Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(14):3625-3649. doi:10.7150/ijms.112724 This issue Cite
Review
Overcoming the Delivery Challenges in CRISPR/Cas9 Gene Editing for Effective Cancer Treatment: A Review of Delivery Systems
1. College of Materials and Engineering, Yangtze Normal University, Chongqing 408100, China.
2. Cancer Center, Department of Pathology, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, Hangzhou 310014, China.
3. Clinical Research Institute, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, Hangzhou 310014, China.
4. Department of Hematology, the Hangzhou First People's Hospital, Hangzhou 310006, China.
#, These authors contributed equally to this work.
Received 2025-2-23; Accepted 2025-6-23; Published 2025-7-28
Abstract
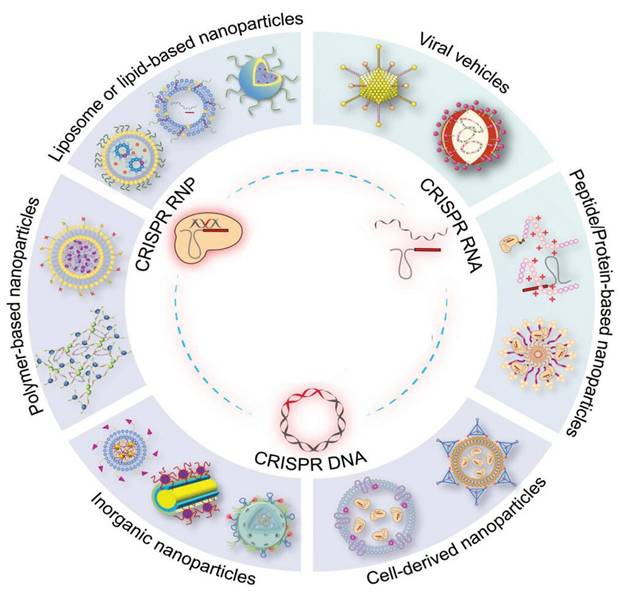
Therapeutic strategies based on gene editing provide the ability to modify faulty genes contributing to the development of diseases such as cancer by directly altering the cellular machinery. The clustered regularly interspaced short palindromic repeats associated nuclease 9 (CRISPR/Cas9) system is currently the primary tool used for gene editing. Several effective Cas9 variants have already been established to address the complex genetic modifications that arise during diseases. Although gene-editing systems have made significant advancements, a primary obstacle that requires attention is the transportation of CRISPR/Cas to diverse target cells, both in vivo and in vitro, to render them suitable for clinical implementation. Various strategies can be utilized to facilitate the transportation of the CRISPR/Cas systems into mammalian cells. Herein, we reviewed contemporary research about delivery systems for gene-editing systems that interact effectively in biological systems. This review explores the benefits and drawbacks of using extracellular vesicles and viral vectors as vehicles for delivering the CRISPR/Cas system in the context of cancer treatment.
Keywords: CRISPR/Cas, cancer treatment, gene-editing systems, extracellular vesicles, viral vectors
Introduction
Cancer is a leading cause of mortality worldwide, giving rise to a multitude of complications across various domains. The origins of cancerous cells are associated with various genetic modifications occurring within these cells [1]. The development of cancer is significantly influenced by the presence of tumor suppressor genes and oncogenes. These two groups of genes exhibit counterbalancing effects. Tumor suppressors are known to trigger apoptosis, while oncogenes are recognized for their ability to stimulate cell proliferation. Thus, the effective utilization of anti-oncogenes and apoptotic genes can be employed in the treatment of cancer. Despite the numerous advancements in screening and therapeutic techniques, the efficacy of treatment remains a formidable challenge [2]. Several therapeutic approaches have been devised, such as gene therapies and small molecules, to combat cancer by regulating the immune system or selectively targeting cancer-causing signaling pathways, resulting in complete remission in certain cases [3, 4]. However, effective therapeutic strategies for the treatment of diverse cancer types are still lacking.
The concept of adaptive immunity was previously believed to be limited to vertebrate organisms [5]. However, the revelation that prokaryotes also exhibit a type of specific defense mechanism has prompted the advancement of technologies with the potential to transform approaches to treating human diseases. The discovery began during the initial stages of sequencing bacterial genomes when scientists observed an atypical arrangement characterized by the presence of concise, repeated DNA sequences within the chromosome of E. coli [6]. Further studies have revealed additional instances of these distinctive structural patterns in various other prokaryotic organisms. In 2005, the intervening sequences within these repeats were thoroughly examined and designated as clustered regularly interspaced short palindromic repeats (CRISPRs). It was subsequently determined that these CRISPR sequences bear an exact match to the bacteriophage's genome [7, 8]. The in-depth analyses of these repetitive-spacer loci have revealed a cluster of functional genes that are located adjacent to the CRISPR arrays. The aforementioned coding genes have been designated as CRISPR-associated proteins (Cas) [9, 10]. In a study conducted in 2007, it was observed that certain bacteria, specifically S. thermophilus, which are involved in the fermentation process of yogurt, exhibited a fascinating defense mechanism against phage infections. These bacteria were found to express Cas proteins and possess a CRISPR array containing spacers that were specifically targeted towards a phage genome. Therefore, the bacteria were effectively shielded from being infected by the phage. Remarkably, the identification of an exclusive protein, known as CRISPR-associated protein 9 (Cas9), has unequivocally demonstrated its exclusive role in facilitating RNA-mediated DNA cleavage within specific bacterial species [11].
The utilization of CRISPR-Cas9 mediated DNA fragmentation presents a viable approach for the precise identification of alterations within cancer-specific genetic sequences. Recently, significant advancements have been achieved in cancer therapy with the introduction of chimeric antigen receptors (CAR). Despite the promising results achieved with CAR T-cell therapy, certain limitations remain that require further investigation. In this regard, the CRISPR/Cas system has emerged as a promising tool for improved effectiveness of CAR T-cell-based cancer immunotherapy [12]. The presence of oncogenic viruses has been a subject of considerable apprehension within the field of gene therapy for an extended period. However, it is interesting to observe that CRISPR technology seems to present a possible solution to the issue by effectively mitigating the risk of these viruses inducing cancer [13]. Recent progress in CRISPR/Cas gene editing has prompted a growing number of review articles exploring its therapeutic potential [14, 15]. However, most existing reviews either provide broad overviews or focus on specific diseases without thoroughly addressing the delivery challenges unique to cancer therapy. This review aims to fill this gap by offering a focused analysis of delivery strategies tailored specifically for CRISPR/Cas-based cancer treatment. In contrast to recent publications, the present work highlights emerging trends in nanocarrier design, explores the critical hurdles in clinical translation, and evaluates novel approaches for tumor-targeted delivery. By combining these aspects, this review provides a timely and comprehensive perspective that distinguishes it from previous reports and contributes meaningful insights to the evolving field. The delivery of the CRISPR-Cas system into mammalian cells can be accomplished through a range of strategies, including the utilization of physical methods, nanocomplexes, extracellular vesicles (EVs), and viral vectors [16]. Herein, we have thoroughly examined the inherent benefits and constraints associated with the utilization of EVs and viral vectors as vehicles for the efficient delivery of the CRISPR/Cas system in the context of diagnosis and cancer treatment.
Development of CRISPR Systems
The CRISPR/Cas system, initially inherent to bacterial immunity, has been subsequently employed for gene editing purposes. This innovative technique allows for the precise engineering of genetic material and greatly enhances the feasibility of conducting genomic studies within mammalian systems. A conventional CRISPR/Cas system typically comprises a guide RNA (gRNA) and an associated Cas RNA-guided nuclease (RGN). The CRISPR genes in bacteria are responsible for encoding a range of short repeats and spacers. The spacers are obtained from foreign DNA sequences that have been collected by bacteria and serve as a "blacklist" within the immune system. The presence of palindromic sequences within the short direct repeats facilitates the formation of a hairpin structure. This hairpin structure can then undergo processing to generate functional trans-activating crRNA (tracr-RNA) and CRISPR RNA (crRNA). CRISPR genes, organized within an operon expression system and positioned adjacent to highly conserved CRISPR/Cas genes, are responsible for target cleavage, crRNA processing, spacer acquisition, and essential functions [17]. The Cas RGN, with the assistance of crRNA and tracrRNA, is capable of identifying and interfering with the foreign sequence to protect against invasive infections [18].
In a broad sense, the CRISPR/Cas system exhibits the ability to identify the intended target, initiate the process of cleavage, and subsequently activate the DNA repair process. The Cas9 RGN is an example of the CRISPR/Cas9 system's capacity to locate a small protospacer adjacent motif (PAM) within the target. Subsequently, it engages in precise base pairing either with a hybrid of crRNA and tracrRNA or with a single gRNA (sgRNA) carrying a 20-nt protospacer [19]. The Cas9 enzyme, serving as an endonuclease, exerts its activity by cleaving DNA's complementary and non-complementary strands, inducing a double-strand DNA break (DSB). The DNA repair process is subsequently activated through precise homology-directed repair (HDR) or error-prone nonhomologous end-joining (NHEJ) [20]. The repair process induced by NHEJ often results in the generation of staggered ends, which can lead to the occurrence of undesired errors such as genetic insertions and deletions. The process of HDR can facilitate the precise introduction of site-specific insertions, rearrangements, nucleotide substitutions, and deletions within the genomic sequence when a homologous donor template is accessible [20]. Hence, the utilization of HDR-mediated CRISPR/Cas editing has become a prevalent method for achieving precise genetic modifications. Since NHEJ is prone to errors while HDR has a greater level of fidelity, further investigation is essential to achieve a deeper understanding of the DNA repair processes that are activated following CRISPR/Cas gene editing. This understanding is crucial to develop a well-designed CRISPR/Cas approach that can effectively serve various editing requirements.
The practical application of CRISPR technology offers new avenues for advancing gene engineering in the direction of clinical applications. The clinical trial (NCT02793856), utilizing the CRISPR/Cas system, was initiated in China during the year 2016. In this ex-vivo study, the T-cells were modified using CRISPR-mediated gene knockout techniques to target programmed death ligand-1 (PD-L1) for treating non-small cell lung cancer (NSCLC). The programmed death-1 (PD-1) and PD-L1 pathways were effectively inhibited through the implementation of gene knockout techniques. Following this intervention, the T-cells were artificially propagated and subsequently reintroduced into the patient. The present clinical findings have indicated the therapeutic potential and safety for conducting large-scale trials [22, 23]. However, in 2019, EDIT-101 was approved to begin clinical studies (NCT03872479). The CRISPR/Cas system, delivered through a virus, was specifically engineered to target and interfere with the CEP290 gene as a therapeutic method for treating LCA10 [24]. The administration of treatment to the initial participant enrolled in the EDIT-101 trial occurred in the early months of 2020, signifying a noteworthy achievement in the clinical application of CRISPR technology.
