining lightweight computational demands [54]. This architecture has already been applied successfully in tasks such as chest X-ray interpretation, ultrasound imaging, and medical anomaly detection, where it has consistently outperformed lightweight models such as MobileNet and ShuffleNet in terms of classification accuracy and inference speed [55-62].
Traditional ultrasound-based DDH diagnosis remains dependent on manual interpretation, which is inherently variable across operators. By contrast, YOLO-based automated classification provides a fast, accurate, and scalable alternative, ideally suited for real-time clinical applications. Building on these advances, the present study leverages YOLOv11 for DDH classification, aiming to address persistent challenges such as dataset imbalance, image noise, and diagnostic inconsistency.
3. Methodology
3.1 Data Acquisition
Ultrasound images used in this study were acquired using a diagnostic ultrasound system. The dataset comprised 6,075 images stored in DICOM format for static frames and AVI format for video sequences. Images were categorized into ten anatomical classes: hip, ankle, soft tissue, wrist, shoulder, finger, knee, elbow, foot, and other.
For the assessment of Developmental Dysplasia of the Hip (DDH), imaging primarily targeted key anatomical structures including the femoral head, acetabulum, and ilium. The Graf classification method, which evaluates hip joint stability through the measurement of the α and β angles, was adopted as the clinical reference standard. Figure 1 illustrates a representative hip joint ultrasound image, highlighting the femoral head's position within the acetabulum and labeling the femoral head, acetabulum, and ilium—structures essential for determining joint alignment and identifying abnormalities.
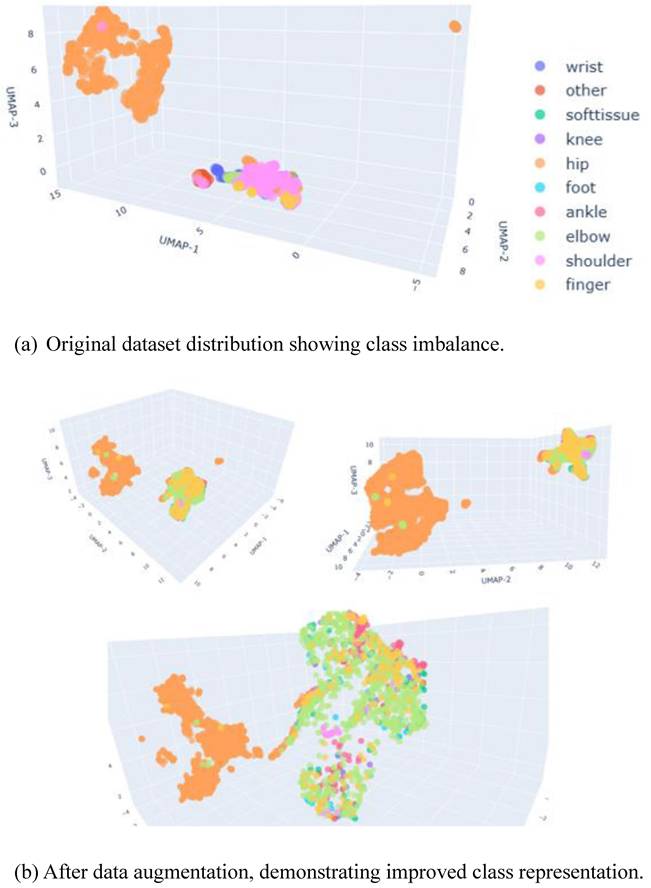
The dataset was collected from multiple clinical sources to capture diversity in patient demographics, imaging protocols, and anatomical variations. However, a marked class imbalance was observed: the hip category contained a disproportionately high number of images (5,159), compared with other categories such as knee (13) and ankle (5). This imbalance necessitated the use of data augmentation strategies, described in Section 3.2.
3.2 Dataset Analysis and Preprocessing
3.2.1 Dataset Imbalance and Visualization
The dataset exhibited pronounced class imbalance, which can bias model training and compromise generalization performance. To evaluate the distribution of samples, we employed Uniform Manifold Approximation and Projection (UMAP), a state-of-the-art dimensionality reduction technique well-suited for high-dimensional medical imaging data [51]. UMAP preserves both local and global data structures, thereby providing a reliable representation of class distribution.
As illustrated in Figure 2(a), the original dataset was dominated by the hip class, which clustered separately from the other anatomical categories. Following the application of data augmentation, the distribution became more balanced, as shown in Figure 2(b). The numerical breakdown of each class before and after augmentation is presented in Table 1.
3.2.2 Data Augmentation Strategy
To mitigate the effects of imbalance and enhance model robustness, multiple augmentation techniques were applied:
- Geometric Transformations: Random rotations (±15°), translations, and scaling to simulate variability in patient positioning.
- Intensity Adjustments: Gamma correction to normalize differences in brightness and contrast across scans.
- Elastic Deformations: Applied to approximate natural soft tissue variability.
- Noise Injection and Bias Field Distortion: Introduced random artifacts to replicate real-world imaging conditions.
These augmentation strategies improved class balance and promoted better generalization, as demonstrated by the more uniform UMAP distribution shown in Figure 2(b).