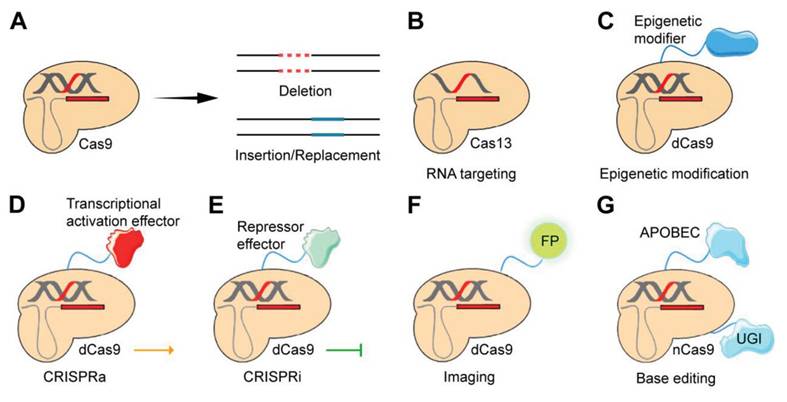
Some applications of the CRISPR/Cas gene-editing system. A) Cas9, as a key player in gene editing, primarily facilitates the precise manipulation of genetic material by enabling targeted gene deletion as well as the insertion or replacement of genetic sequences. B) Certain CRISPR systems, such as Cas13 orthologs, possess the capability to selectively target RNA molecules rather than DNA. C) dCas9 possesses the capacity to be genetically manipulated to incorporate epigenetic modifiers, facilitating the induction of epigenomic editing. D and E) dCas9 can be genetically modified through the incorporation of trans-effectors. This modification enables the establishment of two distinct systems: CRISPRa, which is associated with the activation domain, and CRISPRi, which is associated with the repressor domain. F) A fluorescent protein (FP) can be fused to CRISPR to enable imaging. G) The integration of nCas9 with APOBEC1 and UGI is the foundation of CRISPR/Cas9-based editing [21].
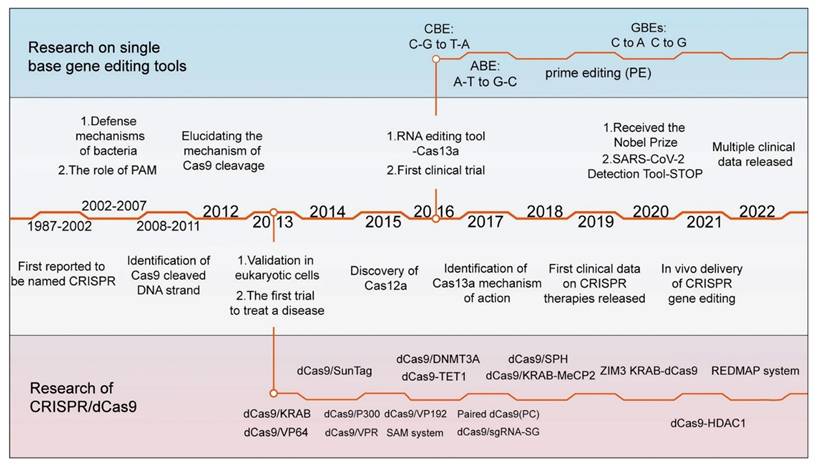
A detailed timeline highlighting major milestones in the development of CRISPR/Cas technology and the key variants of the Cas9 enzyme. The initial documentation of the CRISPR sequence occurred in 1987. In 2012, the mechanism of DNA double-strand cleavage by Cas9 was elucidated, paving the way for its subsequent use in gene editing in mammalian cells. Subsequently, the advancement of CRISPR technology has been swift, leading to the identification of various Cas9 variants that possess distinct functionalities [25].
Types of CRISPR/Cas Systems
Two different variants of CRISPR/Cas systems have been identified, each characterized by distinctive configurations of interference effectors. These variants have been deliberately modified and employed as invaluable toolkits for precise gene editing [26]. The class 1 system has effector complexes with multiple subunits, while the class 2 system, which is the main focus of this review, uses single-protein effectors to edit genes in mammalian cells. Currently, researchers are more interested in class 2 systems and their respective derived variations, such as CRISPR systems that target RNA and DNA. The aforementioned findings effectively clarify the extensive range of functionality and developmental background of CRISPR/Cas [17].
One of the extensively studied DNA-targeting CRISPR/Cas systems is Cas9, which possesses HNH and RuvC nuclease domains. Currently, the utilization of the Cas9 RGN derived from S. pyogenes (Sp-Cas9) is prevalent in the field of DNA gene editing [27]. In addition to the SpCas9 variants, other Cas9 orthologues, including N. meningitidis Cas9 (Nm-Cas9), S. thermophilus Cas9 (StCas9), and S. aureus Cas9 (SaCas9), have also undergone optimization [28].
In addition to the well-known CRISPR/Cas9 system, various other CRISPR/Cas systems exhibiting distinct properties have been evaluated for their potential to edit genes [26]. For instance, Cas12, which is another efficient DNA-targeting subtype, possesses the ability to disrupt sequences of double-stranded DNA. Cas12a, also referred to as Cpf1, and the recently identified Cas12b are both constituents of the Cas12 family [29]. In contrast to Cas9, Cas12 has a physically smaller size with a RuvC domain and is only directed by a short and single crRNA. The preference is for a PAM sequence rich in thymine (T) at the 5' end of the protospacer, resulting in the production of a sticky end located away from the PAM site [17]. Cas12 demonstrates effective trans-cleavage activity, triggering nonspecific and robust ssDNA cleavage following sequence-specific dsDNA recognition. This property, discovered in 2018, has been extensively utilized, particularly in diagnostic applications [30]. CRISPR/Cas12 is capable of inducing double-stranded breaks (DSBs) to enable permanent insertion, gene knockout, or correction. Compared to Cas9, Cas12 offers key advantages, including the ability to generate staggered, or “sticky,” DNA ends, which can promote homology-directed repair and offer greater precision in genome editing [31]. Cas12 also allows for the simultaneous targeting of multiple distinct genomic loci, facilitating strategies for treating diseases that involve multiple genes [32]. Its non-specific single-stranded DNA cleavage activity improves its effectiveness in molecular diagnostics, including the detection of specific DNA sequences linked to infectious diseases and cancers [33].
CRISPR/Cas13, a distinct member of the CRISPR family, is unique in its ability to target RNA instead of DNA. As part of the Class 2, Type VI system, Cas13 features a HEPN domain that facilitates RNA binding and cleavage, guided by a single RNA molecule [34]. The Cas13 system was identified in L. shahii and is characterized by the presence of a pair of nucleotide-binding nuclease domains of prokaryotes and higher eukaryotes [35]. Cas13a, also referred to as C2c2, is the prototypical variant, while subsequent discoveries led to the identification of Cas13b, CasRx (Cas13d), and other related variants, which exhibited a wide array of potential applications [17, 36]. Cas13a, one of its most commonly used subtypes [37], forms a complex with crRNA that becomes activated upon binding to its target RNA, cleaving both the specific target and adjacent non-specific RNAs. This trans-cleavage activity greatly expands the range of detectable targets [38]. Unlike Cas9, which targets DNA, Cas13 specifically interacts with and cleaves single-stranded RNA in a sequence-dependent manner, enabling its use for temporary gene expression regulation without causing permanent changes to the host genome [39, 40]. This characteristic makes Cas13 a valuable tool for identifying RNA sequences associated with viruses and cancer in molecular diagnostics [40]. Unlike Cas9, which exclusively targets specific DNA sequences without any off-target activity, Cas13 exhibits a unique collateral effect. Upon binding to its target RNA and activation, Cas13 can indiscriminately cleave other nearby RNA molecules. This property makes Cas13 highly effective for sensitive diagnostic applications, though it requires careful consideration when used in therapeutic contexts [41].
The simultaneous use of Cas12 and Cas13 enables dual-functionality within a single system, facilitating both precise DNA editing and RNA modulation. This integrated approach significantly broadens the scope of CRISPR technology, allowing researchers to target both the genome and transcriptome. It opens up new avenues for advanced genetic research, functional genomics, and RNA-based therapies, while also enabling more sophisticated diagnostic tools for detecting genetic mutations and RNA alterations in various diseases.
Doudna et al. have recently discovered Cas14, also referred to as Cas12f, which exhibits smaller sizes ranging from 400 to 700 amino acids [42]. This system exhibits the ability to selectively target both single- and double-stranded DNA without displaying any preference for a specific PAM. The utilization of Cas14 has been employed for identifying single-nucleotide polymorphisms, an important component in the early detection of diverse genetic disorders and malignancies with significant clinical implications.
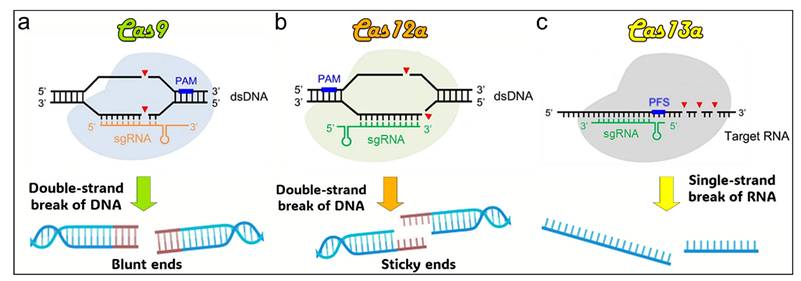
Illustration of DNA strand cleavage tools. (a) Cas9 induces double-strand breaks in DNA, generating blunt ends. (b) Cas12a cleaves DNA double strands, producing sticky ends. (c) Cas13a targets and cleaves RNA strands. PAM: protospacer adjacent motif. sgRNA: single guide RNA. PFS: protospacer flanking sequence [43].
CRISPR/Cas9 base editor
Point mutations, such as SNPs, are responsible for various genetic diseases, including certain cancers. To address these mutations, scientists have inactivated the Cas9 nuclease, allowing it to bind to DNA without causing cleavage. By fusing a base converter to the inactive dCas9, CRISPR/dCas9 systems can precisely convert one base to another, offering a targeted method for gene correction [44, 45]. The CRISPR/Cas base editor is an advanced genome-editing tool derived from CRISPR/Cas9. It enables precise nucleotide changes without causing double-strand breaks by fusing a base converter enzyme to the catalytically inactive dCas9. Two main types of base editors, adenine base editors (ABEs) and cytosine base editors (CBEs), are extensively used in genetic research, including cancer biology and treatment. CBEs convert C•G to T•A base pairs [45, 46] while ABEs facilitate the conversion of A•T to G•C base pairs [44]. To enhance editing efficiency and broaden the spectrum of targetable sites, base editors have been further refined into advanced versions such as BE3, ABEmax, and BE4. These advanced systems incorporate additional functional modules, including uracil glycosylase inhibitors (UGI) and optimized nuclear localization signals, improving both precision and applicability across diverse genetic applications [47]. Currently, by employing various Cas variants such as Cas12a, CRISPR/Cas base editors are capable of targeting approximately 95% of pathogenic transition mutations recorded in the ClinVar database [47, 48]. CRISPR base editors have been frequently utilized in cancer research, including the generation of CRISPR/Cas-modified animal models for investigating various cancer types [47, 49].
CRISPR/Cas9 prime editor
Prime editing, a recently developed CRISPR/Cas-based genome editing technology, enables the precise correction of all types of single-base substitutions as well as the insertion (up to 44 nucleotides) or deletion (up to 80 nucleotides) of genetic sequences [47]. Prime editing provides broader applications in both basic research and clinical gene therapy by allowing precise deletions, insertions, and all forms of point mutations. This technique involves a Cas9 nickase (nCas9 with an inactivated HNH domain) fused to reverse transcriptase, with a prime editing guide RNA (pegRNA) that not only guides Cas9 binding but also encodes the intended edit [47]. Designing an appropriate pegRNA is essential for effective prime editing, and it is more complex than designing a standard sgRNA. Fortunately, several computational tools for pegRNA design have been developed, making prime editing more accessible for both research and practical applications [50-53]. In prime editing, a specially designed prime editing guide RNA (pegRNA) attaches to the target site, directing the nCas9-RTase to cut at the target location. The pegRNA then serves as a template for synthesizing a new DNA strand, which replaces the targeted sequence [47]. The pegRNA includes a primer binding sequence (PBS), which binds to the 3′ end of the nicked target DNA strand, forming a primer-template complex [54]. Prime editing functions as a genome sequence tool for search and replacement without inducing DSBs or requiring donor DNA. Since the desired corrected sequence can be incorporated into the pegRNAs, prime editing is capable of not only correcting point mutations but also introducing small deletions or insertions at specific DNA sites with high accuracy. Prime editing has already been applied to make genomic alterations in various mouse embryos and human cell lines [55]. Prime editing has also been recently utilized to prepare patient-derived disease models, including the replication of mechanisms involved in cancer development [56, 57].