Dataset Distribution Before and After Augmentation.
| Class | Original Training Set | Original Test Set | Augmented Training Set | Augmented Test Set |
|---|---|---|---|---|
| Hip | 5,159 | 573 | 5,159 | 573 |
| Shoulder | 178 | 19 | 890 | 95 |
| Knee | 13 | 4 | 56 | 20 |
| Ankle | 5 | 1 | 25 | 5 |
| Wrist | 31 | 5 | 155 | 25 |
| Finger | 14 | 3 | 70 | 15 |
| Elbow | 12 | 4 | 60 | 20 |
| Foot | 4 | 1 | 20 | 5 |
| Soft Tissue | 5 | 4 | 20 | 20 |
Ultrasound Image of the Hip Joint in Developmental Dysplasia of the Hip (DDH).
UMAP Visualization of Dataset Distribution.
3.3 Proposed YOLOv11-Based Classification Model
The proposed YOLOv11-based framework was designed to automate DDH classification from ultrasound images while maintaining both high inference speed and diagnostic accuracy.
3.3.1 Model Architecture
Figure 3 presents an overview of the YOLOv11 architecture adapted for this study. The framework introduces several key innovations:
(1) Backbone (Feature Extraction)
- C3k2 Blocks: An efficient implementation of Cross-Stage Partial (CSP) bottlenecks, improving gradient flow and feature reuse [52].
- Spatial Pyramid Pooling - Fast (SPPF): Reduces computational cost while retaining multi-scale feature representation.
- C2PSA Attention Mechanism: Enhances spatial feature learning, enabling the model to focus on clinically relevant anatomical regions.
(2) Neck (Feature Aggregation)
- Combines Feature Pyramid Network (FPN) and Path Aggregation Network (PAN) structures.
- C3k2 blocks replace traditional C2f blocks, yielding higher efficiency without compromising accuracy
(3) Head (Prediction)
- Outputs bounding boxes, class probabilities, and confidence scores in a single forward pass, thereby supporting real-time classification.
YOLOv11 Model Architecture.
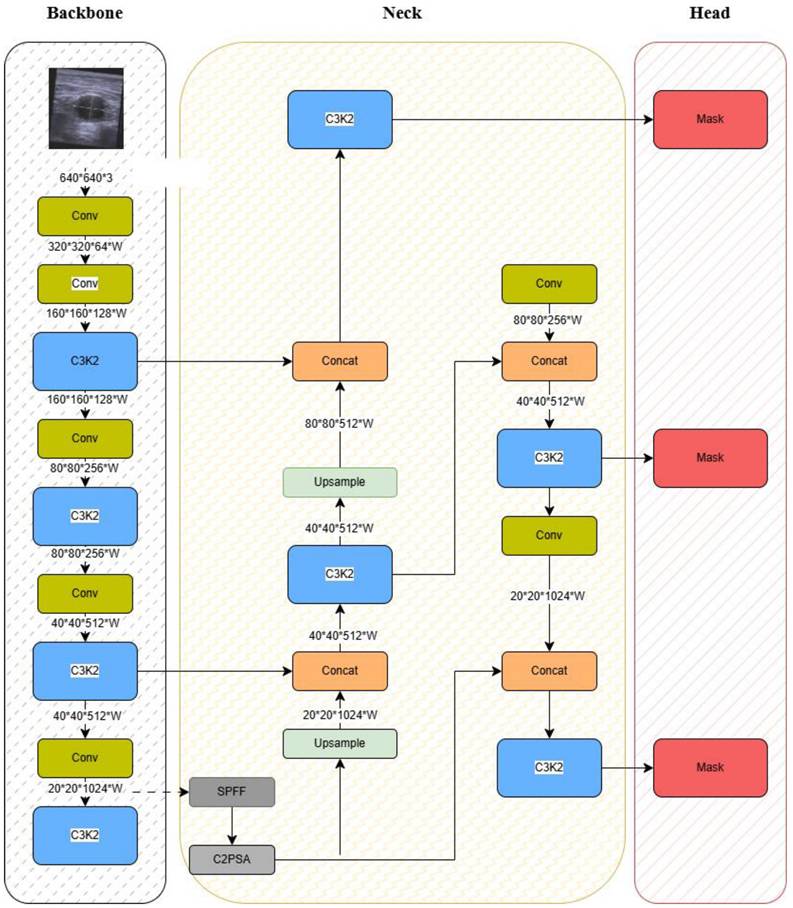
As shown in Figure 3, these components work synergistically to extract discriminative anatomical features, aggregate multi-scale information, and deliver accurate classification under real-time constraints. The integration of CSP and C2PSA modules is particularly critical for modeling complex hip structures in noisy ultrasound environments.
Together, these architectural improvements enable the YOLOv11 framework to achieve both high diagnostic accuracy and computational efficiency, rendering it suitable for deployment in point-of-care ultrasound systems.
3.4 Training Procedure
The training procedure for the YOLOv11-based classification model consisted of data preprocessing, model training, hyperparameter optimization, and performance evaluation. The pipeline was designed to maximize generalization, minimize overfitting, and ensure robust classification across imbalanced classes.
3.4.1 Model Training and Optimization
The model was trained to simultaneously optimize feature extraction, localization, and classification. Training was conducted using the AdamW optimizer, which balances convergence speed and generalization by incorporating weight decay regularization. A cosine annealing learning rate schedule was applied, beginning with an initial learning rate of 0.001 and gradually decaying to stabilize learning and reduce overfitting.
To address class imbalance, Focal Loss was employed, reducing the influence of easily classified samples while emphasizing harder-to-classify cases. For localization refinement, IoU Loss was used to improve bounding box predictions and ensure accurate delineation of hip joint structures. The final training configuration was as follows:
- Initial Learning Rate: 0.001 (cosine decay)
- Batch Size: 16
- Number of Epochs: 100
- Optimizer: AdamW with weight decay = 0.01
- Loss Functions: (1) Focal Loss for classification, (2) IoU Loss for localization
To improve generalization, the dataset was augmented with random rotations, brightness adjustments, and noise injection, as described in Section 3.2.2. Importantly, patient-level data splitting was applied to prevent information leakage: all images from the same patient were assigned exclusively to one partition. An 80/20 split at the patient level was used for development (training/validation), followed by 5-fold cross-validation, also stratified by patient, to ensure robustness. This design eliminated identical-patient overlap across folds, providing an unbiased estimate of model generalization.
3.4.2 Evaluation Metrics
Model performance was evaluated using four standard metrics:
- Accuracy (ACC): Proportion of correctly classified cases among all predictions.
- Precision (P): Proportion of correctly identified positive cases among all predicted positives, reflecting the ability to avoid false positives.
- Recall (R): Proportion of correctly identified positive cases among all actual positives, reflecting sensitivity and the ability to reduce false negatives.
- F1-Score: Harmonic mean of precision and recall, providing a balanced measure of model performance.
The proposed model was benchmarked against lightweight classification architectures, MobileNetV3 and ShuffleNetV2, with comparisons focusing on accuracy, inference time, and computational efficiency. In addition, 5-fold cross-validation was performed to validate the consistency and robustness of results across different dataset partitions.
4. Results and Discussion
4.1 Performance Comparison
The YOLOv11-based DDH classification model was first evaluated on the independent test set and compared with two lightweight baseline architectures, MobileNetV3 and ShuffleNetV2. Performance was assessed using four metrics: Accuracy, Precision, Recall, and F1-Score. The results are summarized in Table 2.
As shown in Table 2, YOLOv11 achieved the highest performance across all metrics, with an overall accuracy of 95.05%. This represents a substantial improvement of nearly 20% in accuracy compared with MobileNetV3 (75.6%) and more than 23% compared with ShuffleNetV2 (71.9%). Precision and recall values for YOLOv11 were also consistently higher, resulting in the highest F1-Score (95.05%).
These results highlight the effectiveness of incorporating CSP and C2PSA modules into the YOLOv11 architecture, which improved the model's capacity to capture clinically relevant anatomical structures in ultrasound images. The superior performance across multiple evaluation metrics demonstrates that the proposed framework not only outperforms lightweight alternatives but also provides reliable classification suitable for real-time clinical deployment.
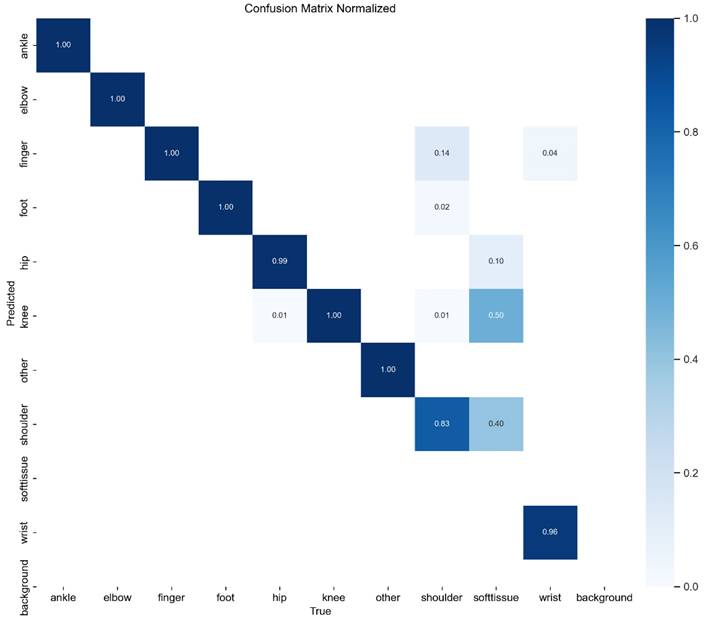
4.2 Confusion Matrix Analysis
To further assess classification performance, a confusion matrix was generated for YOLOv11, as presented in Figure 4. The matrix illustrates the distribution of correct and incorrect predictions across different DDH categories, providing detailed insights into class-specific strengths and weaknesses.
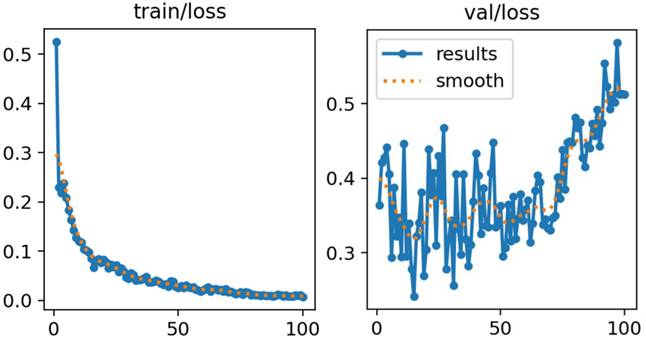
As shown in Figure 5, both training and validation losses decreased steadily throughout the training process, with no evidence of divergence between the two curves. This pattern indicates stable learning dynamics and suggests that overfitting was effectively mitigated. The application of data augmentation and regularization strategies contributed to this stability by improving generalization and reducing susceptibility to noise or class imbalance.