Challenges and safety measures in CRISPR-based gene editing
While CRISPR/Cas9 holds significant promise for cancer diagnosis and treatment, several challenges remain. One major issue is the possibility of CRISPR cutting DNA at unintended sites, causing off-target effects that may lead to harmful genetic alterations. These unintended modifications could increase cancer risks or provoke negative immune responses. Enhancing the CRISPR system's specificity is essential to reducing these risks, and this can be achieved through several approaches: i) Improving HDR efficiency by using small molecules like HDR enhancers (RS-1, a RAD51 activator, which improves DNA repair [58], enhancing the percentage of cells that perform HDR [59]; ii) Utilizing cell cycle regulators, such as checkpoint kinase 1 inhibitors, cyclin-dependent kinase inhibitors, and aurora kinase inhibitors like VX-680 (Tozasertib), to arrest cells in the G2/M phase temporarily. This strategy promotes HDR activity and reduces the effectiveness of CRISPR/Cas9 editing in slowly dividing or non-dividing cells [60]; iii) Enhancing sgRNA design through the incorporation of modified nucleotides or chemical alterations [61] to improve binding stability and specificity [62], as demonstrated in human primary T cells and CD34+ progenitor and hematopoietic stem cells [63]. Utilizing AI-driven bioinformatics tools, such as DeepCRISPR [64] and CRISPR-DO [65, 66], can further optimize sgRNA design. These machine learning (ML) algorithms have been created to predict the specificity and efficiency of guide RNAs, playing a crucial role in designing gRNAs that optimize on-target efficacy while minimizing off-target risks, thus enhancing the safety of CRISPR applications, particularly in therapeutic uses; iv) Enhancing the specificity of enzyme by employing high-fidelity Cas9 variants, such as eSpCas9 [67], HypaCas9 [68] and SpCas9-HF1 [69], which have been engineered with mutations to minimize binding to off-target sites, reducing unintended genomic cuts and preventing undesired genetic alterations [70, 71]; v) Employing base editors, such as CRISPR/Cas9 fused with deaminases, which facilitate the direct chemical modification of individual DNA bases without inducing DSBs, enabling precise single-base editing essential for correcting point mutations [72]; vi) Utilizing prime editing, a method designed can produce specific insertions, deletions, or point mutations without the need for HDR [47]. This approach involves a fusion protein and a prime editing guide RNA, incorporating a Cas9 nickase (a modified form of CRISPR/Cas9 that induces single-strand 'nicks' in DNA, as opposed to the DSBs caused by standard Cas9), along with an inactivated HNH domain and a custom-engineered reverse transcriptase domain [73]. The described approach minimizes the risk of off-target effects and provides a wider scope for genetic modifications [74]; vii) Employing advanced screening methods for off-target detection, such as GUIDE-seq [75] and Digenome-seq [76], which are sequencing-based approaches that map unintended DNA cuts to identify off-target effects. Digenome-seq and GUIDE-seq are used to assess the accuracy of CRISPR edits and optimize gRNA designs to minimize off-target effects. CHANGE-seq is a streamlined, highly sensitive, and automatable tagmentation-based method that assesses the genome-wide activity of Cas9 to evaluate the specificity of genome editing tools [77]. CIRCLE-seq is a newer, faster, and more sensitive method that identifies off-target effects by sequencing circular DNA fragments, offering an accurate assessment of unintended DNA breaks [78, 79]. Moreover, Cas9 derived from bacterial species such as Staphylococcus aureus (SaCas9) and Streptococcus pyogenes (SpCas9) may be recognized as foreign by the human immune system through pattern recognition receptors (PRRs), triggering cytokine release and inflammation, which can lead to undesirable adaptive and innate immune responses [80]. The risk of these adverse immune reactions can be reduced by: (1) employing less immunogenic Cas9 variants derived from rare bacterial strains [81], which can be identified using prediction models [82], or (2) by employing low-immunogenicity Cas9 orthologs from non-pathogenic bacterial species, including Geobacillus stearothermophilus, which has shown remarkable stability in human plasma [83]. 2. Delivering Cas9 transiently, such as in the form of mRNA or protein rather than DNA, shortens its expression time and reduces exposure to the immune system [84, 85]. 3. Humanizing the Cas9 protein by incorporating sequences or motifs that resemble human proteins, decreasing the immune recognition [86, 87]. 4. Utilizing less immunogenic delivery systems, including polymer-based carriers and lipid nanoparticles, to protect Cas9 from immune recognition [88, 89]. Understanding the genotoxic effects linked to therapeutic CRISPR-Cas9 genome editing is essential. Although considerable focus has been directed toward unintended "off-target" DSBs [90], these events can be minimized by employing more precise gRNAs, high-fidelity Cas nucleases, or alternative strategies such as the use of double-nickases [90]. Chromothripsis has recently been identified as an unexpected consequence of on-target Cas9-induced DNA breakage. In cells undergoing active division, the use of Cas9 for genome editing can lead to a substantial rise in the micronuclei and chromosome bridges formation, abnormalities that have the potential to initiate chromothripsi. Beyond contributing to rare human congenital disorders [91, 92], chromothripsis is frequently observed in cancers, where it plays a significant role in tumorigenesis by promoting tumor suppressor loss, generating fusion oncogenes, or amplifying oncogenes through the formation of circular double minute chromosomes [93, 94]. Strategies employing dead Cas9 or Cas9 nickase such as base editing, prime editing, and epigenome editing reduce the formation of DSBs and likely decrease the risk of chromosomal bridge formation and CRISPR-induced chromothripsis. However, since technologies like base and prime editing still involve Cas9 nickase and may occasionally produce DSBs, it remains important to investigate the potential occurrence of these genomic rearrangements whenever evidence of DSBs is detected following Cas9 nickase treatment.
Delivery Systems for CRISPR/Cas9
The delivery methods for CRISPR elements typically encompass both systemic and local administration strategies. In the treatment of cancer, the utilization of CRISPR delivery systems emerges as a pivotal factor in the pursuit of gene editing capabilities and the subsequent attainment of therapeutic efficacy. Several studies have shown the feasibility of employing three distinct variations of the CRISPR/Cas system. These include the use of a highly negatively charged plasmid encoding CRISPR/Cas mRNA and gRNA, the integration of Cas9 protein into an RNP complex with a theoretical charge of approximately +22 mV, and the use of sgRNA containing nearly 100 anionic phosphate groups [95, 96]. So far, researchers have diligently investigated different delivery methods, encompassing physical methods, non-viral vectors, and viral vectors. Physical delivery techniques, including microinjection and electroporation, have been employed to introduce the CRISPR/Cas9 system into cells. Electroporation uses electric currents to prepare temporary pores in the cell membrane, allowing the entry of CRISPR/Cas9 components into the cells [6]. Electroporation offers a cost-effective, convenient, and highly efficient method with good scalability. However, the electrical pulses used can damage cell membranes, resulting in toxicity for many cell types [97]. The level of toxicity varies depending on the type of material introduced, with mRNA causing significantly less toxicity compared to plasmid DNA [98]. Other drawbacks include the complexity of factors affecting transfection outcomes, as multiple biological and physical parameters (such as pulse duration, amplitude, and waveform) must be optimized for each cell type to maximize transfection efficiency while minimizing toxicity [99]. Electroporation requires specialized equipment. Efforts to apply electroporation in vivo face significant challenges, including low efficiency, the requirement for invasive processes, tissue damage caused by electric pulses, and inflammation resulting from the described damage [100]. Furthermore, safety concerns arise due to the necessity of high-voltage sources for electroporator operation [97]. As a result, the primary use of this method remains ex vivo and in vitro cell transfection. However, the process involves variable conditions and can threaten cell viability. Microinjection is a straightforward and widely used technique that allows the direct delivery of plasmid DNA, mRNA (including sgRNA), into the nucleus or cytoplasm of individual cells by piercing the nuclear and cell membrane with a micrometer-scale pipette under visual guidance [101]. Due to its precise control over the quantity, composition, and exact placement of delivered agent, microinjection is often regarded as the "gold standard." It is not constrained by the chemical nature or molecular weight of the cargo and does not require the use of genotoxic and immunogenic vectors [102]. Microinjection, which uses microscopes and fine needles to deliver RNPs into zygotes for generating small animal models, is limited by its high cost, technical complexity, low efficiency, and inability to support in vivo editing [103]. For clear reasons, microinjection is restricted to in vitro and ex vivo use and can only be applied to a limited number of cells, rendering it unsuitable for high-throughput cell engineering. Consequently, this limits its applicability in most clinical settings. These strategies have found application in the context of CRISPR-engineered CAR-T/NK cell therapy [104]. However, it is important to note that current physical methods are constrained by the inherent limitations of cellular function and the challenges associated with their application in vivo [105]. In the subsequent section, we will go into a more thorough review of EVs and viral vectors. However, alternative methodologies, including nanocomplexes and physical methods, were not covered in this review. Readers interested in more information on these methodologies can refer to the cited papers that discuss them [105-107].
Delivery vectors containing CRISPR cargos, such as RNP, mRNA/gRNA, and plasmid DNA, intended for delivery to internal tissues must undergo a sequential process of traversing the vessels and blood circulation, the space of Disse, and finally reaching the hepatocytes [108]. In the context of in vivo administration, successful therapeutic applications of CRISPR systems require overcoming several delivery-related challenges. Firstly, the considerable size of these cargoes poses a significant obstacle. Secondly, their biological stability is limited due to degradation by nucleases found in physiological fluids. Thirdly, their ability to cross cell membranes is restricted by their highly negative charge and hydrophilic characteristics. Lastly, even after being taken up by cells, there is a high likelihood of degradation within lysosomes and endosomes. Hence, it is imperative that the CRISPR/Cas delivery system has sufficient protection and exhibits stability when situated away from the intended site. Further, it should demonstrate proficient internalization into cellular structures and effectively release the cargo for gene editing.
Non-viral vectors
Non-viral delivery, with its transient expression nature, is considered a safer approach and has been widely used for gene delivery. Several non-viral vectors are currently being developed that show promising potential for translation. However, non-viral vectors can be less effective at delivering their payload due to several extracellular and intracellular barriers. In a broad sense, the CRISPR delivery system, when administered at an optimized dosage, should effectively traverse the bloodstream and interstitial space without being eliminated or degraded by the body's protective mechanisms. Subsequently, it effectively traverses the cellular membrane of specific target cell types [106].
Liposomes or nanoparticles composed of lipids, characterized by their hydrophilic heads and hydrophobic tail groups, represent the prevailing vectors employed in gene delivery applications, with numerous commercially available products [108]. Lipid-based vectors have the potential to minimize adverse immune reactions and off-target effects, thus providing a favorable environment for the CRISPR/Cas system to achieve optimal efficiency for gene-editing [109]. Cellular membranes possess efficient encapsulation capabilities and exhibit a strong affinity, rendering them highly desirable for the delivery of CRISPR/Cas systems [106]. Subsequent developments may prioritize physicochemical parameters encompassing molecular structure, size, and rigidity. These vectors exhibit promising characteristics that make them suitable for use as a functional and protective shell. They have the potential to effectively combine the advantages of other biomaterials, serving as versatile platforms for encapsulating CRISPR/Cas systems. For example, researchers have developed a strategy called Selective Organ Targeting, which enables the systematic engineering of nanoparticles for the precise delivery of various cargoes, including sgRNA, mRNA, and Cas9 RNP complexes, to the lungs, spleen, and liver of mice following intravenous administration. In this study, the authors demonstrate that the supplemental component addition, referred to as a selective organ targeting molecule, precisely modulates the RNA delivery profile (in vivo), enabling tissue-specific gene delivery and editing. They provide evidence supporting tissue-specific delivery, establish the applicability of this approach to various nanoparticle systems, and introduce a new method for designing predictable LNPs to target therapeutically relevant cells. The results indicate that the intravenous co-delivery of sgRNA and Cas9 mRNA represents a safe and efficient approach for enabling gene editing [110].