The consistent convergence observed in both curves demonstrates that the model successfully captured discriminative features of hip anatomy without sacrificing generalization capacity. These results further validate the suitability of the proposed training strategy, confirming its robustness in handling heterogeneous ultrasound data.
Performance Comparison of YOLOv11, MobileNetV3, and ShuffleNetV2.
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| YOLOv11 | 95.05 | 94.88 | 95.22 | 95.05 |
| MobileNetV3 | 75.6 | 76.1 | 74.5 | 75.3 |
| ShuffleNetV2 | 71.9 | 72.4 | 70.8 | 71.6 |
Confusion Matrix of YOLOv11 Model.
Training and Validation Loss Curves for YOLOv11.
Overall, the model achieved high classification accuracy across the majority of categories. However, some degree of misclassification was observed. Specifically, overlap occurred between Type IIa and Type IIb hips, reflecting their inherent anatomical similarity and the subtle differences in α angle measurements that challenge even experienced clinicians. In addition, certain Type III cases were misclassified as Type IIc, likely due to similarities in femoral head positioning.
These findings indicate that while the model performs robustly overall, borderline categories remain the most challenging to differentiate. This limitation parallels the clinical reality, where even expert sonographers occasionally encounter difficulties distinguishing between adjacent Graf types. Such results suggest that additional strategies, such as refined preprocessing, multi-view ultrasound integration, or hybrid AI-physician decision-making could further improve classification accuracy in these borderline cases.
4.3 Training and Validation Loss Analysis
The training and validation loss curves for the YOLOv11 model are presented in Figure 5. These curves depict the optimization trajectory across 100 epochs, illustrating both convergence behavior and generalization performance.
4.4 Inference Speed and Computational Efficiency
Inference efficiency is a critical factor for real-time DDH screening. Table 3 compares the number of parameters, FLOPs, and inference time per image for YOLOv11, MobileNetV3, and ShuffleNetV2.
Inference speed and computational efficiency comparison.
| Model | Parameters (M) | FLOPs (B) | Inference Time (ms/image) |
|---|---|---|---|
| YOLOv11 | 12.9 | 49.4 | 11.5 |
| MobileNetV3 | 5.4 | 2.19 | 12.0 |
| ShuffleNetV2 | 2.3 | 1.46 | 11.4 |
Although YOLOv11 contains a larger number of parameters (12.9M) and higher computational complexity (49.4B FLOPs) compared with MobileNetV3 and ShuffleNetV2, its inference speed remained competitive at 11.5 ms per image. This demonstrates that the architectural optimizations—including CSP modules, spatial attention mechanisms, and efficient convolutional blocks—effectively balanced accuracy with efficiency.
The results confirm that YOLOv11 is capable of achieving real-time performance without compromising diagnostic precision. This balance between computational cost and inference speed makes the framework well-suited for integration into point-of-care ultrasound systems, where rapid diagnostic feedback is essential.
4.5 Comparison with Previous Studies
To contextualize the performance of the proposed framework, we compared it against prior deep learning approaches for DDH classification. Table 4 summarizes the results.
As shown in Table 4, YOLOv11 achieved a classification accuracy of 95.05%, outperforming earlier CNN- and ResNet-based models by a margin of 6-8%. This improvement can be attributed to three factors: (i) the use of a larger dataset collected over multiple years, (ii) architectural enhancements such as CSP modules and C2PSA spatial attention, and (iii) robust data augmentation strategies that alleviated class imbalance.
Comparison with Existing DDH Classification Models.
| Study | Model Used | Accuracy (%) | Dataset Size |
|---|---|---|---|
| Sezer et al. (2020) [13] | CNN + Data Augmentation | 87.3 | 2,500 images |
| Chlapoutakis et al. (2022) [24] | ResNet-50 | 89.1 | 3,200 images |
| This Study | YOLOv11 | 95.05 | 6,075 images |
Compared with earlier approaches, which primarily emphasized segmentation or employed conventional CNN backbones, the proposed model provides a more scalable and clinically practical solution. Its superior accuracy, combined with real-time inference speed, underscores its potential for deployment in routine DDH screening workflows.
4.6 Ablation Study
To evaluate the individual contributions of the Cross-Stage Partial (CSP) modules and the C2PSA spatial attention mechanism, an ablation study was conducted under four experimental settings: (1) YOLOv11 without CSP, (2) YOLOv11 without C2PSA, (3) YOLOv11 without both modules, and (4) the full model (CSP + C2PSA). The results are summarized in Table 5.
As shown in Table 5, removal of either CSP or C2PSA resulted in a noticeable decline in performance compared with the full model. Excluding CSP primarily reduced recall, indicating its importance for enhancing sensitivity in detecting dysplastic hips. In contrast, the absence of C2PSA led to lower precision, suggesting that spatial attention was critical for guiding the network toward anatomically relevant regions and minimizing false positives. The combined use of CSP and C2PSA produced the best overall performance, underscoring their complementary roles in improving both feature extraction and anatomical interpretability.
Ablation Study of CSP and C2PSA Modules.
| Model Variant | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| YOLOv11 without CSP | 91.8 | 91.2 | 91.5 | 91.3 |
| YOLOv11 without C2PSA | 92.6 | 92.1 | 92.3 | 92.2 |
| YOLOv11 without CSP + C2PSA | 90.9 | 90.3 | 90.7 | 90.5 |
| Full Model (CSP + C2PSA) | 95.05 | 94.88 | 95.22 | 95.05 |
These findings confirm that the architectural modifications introduced in YOLOv11 are not only computationally efficient but also essential for achieving clinically meaningful performance in DDH classification.
4.7 Explainability Analysis
To improve interpretability and foster clinical acceptance, we generated Gradient-weighted Class Activation Mapping (Grad-CAM) and attention heatmaps for representative cases. As illustrated in Figure 6, the model consistently focused on the acetabular roof and femoral head—key anatomical landmarks central to the Graf classification system.
Grad-CAM visualizations of YOLOv11-based DDH classification. (a) Normal hip: the model predominantly focuses on the acetabular roof and femoral head. (b) Dysplastic hip: stronger activation is observed around the shallow acetabular roof and displaced femoral head. These attention patterns align with Graf classification landmarks, supporting clinical interpretability and acceptance.
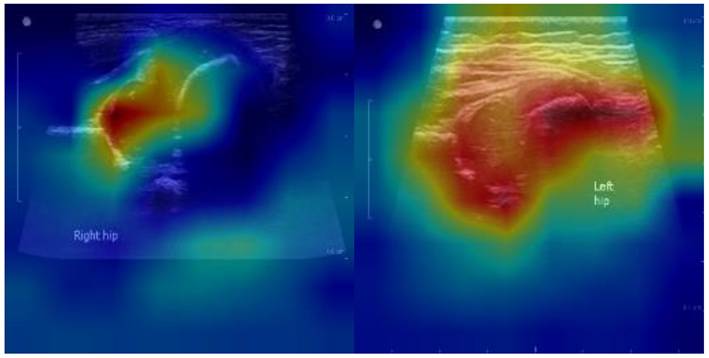
In normal hips, Grad-CAM highlighted concentrated attention on the acetabular roof and femoral head, confirming that the model relies on clinically relevant structures for accurate classification. In dysplastic hips, stronger activations were observed around the shallow acetabular roof and the displaced femoral head, patterns that align closely with radiologists' visual assessments.
These findings suggest that the model's decision-making process is not only data-driven but also anatomically meaningful. By attending to regions routinely evaluated by clinicians, the proposed framework enhances transparency and builds trust, thereby facilitating its potential integration into real-time, point-of-care diagnostic workflows.
4.8 Clinical Workflow Integration
To facilitate clinical adoption, we envision several pathways through which the proposed model could be integrated into routine workflows across different care settings.
Neonatal screening clinics. During routine hip ultrasound examinations, the system can be embedded directly within the ultrasound console or a connected edge device. It provides (i) real-time quality feedback, such as alerts for non-standard planes or probe instability, (ii) live classification overlays to support immediate triage (normal vs. dysplastic/immature), and (iii) structured outputs including key frames and confidence scores for documentation. This integration reduces the need for repeat scans, shortens examination time, and enhances diagnostic efficiency in high-volume screening settings.
Primary healthcare and community hospitals. In resource-limited or non-specialist environments, the framework can function as a decision-support tool. Cases classified with low confidence are flagged for secondary review by pediatric orthopedists, while high-confidence normal cases may be safely discharged with follow-up instructions. This hub-and-spoke model optimizes referral pathways, reduces unnecessary specialist consultations, and ensures that expert attention is focused on the most complex cases.
Training and quality assurance. Explainability features, such as Grad-CAM heatmaps, provide immediate feedback by highlighting clinically relevant structures (e.g., acetabular roof, femoral head). These visualizations can be used as teaching aids for junior sonographers and as part of standardized training programs. From a quality assurance perspective, periodic audits of flagged cases and drift monitoring can be implemented to ensure sustained accuracy after deployment.
Human-in-the-loop safeguards. The model is designed to complement, not replace, physician expertise. Safety features include threshold-based alerts, confidence-calibrated reporting, and mandatory human review for ambiguous or low-confidence cases. These safeguards ensure that final diagnostic responsibility remains with clinicians while leveraging AI to improve efficiency and consistency.
5. Discussion
The proposed YOLOv11-based framework demonstrates both technical innovation and clinical applicability for automated DDH classification.
From a technical perspective, several architectural enhancements directly contributed to the model's superior performance. The incorporation of Cross-Stage Partial (CSP) blocks improved gradient flow and feature reuse, thereby enhancing recall and sensitivity for detecting dysplastic hips. The addition of the C2PSA spatial attention mechanism enabled the model to focus on anatomically meaningful regions, reducing false positives and improving precision. Results from the ablation study confirmed the complementary roles of these modules, with the full model achieving the highest overall accuracy (95.05%) and real-time inference speed (11.5 ms per image). When compared with lightweight baselines such as MobileNetV3 and ShuffleNetV2, the proposed framework consistently demonstrated superior performance across multiple metrics, underscoring the importance of architectural optimization. Moreover, the integration of Focal Loss and IoU Loss effectively addressed challenges related to class imbalance and localization, ensuring stable training and robust generalization.