Emerging and prospective methods of delivering CRISPR/Cas systems using both non-viral and viral systems. The CRISPR/Cas systems are available in three distinct formats: RNP, Cas mRNA/gRNA, and plasmid DNA. These formats can be encapsulated within delivery vectors to facilitate their application in gene therapy, with the ultimate goal of advancing towards clinical applications [21].
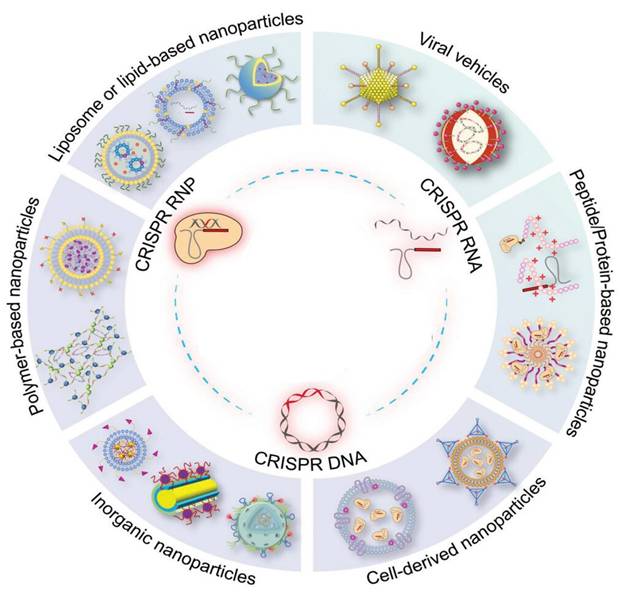
Polymer vectors, which usually consist of cationic groups, have been extensively studied for their ability to carry CRISPR/Cas elements. The cationic nature of these vectors enables them to be electrostatically complex and protect negatively charged genetic cargoes, promoting improved cellular uptake [111]. The delivery potential of polymeric vectors can be significantly influenced by various physicochemical properties such as surface modifications, branching extents, molecular weight, biodegradability, pKa, and charge density [112]. The effective delivery of CRISPR cargo relies heavily on the relational development of polymeric nano-carriers [113]. Several different types of functional polymers can be used to transport gene-editing machinery. The physicochemical characteristics of these materials can be adjusted, making them suitable for use at the animal and/or cellular level. However, cytotoxicity is often encountered during administration with polymeric nano-carriers, particularly when the monomers are not adequately removed [106].
Inorganic nanoparticles, with a particular emphasis on gold nanoparticles, are currently undergoing thorough investigation for their potential as carriers for CRISPR delivery. Zhang et al. effectively engineered a gold nanocluster (GNC) by incorporating the HIV-1 trans-activating transcriptor (TAT) peptide. This innovative modification allowed for the seamless integration of a Cas9-encoding plasmid. To enhance liver targeting, the researchers then successfully enveloped the GNC-TAT-Cas9 complex with a galactose-based liposome layer [114]. The in vitro findings have yielded a remarkable 57% efficiency in gene editing, resulting in a concomitant reduction in protein expression. Furthermore, the in vivo investigations have unequivocally showcased a substantial decline in PSCK9 protein levels, coupled with a noteworthy 30% reduction in serum levels of low-density lipoprotein cholesterol. In addition to other inorganic nanoparticles, black-phosphorus nanosheets, graphene oxide nanoparticles, and mesoporous silicon material have emerged as potential carriers for gene-editing tools [115].
A summary of current research focused on delivering CRISPR/Cas into cancer cells by employing EVs or viral vectors as delivery systems.
| Cancer type | Vector | Approach | Outcome | Ref |
|---|---|---|---|---|
| B-cell malignancies | EVs | EVs with anti-CD19-CAR loaded with the MYC oncogene-targeting CRISPR/Cas9 system. | The MYC gene was subjected to loss-of-function mutations induced by CRISPR/Cas9 in CD19 + cells. | [116] |
| Pancreatic cancer | Exosome | CRISPR/Cas9-loaded exosomes that target mutant KrasG12D oncogene | Suppressing tumor growth and inhibiting proliferation in orthotopic and syngeneic models of pancreatic carcinoma. | [117] |
| Ovarian cancer | Exosome | PARP-1-sgRNA and Cas9-loaded tumor exosomes | Ovarian cancer cells undergo apoptosis and enhanced sensitivity to cisplatin after PARP-1 suppression by CRISPR/Cas9. | [118] |
| Recipient cells | Exosome | CRISPR/dCas9 or miR-155 loaded CD9-HuR exosomes | Enhanced loading of RNA cargo into exosomes | [119] |
| Mesenchymal stem cells (MSCs) | Exosome | CRISPR/Cas9 loaded hybrid liposomes-exosomes | Hybrid exosomes were used to effectively deliver CRISPR/dCas9 and inhibit the hCTNNB1 and mRunx2 expression in MSCs. | [57] |
| AML-M5 | RBC-Evs | gRNA, mRNA, and HA-tagged Cas9-loaded RBC-EVs | Exosomes were found to effectively transfect both xenograft mice and human cells, without any significant cytotoxic effects. | [54] |
| Pancreatic cancer | Adenovirus and Lentivirus | Genomic tampering of cancerous cells of the pancreas through the utilization of CRISPR/Cas9 for producing genetically modified mouse lines | This approach facilitates the examination of molecular modifications that underlie the progression of pancreatic cancer at every stage. | [120] |
| Patient-derived xenograft (PDX) cancer model | Adenovirus | Targeting translocated genes' introns using CRISPR/Cas9 | The knockout cancer-causing mutation in cancerous cells results in a targeted and proficient mechanism for the eradication of malignant cells. | [121] |
| Gastric cancer | Lentivirus | Genomic-scale gastric cancer CRISPR-Cas knockout library | Out of the 184 different genes associated with gastric cancer, inhibiting methyltransferase-1 has been the most extensively validated method for targeted treatment against cancer. | [122] |
| Hepatocellular Carcinoma (HCC) | Lentivirus | CRISPR-Cas9-based genomic screening for the identification of therapeutic factors that are responsible for the survival of hypoxia in HCC | The knockout of PTPMT1 leads to the generation of ROS and triggers cell death in hypoxic cells of HCC. | [123] |
| Prostate cancer | Lentivirus | CRISPR_Cas9-based screening to identify genes that have a role in metformin insensitivity in prostate cancer | The activation of NIPSNAP1, RAD9A, EEF1A1, BPY2, ABCA12, and ECE1 linked to metformin resistance. | [124] |
| Colon cancer | Lentivirus | CRISPR_Cas9-based screening to identify genes that have a role in the modulation of oxidative stress. | The overexpression of Gal2 results in a reduction in the growth of colon tumors in humans. | [125] |
| Cervical cancer | AAVs | Inducing several changes in the HPV E6 gene within cervical carcinoma cells utilizing the CRISPR/Cas9 | Inhibition of tumor growth by overexpression of tumor suppressors and an increase in apoptosis within cancerous cells. | [126] |
| Anal cancer | AAVs | CRISPR/Cas9-mediated HPV16-E6 or -E7 gene cleavage in human anal tumor cells | Effective and targeted inhibition of tumor growth. | [127] |
Extracellular vesicles (EVs)
EVs are submicron-sized non-viral carriers that possess the potential for diverse applications, including their utilization as a targeted delivery platform. EVs are membranous structures enveloped in lipids that are synthesized by various cells with the inherent purpose of facilitating intercellular transport of cargo, including proteins and genetic material [128]. EVs can be classified into three primary classifications: apoptotic bodies, exosomes, and microvesicles (MVs). They are distinct from one another in terms of their ability to function, biogenesis, packaging, and the process by which they are released. Among these, exosomes with diameters ranging from 30 to 150 nm are real choices for the selective transport of genetic materials and proteins, such as the CRISPR/Cas system [129, 130].
Exosomes are gaining significant interest due to their potential applications in the fields of cancer diagnosis and treatment. Exosomes derived from cancerous cells possess considerable potential as a promising modality for cancer-targeted treatment due to their inherent similarity to the parent cells, enhancing their cellular uptake capability. The implementation of a cell-specific affinity approach in cancer treatment can be enhanced by regulating this characteristic and incorporating desired payloads into exosomes derived from cancerous cells. It was found that the systemic injection of tumor cell-derived exosomes loaded with Doxil into the tissue of origin can lead to enhanced tumor suppression compared to the administration of the drug in isolation [131]. Researchers developed a novel approach by encapsulating doxorubicin (DOX@E-PSiNPs) within biocompatible porous silicon nanoparticles (PSiNPs) and introducing them to isolated tumor cells. These DOX-loaded exosomes were then administered intravenously to mice, where they were successfully taken up by both cancer stem cells and tumor cells, leading to a substantial inhibition of tumor growth [132]. The exosome's potential for delivering CRISPR/Cas to cancer cells can be inferred from their capacity to transport targeted and effective delivery systems. In light of the exosomal potency, a recent investigation employed exosomes derived from tumors to facilitate the in vivo administration of CRISPR/Cas9 into SKOV3 xenograft mice cells afflicted with ovarian cancer. They compared the potency of exosomes derived from cancer and epithelial cells in targeting the delivery of CRISPR/Cas9 and downregulating the poly(ADP-ribose) polymerase-1 (PARP-1) expression. The findings revealed a notable occurrence of apoptosis in the ovarian cancer cells, accompanied by a synergistic augmentation of chemosensitivity towards cisplatin. The confluence of both therapeutic methodologies led to a 57% suppression of cancer cell growth, which is nearly double the level of inhibition observed with either cisplatin or exosome treatment alone [133]. A significant issue regarding the utilization of exosome-mediated gene delivery, especially for CRISPR/Cas delivery, is the potential for adverse effects on non-targeted distant and peripheral tissues.