From a clinical perspective, the framework represents an important step toward standardizing DDH screening, which remains subject to significant inter-observer variability under the Graf classification system. By enabling real-time automated classification, the model can support clinicians in neonatal screening clinics and community healthcare settings, where operator expertise is often limited. Potential applications include immediate feedback during scanning, triage support through abnormal case flagging, and automated report generation to reduce documentation burden. Importantly, explainability analyses such as Grad-CAM demonstrated that the model consistently focused on the acetabular roof and femoral head, key anatomical landmarks used in clinical practice. This alignment with established diagnostic criteria enhances transparency, fosters clinician trust, and strengthens the case for clinical adoption.
Despite these strengths, several limitations must be acknowledged. First, although the dataset comprised more than 6,000 ultrasound images, all data were obtained from a single institution. This may restrict generalizability due to variations in imaging equipment, acquisition protocols, and patient demographics. To address this limitation, future work will emphasize external validation across multiple centers and populations. Second, although patient-level data splitting was applied to eliminate information leakage, prospective clinical validation remains necessary to fully assess performance in real-world workflows. Finally, while ultrasound is the gold standard for infant DDH screening, incorporating multimodal imaging modalities such as X-ray and MRI could broaden diagnostic capability and improve precision.
In summary, the proposed YOLOv11 framework integrates ablation-validated architectural innovations, real-time feasibility, and clinically meaningful explainability. Its potential applications extend beyond technical accuracy to address practical challenges in neonatal screening and primary care environments. Future research should prioritize multi-center validation, prospective deployment in point-of-care ultrasound systems, and multimodal integration to ensure robust clinical translation and maximize the framework's impact in standardized DDH diagnosis.
6. Conclusion
In this study, we developed an attention-enhanced YOLOv11 framework for the automated classification of developmental dysplasia of the hip (DDH) from ultrasound images. By integrating Cross-Stage Partial (CSP) modules and C2PSA spatial attention, the model achieved superior performance, with an accuracy of 95.05% and an inference speed of 11.5 ms per image. Ablation experiments confirmed the complementary roles of CSP and C2PSA, demonstrating their collective impact on improving both sensitivity and precision.
Beyond technical performance, the framework provides tangible clinical benefits. Real-time classification and interpretable visualizations, supported by Grad-CAM heatmaps, align with established diagnostic landmarks such as the acetabular roof and femoral head. These features enhance transparency, reduce inter-observer variability, and facilitate integration into neonatal screening clinics and community healthcare settings, particularly where operator expertise may be limited.
To ensure unbiased evaluation, patient-level data splitting was employed to prevent information leakage, providing a reliable estimate of clinical performance. Nevertheless, the reliance on a single-institution dataset remains a limitation. Future work will focus on multi-center external validation, prospective deployment within point-of-care ultrasound systems, and multimodal integration (e.g., X-ray and MRI) to further expand diagnostic capability and generalizability.
In conclusion, this work demonstrates both technical innovation and clinical practicality. By combining ablation-validated architectural improvements, explainability, and workflow-oriented design, the proposed YOLOv11 framework establishes a foundation for clinically deployable, AI-assisted DDH screening. Future research directed toward multi-center validation and multimodal expansion will be essential for translating this framework into standardized clinical practice and maximizing its impact in pediatric orthopedic care.
Acknowledgements
We thank the clinical staff and data engineers at Changhua Christian Hospital for their support and contributions to image labeling and system testing.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (IRB) of Changhua Christian Hospital (IRB No. 230317). All data were anonymized, and ultrasound images were collected between January 1, 2020 and March 31, 2023.
Consent for publication
All authors reviewed and approved the final version of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request, subject to ethical restrictions.
Author contributions
All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Agrawal K, Bishnoi K, Chow G, Armstrong T, Van den Wyngaert T. Developmental dysplasia of the hip (DDH). In: Van den Wyngaert T, Gnanasegaran G, Strobel K, editors. Clinical Atlas of Bone SPECT/CT. Cham: Springer. 2023 p. 1-6
2. Paton RW, Choudry Q. Developmental dysplasia of the hip (DDH): Diagnosis and treatment. Surgery (Oxford). 2020;38(9):536-543
3. Gold M, Munjal A, Varacallo MA. Anatomy, bony pelvis and lower limb, hip joint. StatPearls Publishing. 2023
4. Jordan JA, Burns B. Anatomy, abdomen and pelvis: Hip arteries. StatPearls. 2023
5. Ibrahim MM, Smit K. Anatomical description and classification of hip dysplasia. In: Beaulé P, editor. Hip Dysplasia. Cham: Springer. 2020 p. 1-10
6. Nandhagopal T, Tiwari V, De Cicco FL. Developmental dysplasia of the hip. Kern Medical Center; Ross Medical University; Apollo Sage Hospital; Hospital Italiano de Buenos Aires. 2024
7. Harsanyi S, Zamborsky R, Krajciova L, Kokavec M, Danisovic L. Developmental dysplasia of the hip: A review of etiopathogenesis, risk factors, and genetic aspects. Medicina (Kaunas). 2020;56(4):153
8. Wen J, Ping H, Kong X, Chai W. Developmental dysplasia of the hip: A systematic review of susceptibility genes and epigenetics. Gene. 2023;853:147067
9. Placzek S, Bornemann R, Skoulikaris N. Development of the hip joint: Embryology and anatomy of the neonatal hip joint. In: O'Beirne J, Chlapoutakis K, editors. Developmental Dysplasia of the Hip. Cham: Springer. 2022
10. Bakarman K, Alsiddiky AM, Zamzam M, Alzain KO, Alhuzaimi FS, Rafiq Z. Developmental dysplasia of the hip (DDH): Etiology, diagnosis, and management. Cureus. 2023;15(8):e43207
11. Zídka M, Džupa V. National register of joint replacement reflecting the treatment of developmental dysplasia of the hip in newborns. Acta Chir Orthop Traumatol Cech. 2019;86(5):324-329
12. Zaghloul A, Elalfy M. Hip joint: Embryology, anatomy, and biomechanics. Biomed J Sci Tech Res. 2019;12(3):7
13. Sezer A, Sezer HB. Deep convolutional neural network-based automatic classification of neonatal hip ultrasound images: A novel data augmentation approach with speckle noise reduction. Ultrasound Med Biol. 2020;46(3):735-749
14. Chavoshi M, Mirshahvalad SA, Mahdizadeh M, Zamani F. Diagnostic accuracy of ultrasonography method of Graf in the detection of developmental dysplasia of the hip: A meta-analysis and systematic review. Arch Bone Jt Surg. 2021;9(3):297-305
15. Loeber JG. Neonatal screening in Europe: The situation in 2004. J Inherit Metab Dis. 2007;30(4):430-438
16. Jacobino BCP, Galvão MD, da Silva AF, de Castro CC. Using the Graf method of ultrasound examination to classify hip dysplasia in neonates. Autops Case Rep. 2012;2(2):5-10
17. Graf R. The diagnosis of congenital hip-joint dislocation by the ultrasonic compound treatment. Arch Orthop Trauma Surg. 1980;97(2):117-133
18. Hareendranathan AR, Mabee M, Punithakumar K, Noga M, Jaremko JL. Toward automated classification of acetabular shape in ultrasound for diagnosis of DDH: Contour alpha angle and the rounding index. Comput Methods Programs Biomed. 2016;129:89-98
19. Sezer HB, Sezer A. Automatic segmentation and classification of neonatal hips according to Graf's sonographic method: A computer-aided diagnosis system. Appl Soft Comput. 2019;82:105516
20. Graf R, Scott S, Lercher K. et al. Hip joint ultrasound examination. Berlin: Springer. 2006
21. Kolovos S, Sioutis S, Papakonstantinou ME. et al. Ultrasonographic screening for developmental dysplasia of the hip: The Graf method revisited. Eur J Orthop Surg Traumatol. 2024;34:723-734
22. Chen YP, Fan TY, Chu CJ, Lin JJ, Ji CY, Kuo CF, Kao HK. Automatic and human level Graf's type identification for detecting developmental dysplasia of the hip. Biomed J. 2024;47(2):100614
23. Yadav K, Yadav S, Dubey PK. Importance of ultrasonic testing and its metrology through emerging applications. In: Aswal DK, Yadav S, Takatsuji T, Rachakonda P, Kumar H, editors. Handbook of Metrology and Applications. Singapore: Springer. 2023 p. 737-754
24. Chlapoutakis K, Kolovos S, Tarrant A, Maizen C. Hip sonography according to Graf. In: O'Beirne J, Chlapoutakis K, editors. Developmental Dysplasia of the Hip. Cham: Springer. 2022 p. 131-141
25. Liu D, Mou X, Yu G, Liang W, Cai C, Li X, Zhang G. The feasibility of ultrasound Graf method in screening infants and young children with congenital hip dysplasia and follow-up of treatment effect. Transl Pediatr. 2021;10(5):1333-1339
26. Omeroğlu H. Use of ultrasonography in developmental dysplasia of the hip. J Child Orthop. 2014;8(2):105-113
27. Chen T, Zhang Y, Wang B, Wang J, Cui L, He J, Cong L. Development of a fully automated Graf standard plane and angle evaluation method for infant hip ultrasound scans. Diagnostics (Basel). 2022;12(6):1423
28. Mienye ID, Swart TG. A comprehensive review of deep learning: architectures, recent advances, and applications. Information. 2024;15(12):755
29. Alzubaidi L, Zhang J, Humaidi AJ. et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8:53
30. Khoei TT, Aissou G, Al Shamaileh K, Devabhaktuni VK, Kaabouch N. Supervised deep learning models for detecting GPS spoofing attacks on unmanned aerial vehicles. In: 2023 IEEE International Conference on Electro Information Technology (eIT). 2023 May; Romeoville, IL, USA. p. 340-346
31. Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. A survey on deep transfer learning. In: International Conference on Artificial Neural Networks. Berlin: Springer. 2018 p. 270-279