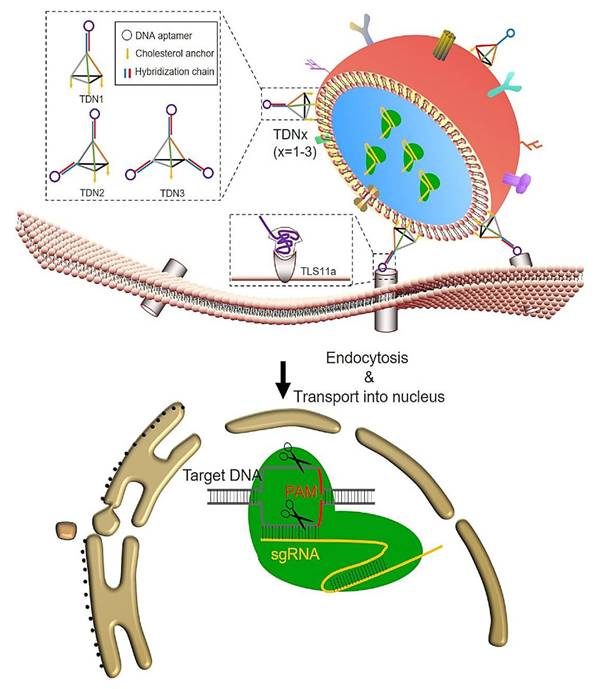
The development of exosomes as targeted carriers is an important objective, alongside their existing packaging capabilities. Alvarez-Erviti et al. produced short interfering RNA (siRNA)-loaded exosomes with a specific affinity for the brain by using dendritic cells that were genetically modified to express Lamp2b, a membrane protein present on exosomes. To enhance their brain-targeting capabilities, the researchers fused the Lamp2b protein with the rabies viral glycoprotein (RVG), which is known to have a specific affinity for the nervous system. The findings from the in vivo administration of these exosomes in mice demonstrated robust therapeutic capabilities and the absence of non-specific absorption by surrounding tissues [134]. Another investigation employed a comparable methodology to deliver miRNA within the cartilaginous tissue as a therapeutic intervention for osteoarthritis, exhibiting an interesting capability to selectively target hard-penetrating tissues [135, 136]. Exosomes also have the potential to be modified through the incorporation of DNA aptamers, which are short synthetic oligonucleotides. This modification allows for the use of exosomes as a precise and targeted delivery system. DNA aptamers exhibit advantages such as their non-stimulatory nature towards the immune system, being readily available, and being cost-effective. Consequently, they emerge as a viable substitute for antibodies or other probing agents [137, 138]. Zhuang et al. employed cholesterol-anchored, valency-controlled tetrahedral DNA nanostructures (TDNs) that were coupled with DNA aptamers on the outer layer of exosomes [139]. This approach was used to specifically carry the CRISPR/Cas system into HCC to suppress the WNT10B gene. The experiments were conducted ex vivo, in vitro, and even in vivo. The method demonstrated promising outcomes in the selective inhibition of genes in HCC, exhibiting the potential of EVs as effective carriers for the precise delivery of the CRISPR/Cas system.
Hybrid exosomes, which are a fusion of exosomes derived from cells and liposomes synthesized in the laboratory, have been engineered for transporting substantial payloads such as the CRISPR/Cas system [140]. Hybrid exosomes exhibit an enhanced packaging capacity, which can be attributed to their unique characteristics. Owing to the positively charged nature of liposomes, these hybrid exosomes demonstrate improved interactions with negatively charged genetic material. This interaction facilitates their uptake through membrane fusion processes. This approach avoids the necessity of introducing genetic material into exosomes through mechanical processes like electroporation. It is of significance to note that the modification applied to exosomes did not exhibit any apparent effect on their ability to selectively target specific cell types and be internalized by them [141]. The approach described by Lin et al. has been employed to effectively introduce CRISPR/Cas9 expression vectors into MSCs, a cell type known for its low transfection efficiency [142].
Viral vectors
Viruses act as reliable vehicles for delivering genetic material. The use of pseudo-typed and recombinant viral vectors has made significant progress in the field of mammalian gene therapy. The adeno-associated viruses, adenoviral vectors, and lentiviral vectors are the predominant viral vectors employed for the CRISPR/Cas system's delivery. These vectors have gained significant attention in ongoing clinical trials [127, 143, 144].
A strategy utilizing TDNs for targeting exosomes to deliver CRISPR/Cas9 specifically to liver cancer cells [139].
Adeno-associated viruses (AAVs)
AAVs are diminutive, non-enveloped entities containing a single strand of DNA. These viruses do not exhibit any pathogenicity toward the human host. The gene delivery systems derived from members of the Parvoviridae family have drawn significant interest among researchers [126]. Although approximately 80% of the overall population tests positive for these viruses, there is currently no reported association between human diseases and AAVs. AAVs possess several important features that make them highly suitable as delivery vehicles for CRISPR/Cas, principally in vivo applications. These features include their relatively low cytotoxicity, immunogenicity, and probability of chromosomal integration [145, 146]. Furthermore, a diverse range of AAVs can be employed to introduce genes into various cell types, including those found in the muscles, heart, lungs, and neurons. This characteristic makes AAVs highly effective as tropism vectors, particularly in applications requiring targeting specific tissues [147].
To a certain extent, AAVs still possess the capability of inserting their genetic material into the genome of the host. However, to address this constraint, researchers have devised a solution known as recombinant AAV [148]. The gene Rep found within AAVs is involved in the synthesis of Rep proteins (i.e., Rep40, 52, 68, and 78). These Rep proteins play pivotal roles in various essential processes such as viral genome packaging, gene expression, genetic material integration, and replication [149]. AAVs rely on Rep proteins, specifically Rep68 and 78, for precise site-specific integration. In genetically engineered versions of AAVs, the Rep gene is deliberately removed to facilitate safe and efficient gene delivery [150]. In general, the use of recombinant AAVs has demonstrated efficacy as gene delivery systems. For instance, the successful delivery of CRISPR/Cas9 to mice with a mutation in the low-density lipoprotein receptor (LDLR) gene resulted in therapeutic outcomes [151]. To date, numerous gene therapies using AAVs have received approval from the FDA. The examples include treatments for inherited retinal diseases and Pompe disease [152]. These successes serve as evidence for the efficacy of AAV vectors. Moreover, the proficient administration of the CRISPR/Cas9 system through AAVs has opened up new avenues for the establishment of disease models encompassing sickle cell disease, hepatic ailments, muscular dystrophy, and neurodegenerative disorders [123, 124].
Certain genes exceed the size limit of dual AAV vectors, which have a capacity of 9 kb. For instance, the DMD gene linked to muscular dystrophy and the CDH23 gene associated with Usher syndrome are approximately 11.1 kb and 10 kb, respectively. To facilitate the transduction of these specific genes into recipient cells, researchers have devised a triple AAV system [117, 118]. The use of CRISPR/Cas-mediated prime and base editing, employing non-functional or Cas9 nickase (nCas9) enzymes, presents a promising strategy in the field. This innovative approach allows for modification of the CRISPR-Cas system, enabling the utilization of a single vector delivery system. Consequently, this method effectively addresses the barriers associated with the constrained packaging capacity of viral vectors [116]. Additional features that distinguish this system include exhibiting outstanding proficiency in modifying non-dividing cells, avoiding the requirement for a DNA donor template, and the absence of DSB induction [54]. AAVs have demonstrated their potential as viable platforms for the delivery of CRISPR DNA base-editing tools. For example, a study showed the successful in vivo administration of the CRISPR/Cas-based cytidine-base editor using dual AAVs, demonstrating its therapeutic efficacy in addressing amyotrophic lateral sclerosis (ALS) within an animal model [126]. The efficacy of the AAV-mediated prime editor has been demonstrated in the correction of pathogenic alleles and cancer modeling in adult mice. It exhibits a particularly reduced off-target impact compared to the base editor based on the CRISPR/Cas system [57]. While it is genuine that AAV-based and prime editing systems have shown advancements in addressing certain limitations associated with AAV-mediated CRISPR/Cas9 delivery, it is important to keep in mind that certain challenges persist to a certain degree. These challenges encompass off-target activity, vector persistence, and viral-induced immune responses [116].
Adenoviral vectors (AdVs)
The AdVs are double-stranded DNA viruses. These viruses can infect a wide range of cells, including both dormant and actively dividing cells. The maximum genetic payload that may be carried by one of these vectors is 37 kb. Upon transduction, AdVs generate episomal DNA that remains adjacent to the host genome instead of being integrated into it. Notably, the episomal nature of gene expression in AdVs helps mitigate off-target effects, a common limitation associated with the CRISPR/Cas system [153].
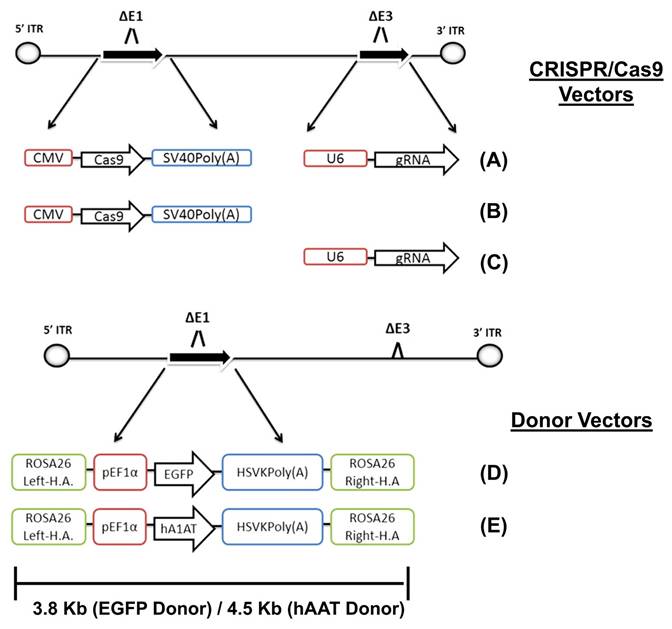
There are multiple generations of AdVs. In the first generation, the E1 gene was removed. However, it is important to understand that using this generation may lead to chronic as well as acute immune responses [154]. The immunological response was reduced by eliminating the AdVs E2 and E4 genes in the subsequent generation. The second generation has a significantly higher capacity (~14 kb) for incorporating transgenes compared to the first one (~8 kb) [155]. The gutted vectors, also known as helper-dependent vectors, are a more advanced generation of AdVs. These vectors lack viral genes, enabling them to accommodate larger DNA fragments of up to 35 kb. Consequently, they overcome the size limitation obstacle associated with carrying the CRISPR/Cas9. It is important to note that this generation of AdVs, like the second one, does not cause any chronic or acute immunological responses [156].
Recombinant AdV5 has been shown to enable in vivo CRISPR/Cas9-mediated knock-in of the human alpha-1-antitrypsin gene in mice, with stable gene expression for over 200 days [157]. Tsukamoto et al. used AdVs as a delivery system for CRISPR/Cas12a in human hepatocytes and demonstrated promising prospects for gene editing in human cells. However, an immune response against the Cas protein and vector was observed [158]. The structural proteins of AdVs, including fiber, penton, and hexon, exhibit a high degree of manipulability, offering significant benefits in the development of AdVs with the ability to selectively target specific tissues. AdVs have significant potential for CRISPR/Cas delivery because of their characteristics, such as being amenable to mass production, manufactured cost-efficiently, and safe for use in clinical trials [159]. The advantageous properties of AdVs were demonstrated by their selection as the preferred platform for the development of mRNA-based COVID-19 vaccines [160].
Adenoviral-mediated delivery of CRISPR/Cas9 enables targeted gene editing. This approach involves genetic designs based on CRISPR/Cas9 for precise homologous recombination [157].
Lentiviral vectors (LVs)
The LVs derived from HIV-1 are a type of ssRNA virus that are primarily employed to incorporate the desired transgene into both dormant and actively dividing cells. These vectors can accommodate a maximum genetic payload of approximately 9 kb. The LVs can be categorized into four generations, the last two generations being predominantly considered suitable for clinical applications [161].
The efficacy of CRISPR/Cas-mediated gene editing is intricately linked to the successful translocation of its constituent components into the cellular environment of the host organism. LVs have drawn significant attention due to their effectiveness in gene delivery [162]. Similar to short hairpin RNAs (shRNAs), LVs containing CRISPR/Cas9 systems were initially developed in a library format, incorporating a multitude of sgRNAs [163]. Shalem et al. reported that the use of an LV-based genome-scale CRISPR-Cas9 knockout library, which targets over 18,000 human genes, has proven to be highly advantageous for conducting positive and negative selection screening [162]. These screening methods are commonly employed in in vitro assays to identify cellular disruptions, particularly in cancer cells that have been influenced by diverse stimuli. This database has been utilized specifically to delineate genes that are essential for the survival of pluripotent stem cells and cancer cells. Furthermore, this technique facilitated the examination of gene loss-of-function mutations that confer resistance to the chemotherapy drug vemurafenib in tumor cells [162]. Studies have been conducted to enhance LV-based CRISPR/Cas libraries, leading to the discovery of previously unidentified tumor-suppressor genes implicated in the development of myeloid leukemia (Runx1, Tet2, Dnmt3a, Ezh2, and Nf1), as well as the reinduction of fetal hemoglobin (BCL11A) [164, 165].
For temporal advancement, the functionalities of the LV-delivered CRISPR/Cas9 system are progressively broadening their perspective within the field of targeted therapeutic interventions for HBV and HIV-1 infections, alongside improving the symptoms of genetically flawed diseases, including neurodegenerative disorders and cystic fibrosis [166]. Furthermore, the adoption of alternative techniques such as epigenetic modifiers and base-prime editing CRISPR systems has been observed in conjunction with LVs [167]. LVs can integrate these systems into the host genome effectively. However, the probability of off-target effects might be higher due to their sustained gene expression. To minimize this potential concern, researchers developed integration-deficient LVs as a means to offer a transient expression of CRISPR/Cas [168].
In the setting of unresectable HCC, the HIF-1α expression has the potential to adversely impact the prospects of cancer and impede the overall progress of patients. Liu et al. utilized an LV-based CRISPR/Cas9 system to knock out the HIF-1α gene in mice, demonstrating a significant decrease in HIF-1α expression in tumor tissues just three days post-injection. These findings highlight the promising antitumor effects associated with this approach [169]. Moreover, chromosomal translocations leading to the formation of fusion oncogenes have been observed in specific types of cancer. These translocations play a significant role in the progression and initiation of tumorigenesis. An instance of tyrosine kinase, which is generated as a result of BCR-ABL rearrangement, has the potential to give rise to chronic myeloid leukemia (CML) [170]. Imatinib, a tyrosine kinase inhibitor, has demonstrated substantial efficacy in inhibiting the expression of BCR-ABL. However, there have been reported cases of drug resistance emerging against these tyrosine kinase inhibitors [171]. Martinez-Lage et al. effectively employed the use of a lentiviral CRISPR/Cas9 platform to target intronic regions of the BCR-ABL fusion gene, effectively disrupting the BCR transactivation domain and introducing a frameshift mutation in the ABL DNA-binding domain [172].
Vexosomes
The application of exosomes as a gene delivery vehicle, specifically in the form of exosome-enveloped viral vectors, or vexosomes, represents an innovative approach in the field. This strategy draws on the inherent characteristics of exosomes, such as their ability to interact with a wide range of cells, their minimal immunogenicity, and their suitability for large-scale production. By incorporating these exosome features into viral vectors, the potential of gene therapy can be significantly enhanced [173]. The process of vexosome production requires the introduction of plasmids encoding the viral genome into the packaging cells through infection. The cytoplasm of the cells undergoes an enrichment process as the viral genome and proteins are generated. These components are then identified by the cell's receptors located in the phagosomes, endosomes, and plasma membrane. Hence, EVs effectively encapsulate the viral elements during their release from the cellular environment. The method employed by Saari et al. involved the generation of capsid-free EV-based vectors that contained oncolytic adeno-associated virus components. These AAV/EVs were then introduced into cancer cells [174]. The utilization of vexosomes for the delivery of the CRISPR/Cas system has not yet been explored. However, their potential to use the advantageous attributes of both EVs and viral vectors holds promise for improving the efficiency of gene editing procedures.
Targeted delivery of genes can be achieved through the modification of AAV/EVs, wherein targeting peptides are incorporated onto their surfaces. Several studies have reported the advantageous findings associated with the use of targeted AAV/EVs both in vivo and in vitro. For instance, Wood et al. conducted research indicating that these vectors possess the ability to traverse the blood-brain barrier and efficiently introduce genetic material into neural cells [175]. Various serotypes of AAV, such as AAV6, AAV1, and AAV9, have been employed to achieve specific transgene delivery into distinct cell types within the vestibular and cochlear systems. AAV1 has been employed for targeting vestibular and cochlear hair cells; AAV6 for neurons; and AAV9 for oligodendrocytes. These serotypes have demonstrated efficacy in facilitating targeted gene transfer in these specific cell populations [176, 177]. This observation suggests that the proposed methodology is highly compatible with the delivery of genetic material across a wide variety of cell types.
The EVs that encapsulate the AAVs possess the potential to function as Trojan horses, enabling evasion of the pre-existing immune response directed towards the viral vectors. Meliani et al. systematically administered AAV9/EVs into mice that possessed pre-existing immunity to AAV9. The findings of this study revealed a notable achievement in evading the immune system [178]. Moreover, this particular methodology has demonstrated its capability to decrease the frequency of injections necessary for optimal vector administration, mitigating the potential for AAV-induced cytotoxicity [179, 180]. Despite the potential advantages associated with this particular mode of delivery, the use of this delivery method for the CRISPR/Cas system has not yet been thoroughly investigated.
Clinical Trials Utilizing CRISPR Technology in Cancer Therapy
The in vivo application of CRISPR-based therapeutics requires rigorous assessment of safety, including off-target effects, immune responses, and long-term genomic stability. Advances in CRISPR-Cas9 delivery systems have significantly improved the precision and safety of these therapies in clinical applications. Encouraging results from preclinical in vitro and in vivo studies have demonstrated favorable safety profiles, justifying progression into human trials [181]. Frangoul H et al. conducted electroporation of CD34+ hematopoietic stem and progenitor cells from healthy donors, using CRISPR-Cas9 to target the BCL11A erythroid-specific enhancer. Around 80% of the alleles at this locus were successfully modified, with no indications of off-target editing [182]. The CRISPR/Cas technology represents a fundamental tool for gene editing; however, its impact in the field of clinical cancer therapy has been limited [183]. However, it is interesting to acknowledge the considerable potential that CRISPR-based genome editing holds for the advancement of cancer therapies in the near future (as indicated in Table 2). In 2016, the CRISPR/Cas9 system was applied for the treatment of NSCLC at West China Hospital, Sichuan University [22]. In the aforementioned study, it was observed that the T-cells of the patients were subjected to genetic modification, specifically targeting the suppression of PD-1, a T-cell activation inhibitor. This approach has been employed for the treatment of cancer in various tissues, such as the esophageal, renal, prostate, and bladder [184]. Stadtmauer et al. conducted a Phase I clinical trial evaluating the safety and practicality of CRISPR-Cas9-mediated engineering of T cells [185]. In this study, three patients with refractory cancers were enrolled, and CRISPR/Cas9 was utilized to delete two endogenous TCR genes, aiming to minimize TCR mispairing along with improved expression of a cancer-specific TCR transgene. The gene encoding PD-1 was removed to boost antitumor immunity. The edited T cells successfully engrafted and remained detectable for the overall duration of nine months, highlighting the promise of CRISPR/Cas9 technology in enhancing immunotherapeutic strategies. This groundbreaking trial set the stage for further research into targeted therapies and improving immunotherapy efficacy.
Two Phase I trials have demonstrated the safety and effectiveness of CRISPR/Cas9-based T cell editing in lung cancer. In a study by Lu et al., 22 patients with advanced NSCLC were enrolled, with 12 receiving T cells edited via CRISPR/Cas9 to target PD-1 [23]. Following infusion, edited T cells were found in the peripheral blood without causing any major adverse events. The median progression-free survival was determined to be 7.7 weeks, with 42.6 weeks of overall survival. Off-target events were evaluated using next-generation sequencing, which revealed 0.05% of median mutation frequency, providing additional evidence for the feasibility and safety of CRISPR/Cas9-edited T cells. Recently, Wang et al. enrolled 15 patients with mesothelin-positive solid tumors and employed CRISPR/Cas9 to create CAR-T cells targeting mesothelin, with PD-1 and TCR deficiencies. They assessed the response to escalating doses of the therapy [186]. Two patients experienced stable disease, with edited T cells circulating at their highest levels between days 7 and 14, before becoming undetectable after a month. No significant adverse effects or toxicities were reported, further supporting the safety and feasibility of CRISPR/Cas9-edited T cells. Liao et al. also identified PD-L1 as a promising target for CRISPR/Cas9-mediated knockout in osteosarcoma patients [187]. These results mark an important first step in establishing the safety and efficacy of CRISPR/Cas9 for treating NSCLC, sarcoma, and potentially other cancers, given the pivotal role of the PD-1/PD-L1 axis in cancer immune evasion and therapy. Various Phase I and II clinical trials are actively exploring the potential of CRISPR/Cas9 technologies for cancer therapy. One ongoing Phase I trial focuses on autologous T cells modified to target CD19, using CRISPR to knockout the HPK1 gene in CD19+ leukemia or lymphoma (NCT04037566). The use of CRISPR/Cas9 enables site-specific, consistent integration, reducing the risk of heterogeneous transgene expression, which is more common with traditional retroviral or lentiviral transduction. CTX110 and CTX112, designed to target CD19, are currently under evaluation in clinical trials for the treatment of relapsed or refractory B-cell malignancies (NCT05643742). Another construct, CTX130, is being tested for advanced, relapsed, or refractory renal cell carcinoma with clear cell differentiation, targeting CD70. Ongoing clinical investigations include Phase I trials focusing on PD-1 targeting in EBV-related cancers [188], along with Phase II studies evaluating therapies for CD19-positive leukemia and lymphoma (NCT03166878), as well as treatment-resistant or relapsed cases of these diseases[189]. It is especially important to note that certain clinical trials have opted to retract their investigations [190].
The advancement in the field of CAR T cell receptors has demonstrated interesting progress in the field of cancer therapy. The FDA has authorized Yescarta and Kymriah, two CAR T-cell therapies directed against CD19, for use in treating B-cell lymphoma and leukemia [191]. In light of the promising outcomes observed with autologous CAR T-cell therapy, it is imperative to understand the existing barriers that require further attention and resolution. In some cases, such as with infants, there may be an inadequate supply of T-cells to produce CAR-T cells for autologous transplantation. The incorporation of the CRISPR/Cas into CAR T-cell engineering enables the creation of universal CAR T-cells sourced from healthy donors [192]. The integration of CAR genes into T-cells by viral vectors occurs in a non-targeted manner, lacking site-specificity, potentially leading to unfavorable consequences on the genome. Eyquem et al. utilized CRISPR/Cas9 to precisely insert the CD19-specific CAR gene into the T-cell receptor α constant (TRAC) locus. The resulting modified T-cells exhibited enhanced safety, precision, and therapeutic effectiveness in selectively eliminating acute lymphoblastic leukemia cells compared to earlier CAR T-cell designs [193].
Considering the potential of introducing the CRISPR/Cas system into human cells, it appears to possess significant efficacy as a tool for safeguarding against cancer-causing viruses [194]. Certain viruses can induce the development of cancer in humans. Viruses such as human papillomavirus (HPV), Epstein-Barr virus (EBV), and hepatitis B and C viruses (HBV and HCV) have been linked to the emergence of certain cancers, including Burkitt lymphoma, cervical cancer, and hepatocellular carcinoma (HCC), respectively [195-197]. Wang et al. reported that targeting EBV genes such as EBNA-3C, LMP-1, and EBNA-1 with CRISPR/Cas9 in Burkitt's lymphoma cells from a patient with latent EBV infection resulted in a significant decrease in both cell proliferation and viral load [198]. This particular approach has garnered support from various studies. Since EBV contributes to the onset of various malignancies, CRISPR/Cas9-based targeted therapies offer considerable potential for preventing the progression of these diseases.