32. Yang Y, Xia X, Lo D, Grundy J. A survey on deep learning for software engineering. ACM Comput Surv. 2022;54(10s):Article 206, 73 pages
33. Nguyen TT, Nguyen QVH, Nguyen DT. et al. Deep learning for deepfakes creation and detection: a survey. Comput Vis Image Underst. 2022;223:103525
34. Dong S, Wang P, Abbas K. A survey on deep learning and its applications. Comput Sci Rev. 2021;40:100379
35. Ni J, Young T, Pandelea V, Xue F, Cambria E. Recent advances in deep learning based dialogue systems: a systematic survey. Artif Intell Rev. 2023;56:1-101
36. Younesi A, Ansari M, Fazli M. et al. A comprehensive survey of convolutions in deep learning: applications, challenges, and future trends. IEEE Access. 2024;12:41180-41218
37. Indolia S, Goswami AK, Mishra SP, Asopa P. Conceptual understanding of convolutional neural network—A deep learning approach. Procedia Comput Sci. 2018;132:679-688
38. Li Z, Liu F, Yang W, Peng S, Zhou J. A survey of convolutional neural networks: analysis, applications, and prospects. IEEE Trans Neural Netw Learn Syst. 2022;33(12):6999-7019
39. Mohammed FA, Tune KK, Assefa BG. et al. Medical image classifications using convolutional neural networks: a survey of current methods and statistical modeling of the literature. Mach Learn Knowl Extr. 2024;6(1):699-735
40. Heim E, Roß T, Seitel A. et al. Large-scale medical image annotation with crowd-powered algorithms. J Med Imaging (Bellingham). 2018;5(3):034002
41. Aljabri M, AlAmir M, AlGhamdi M. et al. Towards a better understanding of annotation tools for medical imaging: a survey. Multimed Tools Appl. 2022;81:25877-25911
42. Zhang Y, Chen J, Ma X, Wang G, Bhatti UA, Huang M. Interactive medical image annotation using improved Attention U-Net with compound geodesic distance. Expert Syst Appl. 2024;237(Part A):121282
43. Razzak MI, Naz S, Zaib A. Deep learning for medical image processing: overview, challenges and the future. In: Dey N, Ashour A, Borra S, editors. Classification in BioApps. Lecture Notes in Computational Vision and Biomechanics, vol 26. Cham: Springer. 2018
44. Salehi AW, Khan S, Gupta G. et al. A study of CNN and transfer learning in medical imaging: advantages, challenges, future scope. Sustainability. 2023;15(7):5930
45. Shahinfar S, Meek P, Falzon G. “How many images do I need?” Understanding how sample size per class affects deep learning model performance metrics for balanced designs in autonomous wildlife monitoring. Ecol Inform. 2020;57:101085
46. Szyc K. Determining the minimal number of images required to effectively train convolutional neural networks. In: Zamojski W, Mazurkiewicz J, Sugier J, Walkowiak T, Kacprzyk J, editors. Theory and Applications of Dependable Computer Systems. DepCoS-RELCOMEX 2020. Advances in Intelligent Systems and Computing, vol 1173. Cham: Springer. 2020
47. Momeny M, Latif AM, Sarram MA, Sheikhpour R, Zhang YD. A noise robust convolutional neural network for image classification. Results Eng. 2021;10:100225
48. Ghoben MK, Noor Muhammed LA. Exploring the impact of image quality on convolutional neural networks: a study on noise, blur, and contrast. In: 2023 Int Conf of Computer Science and Information Technology (ICOSNIKOM), Binjia, Indonesia. 2023 p. 1-7
49. Qi S, Zhang Y, Wang C. et al. Representing noisy image without denoising. IEEE Trans Pattern Anal Mach Intell. 2024;46(10):6713-6730
50. Kalaiselvi T, Anitha T, Sriramakrishnan P. Data preprocessing techniques for MRI brain scans using deep learning models. In: Chaki J, editor. Brain Tumor MRI Image Segmentation Using Deep Learning Techniques. Academic Press. 2022 p. 13-25
51. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IW, Ng LG. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37(1):38-44
52. Wang CY, Bochkovskiy A, Liao HYM. YOLOv4: Optimal speed and accuracy of object detection. arXiv preprint arXiv:2004.10934. 2021
53. Ma N, Zhang X, Zheng HT, Sun J. ShuffleNet V2: Practical guidelines for efficient CNN architecture design. In: Proceedings of the European Conference on Computer Vision (ECCV). 2018 p. 122-138
54. Howard AG, Zhu M, Chen B. et al. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861. 2017
55. Howard A, Sandler M, Chu G. et al. Searching for MobileNetV3. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV). 2019 p. 1314-1324
56. Zhang X, Zhou X, Lin M, Sun J. ShuffleNet: An extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2018 p. 6848-6856
57. Wang CY, Liao HYM, Wu YH. et al. CSPNet: A new backbone that can enhance learning capability of CNN. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). 2020 p. 390-391
58. Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2018 p. 7132-7141
59. Tan M, Le QV. EfficientNet: Rethinking model scaling for convolutional neural networks. In: Proceedings of the International Conference on Machine Learning (ICML). 2019 p. 6105-6114
60. Woo S, Park J, Lee JY, Kweon IS. CBAM: Convolutional block attention module. In: Proceedings of the European Conference on Computer Vision (ECCV). 2018 p. 3-19
61. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016 p. 770-778
62. Dosovitskiy A, Beyer L, Kolesnikov A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. In: International Conference on Learning Representations (ICLR). 2021
Author contact
![]() Corresponding author: wangweihsunnchu.edu.tw.
Corresponding author: wangweihsunnchu.edu.tw.

 Global reach, higher impact
Global reach, higher impact