Limitations and Future Directions
Using viral vectors for delivery raises concerns about potential side effects linked to the presence of viral particles. The systemic application of viral vectors like adenoviruses is often restricted due to the existence of innate immunity against specific serotypes in humans, acquired through natural vaccinations or infections. Therefore, detecting and reducing immunogenicity is one of the most challenging aspects of viral delivery for CRISPR/Cas9 components. Several strategies, utilizing non-human AdV vectors and AdV engineering via copolymer encapsulation, are currently being employed to minimize the impact of cross-reactive immunity [199, 200]. Besides this, an alternative strategy involves employing a heterologous prime-boost regimen, delivering the same antigens through different vectors [201]. Repeated administration of viral vectors can induce the formation of neutralizing antibodies, potentially reducing the overall efficacy of the therapy [202].
Clinical trials studying the efficacy of cancer treatment utilizing CRISPR/Cas technology.
| NTC Number | Interventions | Disease or condition | Phases | Age (Years) | Gender | Status of trial |
|---|---|---|---|---|---|---|
| 02793856 | PD-1 Knockout T-Cells (Biologic) | Cyclophosphamide (Drug) | NSCLC | Phase-1 | 18-70 | All | Completed |
| 03044743 | Interleukin-2 (Drug) | Cyclophosphamide (Drug) | Fludarabine (Drug) | Lymphoma (Stage IV)|Nasopharyngeal Carcinoma (Stage IV)|Gastric Carcinoma (Stage IV) | Phase-1 | Phase-2 | 18-75 | All | Unknown status |
| 03081715 | PD-1 Knockout T-Cells | Esophageal Cancer | N/A | 18-80 | All | Completed |
| 03166878 | UCART019 (Biologic) | B-Cell Lymphoma|B-Cell Leukemia | Phase-1 | Phase-2 | 12-75 | All | Unknown status |
| 04037566 | XYF19 CAR-T cells (Genetic) | Fludarabine (Drug) | Cyclophosphamide (Drug) | CD19 Positive|B-Cell Lymphoma|Acute Leukemia Lymphocytic (ALL) in Refractory or Relapsed | Phase-1 | 18-55 | All | Recruiting |
| 04244656 | CTX120 (Biologic) | Multiple Myeloma | Phase-1 | 18< | All | Recruiting |
| 04417764 | PD-1 knockout T-cells (Biologic) | Hepatocellular Carcinoma (Advanced stage) | Phase-1 | 18-70 | All | Unknown |
| 04438083 | CTX130 (Biologic) | Adult Kidney Cancer | Phase-1 | 18< | All | Recruiting |
| 04502446 | CTX130 (Biologic) | T-Cell Lymphoma | Phase-1 | 18< | All | Recruiting |
| 04557436 | PBLTT52CAR19 (Drug) | B-Cell ALL | Phase-1 | 0.5-18 | All | Recruiting |
| 04637763 | CB-010 (Genetic) | Fludarabine (Drug) | Cyclophosphamid (Drug) | Lymphoma | Phase-1 | 18< | All | Recruiting |
| 04976218 | TGF²R-KO CAR-EGFR T-Cells (Biologic) | EGFR Over-expression | Solid Tumor | Phase-1 | 18-75 | All | Recruiting |
| 05066165 | Arm 1, NTLA-5001 & Arm 2, NTLA-5001 (Genetic) | Acute Leukemia | Phase-1 | Phase-2 | 18< | All | Recruiting |
However, this issue can be alleviated by employing temporary immunosuppression strategies [203]. An alternative method to evade serological recognition includes engineering adenoviral capsid chimeras by replacing the fibers of the Ad5 vector with less immunogenic serotypes, such as Ad45 [204]. Virus-based gene therapy primarily relies on the interaction between the fiber proteins of the viral vector and host cell receptors specific to each virus strain or serotype. When target tissues lack sufficient compatible receptors, the efficiency of gene delivery and viral infection is significantly reduced. A key challenge with using viruses as vectors is the concern that therapeutic genes may be transferred to non-target cells or tissues. To address this issue, transductional retargeting is commonly employed in viral gene therapy. This approach involves modifying the surface proteins of viral vectors to selectively bind to receptors present on the intended target cells, such as cancer cells [205]. Consequently, advancing the safety and effectiveness of viral vector-based gene therapy will require future research to prioritize innovative strategies, including heterologous prime-boost regimens, transductional retargeting, and temporary immunosuppression.
In addition to VVs, EVs are gaining recognition as promising vehicles for CRISPR delivery due to their superior biocompatibility, minimal immunogenicity, and outstanding tissue penetration. These beneficial characteristics position EVs as strong contenders for future clinical applications. Their potential underscores the growing importance of EVs in gene therapy research, especially as CRISPR technology continues to advance. Various types of EVs, including engineered, unmodified, and exosome-liposome hybrids, have been developed for CRISPR delivery. Despite their potential, challenges remain when applying EVs to clinical CRISPR delivery, especially regarding stability, targeting accuracy, and safety. To overcome these obstacles, it is crucial to choose suitable parental cell lines and establish rigorous post-production protocols. Although challenges persist, the potential to optimize EVs for the efficient and safe delivery of CRISPR systems to targeted cells holds great promise, has profound effects on precision medicine.
Although EV-based therapies hold considerable potential for advancing healthcare, they also present critical ethical challenges and risks, including issues associated with informed patient consent, fair accessibility, safety assurance, and data privacy. At present, there are no well-established ethical frameworks for the clinical use of EVs. Effectively addressing these ethical challenges and minimizing associated risks requires a comprehensive strategy that includes robust informed consent practices, strict regulatory compliance, clear communication, strong data privacy protections, and ethical oversight. Beyond these ethical considerations, the inherent heterogeneity of EVs represents a significant barrier to their standardization and clinical application.
Important issues that are frequently disregarded in studies include EV safety and possible toxicity. Tumor-derived EVs possess an innate ability to specifically target tumor cells, a characteristic that has made them promising candidates for targeted cancer therapies in preclinical studies. However, other studies have shown that tumor-derived EVs can suppress CD8+ T cell proliferation by reducing IL-2 levels or induce CD8+ T cell apoptosis through mechanisms like the FasL-TRAIL and Galectin 9 pathways [206, 207]. Furthermore, tumor-derived EVs can trigger signaling pathways that support tumor growth, metastasis, and angiogenesis, while also aiding tumors in evading immune recognition by vaccines, therefore enhancing resistance to treatment [208]. Hence, it is crucial to address the balance between the anti-tumor and pro-tumor effects of EVs to ensure effective therapeutic outcomes [209]. M1-type macrophage-derived EVs, recognized for their pro-inflammatory properties, have been used in anti-tumor therapies. However, they also carry safety risks, such as the potential to stimulate macrophages in the liver and other organs, promoting a pro-inflammatory phenotype that could result in organ damage [210, 211]. Immune concerns could arise from using heterogeneous EVs to treat diseases. In general, further research is necessary to evaluate the toxicity and potential side effects of EVs before their clinical application. Furthermore, increasing the modified EVs' time in circulation could trigger immune responses and introduce safety concerns. A comprehensive evaluation of their functionality and immunogenicity is essential for their safe clinical application.
Ideally, EVs would efficiently encapsulate CRISPR/Cas9 ribonucleoprotein (RNPs) and target specific tissues or cells, offering a gene editing approach that maximizes both efficacy and safety. Multiple studies have demonstrated the ability of EVs to transport and release the CRISPR/Cas9 system for gene modifications. However, challenges persist, particularly in ensuring the safety, capacity, consistency, efficiency, and precise targeting of EVs for therapeutic applications. A primary concern arises from the biogenesis of EVs, as they contain biomacromolecules that could potentially affect the physiological functions of recipient cells, leading to unintended biological consequences. Although tumor-derived EVs can target tumors effectively, they also carry molecules associated with tumor growth and metastasis, necessitating careful evaluation. This highlights the importance of further research and development to ensure the safety and authenticity of EVs used in therapeutic applications.
The artificial intelligence (AI) integration into CRISPR diagnostics offers substantial benefits in overcoming several challenges. AI improves the speed, scalability, and accuracy of CRISPR applications by accelerating data analysis and refining target recognition. For example, ML models can identify genetic sequences linked to diseases, predict the most effective guide RNAs (gRNAs), and reduce off-target effects, improving the precision and efficiency of CRISPR-based technologies [212, 213]. This progress enhances both the specificity and activity of CRISPR-based diagnostic systems, enabling their use in personalized medicine. By customizing tools to identify patient-specific infections or mutations, AI integration ensures more accurate and individualized diagnoses, ultimately improving treatment outcomes and therapeutic strategies [212, 213]. Furthermore, the integration of AI into drug delivery systems (DDSs), especially exosome-based therapies and diagnostics, is revolutionizing precision medicine, especially in oncology. When integrated with AI-driven data analytics, exosomes improve therapeutic accuracy, optimize therapies, and facilitate extensive patient monitoring. This convergence marks a turning point in the treatment of cancer and could have wider implications for other areas of medicine. [214, 215]. AI simulations have transformed the design of hybrid exosomes by allowing for the precise modification and selection of surface ligands, improving cargo loading efficiency, and customizing structural characteristics to meet specific therapeutic needs. These developments have enhanced the precision of drug release, allowing for controlled and prolonged delivery. They have facilitated the design of exosomes that respond to specific environmental factors, such as temperature, pH, or enzyme activity, to release therapeutic agents at targeted locations with high spatial and temporal accuracy [214, 215]. Recent advancements highlight the growing potential of AI in diagnostics. For instance, Yin et al. used ML to examine serum-derived exosomes in colorectal cancer (CRC), employing 4D-DIA proteomics to detect biomarkers like AACT and PF4. Their AI-based random forest model showed significantly better sensitivity and specificity compared to traditional biomarkers such as CA19-9 and CEA. This demonstrates the efficacy of exosome delivery devices enhanced by AI in noninvasive CRC diagnosis [216].
Therefore, AI is an effective tool in exosome biomarker discovery, diagnostic development, and therapeutic optimization. Moving forward, research should prioritize training algorithms on diverse, globally representative datasets to minimize biases and improve their generalizability across different populations [217, 218]. Applying AI to simulate exosome behavior in the TME and predict therapeutic delivery outcomes could greatly accelerate the advancement of engineered exosomes toward clinical use, improving their targeting accuracy and therapeutic impact [214, 219]. While AI holds significant promise for revolutionizing DDSs, there is still much progress to be made before it can be fully integrated into real-world applications. Future efforts should focus on enhancing interpretability, offering clear guidance on selecting ML models, and improving data quality to unlock the full potential of AI in drug delivery.
Despite the challenges that have hindered the clinical application of these delivery systems, recent advancements in drug delivery hold significant promise. Achieving their full potential will require a collaborative effort across academic research, laboratory experimentation, medicine, pharmaceuticals, and continuous innovation. This interdisciplinary approach is essential for translating laboratory findings into effective treatments for patients [220]. Vargason et al. [221] suggest that cell therapies could significantly address the bio-acceptability challenges encountered by DDSs. They propose that cell therapies may enable the development of single, effective doses while minimizing excessive drug accumulation in the body. Furthermore, cell therapies offer the potential for a sustained supply of complex biologics, the ability to overcome inherent biological barriers, and the induction of responses that closely mimic natural physiological processes. Adepu [222] has proposed the use of inorganic mesoporous nanoparticles, microfluidics, and molecular imprinting polymers as potential solutions to address some of the challenges faced by DDSs. Cell-based DDSs, which integrate cells with nanobiomaterials, should be explored further in the field of biomaterials. Since cells are native to the human body, this innovative approach holds great potential, although it remains largely theoretical. It represents a creative and promising direction for drug delivery, to achieve optimal drug delivery patterns. However, significant clinical trials and research are still necessary to improve the efficiency of these modern DDSs and address the challenges that currently hinder their widespread use.
Conclusion and Outlook
Despite being a relatively recent innovation, the CRISPR system has already demonstrated significant advancement in the fields of cancer detection and therapy. CRISPR/Cas technology allows researchers to modulate gene expression in-vivo, in-vitro, or ex-vivo through inhibition or induction. Despite facing certain obstacles, CRISPR has attracted significant interest in recent times owing to its potential to deliver accurate cancer therapy and immunotherapy. The delivery of the CRISPR/Cas system to specific cells, whether in-vivo or in-vitro, shows certain inherent challenges. Various delivery systems are being used to tackle these issues, such as physical modalities, nanocomplexes, EVs, viral vectors, and so on. EVs and viral vectors are two naturally occurring systems that are utilized for the delivery of genetic material. In recent times, the combination of two authentic systems has resulted in the emergence of a new vector known as vexosomes. In general, the progress made in delivery systems holds great potential for the effective implementation of CRISPR/Cas-mediated cancer treatment.
Despite the wide range of potential uses of CRISPR in the diagnosis and treatment of cancer, there remain certain limitations and issues that require further attention in the future. One factor to consider is the likelihood of CRISPR/Cas-mediated DSBs resulting in undesired extensive genomic deletions. In the most severe cases, this can result in chromothripsis, which has the potential to interfere with cell regulatory systems and tumor suppressors [198, 223]. An additional concern pertains to the non-specific impacts of the CRISPR/Cas mechanism. However, this literature analysis has outlined some ways to circumvent such effects. Off-target effects have been a subject of concern in regard to CRISPR technology, as they have the potential to induce the proliferation of cancer cells. However, it is currently reassuring that no evidence has been found to support this claim. It is worth noting that the aforementioned effects have the potential to be mitigated through the implementation of meticulous procedures and regulations [224, 225]. The CRISPR/Cas system may be limited by pre-existing immunity to the widely utilized Cas9, which originates from bacteria [226, 227]. The manipulation of Cas enzymes through genetic modification, along with the production of diverse variants exhibiting distinct antigenic characteristics, could provide a viable solution in this context [228].
Acknowledgements
This study was supported by grants from Zhejiang medical Health Science and Technology Project (No. 2021KY458).
Author contributions
LZ and ST conceived the idea, Material collection ST, XC, XT, writing—original draft preparation ST and XC, writing—review and editing ST, XC, XT, and LZ. Correspondence XT and LZ. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nam AS, Chaligne R, Landau DA. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nature Reviews Genetics. 2021;22:3-18
2. Cao W, Chen H-D, Yu Y-W, Li N, Chen W-Q. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chinese Medical Journal. 2021;134:783-91
3. Zahavi D, Weiner L. Monoclonal antibodies in cancer therapy. Antibodies. 2020;9:34
4. Belete TM. The current status of gene therapy for the treatment of cancer. Biologics: Targets and Therapy. 2021:67-77
5. Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815-22
6. Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology. 1987;169:5429-33
7. Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Molecular Microbiology. 2000;36:244-6
8. Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551-61
9. Jansen R, Embden JDv, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular microbiology. 2002;43:1565-75
10. Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Computational Biology. 2005;1:e60
11. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709-12
12. Li C, Mei H, Hu Y. Applications and explorations of CRISPR/Cas9 in CAR T-cell therapy. Briefings in Functional Genomics. 2020;19:175-82
13. Balon K, Sheriff A, Jacków J, Łaczmański Ł. Targeting Cancer with CRISPR/Cas9-Based Therapy. International Journal of Molecular Sciences. 2022;23:573
14. Youssef E, Palmer D, Fletcher B, Vaughn R. Exosomes in Precision Oncology and Beyond: From Bench to Bedside in Diagnostics and Therapeutics. Cancers. 2025;17:940
15. Youssef E, Fletcher B, Palmer D. Enhancing precision in cancer treatment: the role of gene therapy and immune modulation in oncology. Frontiers in Medicine. 2025;11:1527600
16. Song X, Liu C, Wang N, Huang H, He S, Gong C. et al. Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Advanced Drug Delivery Reviews. 2021;168:158-80
17. Pyzocha NK, Chen S. Diverse class 2 CRISPR-Cas effector proteins for genome engineering applications. ACS Chemical Biology. 2018;13:347-56
18. Nasko DJ, Ferrell BD, Moore RM, Bhavsar JD, Polson SW, Wommack KE. CRISPR spacers indicate preferential matching of specific virioplankton genes. MBio. 2019;10:e02651-18
19. Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57-63
20. Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096
21. Kong H, Ju E, Yi K, Xu W, Lao YH, Cheng D. et al. Advanced nanotheranostics of CRISPR/Cas for viral hepatitis and hepatocellular carcinoma. Advanced Science. 2021;8:2102051
22. Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016 539
23. Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L. et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nature medicine. 2020;26:732-40
24. Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS. et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nature Medicine. 2019;25:229-33
25. Li T, Yang Y, Qi H, Cui W, Zhang L, Fu X. et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduction and Targeted Therapy. 2023;8:36
26. Adli M. The CRISPR tool kit for genome editing and beyond. Nature Communications. 2018;9:1911
27. Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172:1239-59
28. Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481-5
29. Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nature Reviews Molecular Cell Biology. 2019;20:490-507
30. Li S-Y, Cheng Q-X, Liu J-K, Nie X-Q, Zhao G-P, Wang J. CRISPR-Cas12a has both cis-and trans-cleavage activities on single-stranded DNA. Cell research. 2018;28:491-3
31. Li P, Zhang L, Li Z, Xu C, Du X, Wu S. Cas12a mediates efficient and precise endogenous gene tagging via MITI: microhomology-dependent targeted integrations. Cellular and Molecular Life Sciences. 2020;77:3875-84
32. Guo LY, Bian J, Davis AE, Liu P, Kempton HR, Zhang X. et al. Multiplexed genome regulation in vivo with hyper-efficient Cas12a. Nature cell biology. 2022;24:590-600
33. Srivastava S, Upadhyay DJ, Srivastava A. Next-generation molecular diagnostics development by CRISPR/Cas tool: rapid detection and surveillance of viral disease outbreaks. Frontiers in molecular biosciences. 2020;7:582499
34. Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ. et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280-4
35. Smargon AA, Shi YJ, Yeo GW. RNA-targeting CRISPR systems from metagenomic discovery to transcriptomic engineering. Nature Cell Biology. 2020;22:143-50
36. Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438-42
37. Zhang Y, Li S, Li R, Qiu X, Fan T, Wang B. et al. Advances in application of CRISPR-Cas13a system. Frontiers in Cellular and Infection Microbiology. 2024;14:1291557
38. Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573
39. Dubey S, Chen Z, Jiang YJ, Talis A, Molotkov A, Ali A. et al. Small extracellular vesicles (sEVs)-based gene delivery platform for cell-specific CRISPR/Cas9 genome editing. Theranostics. 2024;14:2777
40. Huang Z, Fang J, Zhou M, Gong Z, Xiang T. CRISPR-Cas13: A new technology for the rapid detection of pathogenic microorganisms. Frontiers in Microbiology. 2022;13:1011399
41. Hillary VE, Ceasar SA. A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Molecular biotechnology. 2023;65:311-25
42. Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP. et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839-42
43. Di Carlo E, Sorrentino C. State of the art CRISPR-based strategies for cancer diagnostics and treatment. Biomarker Research. 2024;12:156
44. Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI. et al. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature. 2017;551:464-71
45. Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420-4
46. Pfeiffer LS, Stafforst T. Precision RNA base editing with engineered and endogenous effectors. Nature Biotechnology. 2023;41:1526-42
47. Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149-57
48. Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic acids research. 2016;44:D862-D8
49. Pal M, Herold MJ. CRISPR base editing applications for identifying cancer-driving mutations. Biochemical Society Transactions. 2021;49:269-80
50. Chow RD, Chen JS, Shen J, Chen S. A web tool for the design of prime-editing guide RNAs. Nature biomedical engineering. 2021;5:190-4
51. Hsu JY, Grünewald J, Szalay R, Shih J, Anzalone AV, Lam KC. et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nature Communications. 2021;12:1034
52. Hwang G-H, Jeong YK, Habib O, Hong S-A, Lim K, Kim J-S. et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Research. 2021;49:W499-W504
53. Li C, Chu W, Gill R, Sang S, Shi Y, Hu X. et al. Computational tools and resources for CRISPR/Cas genome editing. Genom Proteom Bioinform. 2022
54. Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nature biotechnology. 2020;38:824-44
55. Liu Y, Li X, He S, Huang S, Li C, Chen Y. et al. Efficient generation of mouse models with the prime editing system. Cell discovery. 2020;6:27
56. Geurts MH, de Poel E, Pleguezuelos-Manzano C, Oka R, Carrillo L, Andersson-Rolf A. et al. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life science alliance. 2021 4
57. Liu P, Liang S-Q, Zheng C, Mintzer E, Zhao YG, Ponnienselvan K. et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nature communications. 2021;12:2121
58. Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9-and TALEN-mediated knock-in efficiency. Nature communications. 2016;7:10548
59. Lee AB, Tan M-H, Chai CL. Small-molecule enhancers of CRISPR-induced homology-directed repair in gene therapy: A medicinal chemist's perspective. Drug Discovery Today. 2022;27:2510-25
60. Li G, Yang X, Luo X, Wu Z, Yang H. Modulation of cell cycle increases CRISPR-mediated homology-directed DNA repair. Cell & Bioscience. 2023;13:215
61. Ryan DE, Taussig D, Steinfeld I, Phadnis SM, Lunstad BD, Singh M. et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic acids research. 2018;46:792-803
62. Guo C, Ma X, Gao F, Guo Y. Off-target effects in CRISPR/Cas9 gene editing. Frontiers in bioengineering and biotechnology. 2023;11:1143157
63. Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature biotechnology. 2015;33:985-9
64. Chuai G, Ma H, Yan J, Chen M, Hong N, Xue D. et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome biology. 2018;19:1-18
65. Ma J, Köster J, Qin Q, Hu S, Li W, Chen C. et al. CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics. 2016;32:3336-8
66. Barozzi I, Slaven N, Canale E, Lopes R, Amorim Monteiro Barbosa I, Bleu M. et al. A functional Survey of the Regulatory Landscape of estrogen Receptor-Positive Breast Cancer evolution. Cancer discovery. 2024;14:1612-30
67. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84-8
68. Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407-10
69. Tsai SQ, Nguyen N, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature. 2016;529:490-5
70. Park J, Yoon J, Kwon D, Han M-J, Choi S, Park S. et al. Enhanced genome editing efficiency of CRISPR PLUS: Cas9 chimeric fusion proteins. Scientific reports. 2021;11:16199
71. Wang L, Han H. Strategies for improving the genome-editing efficiency of class 2 CRISPR/Cas system. Heliyon. 2024 10
72. Lahr WS, Sipe CJ, Skeate JG, Webber BR, Moriarity BS. CRISPR-Cas9 base editors and their current role in human therapeutics. Cytotherapy. 2023;25:270-6
73. Pacesa M, Pelea O, Jinek M. Past, present, and future of CRISPR genome editing technologies. Cell. 2024;187:1076-100
74. Zeng H, Daniel TC, Lingineni A, Chee K, Talloo K, Gao X. Recent advances in prime editing technologies and their promises for therapeutic applications. Current Opinion in Biotechnology. 2024;86:103071
75. Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology. 2015;33:187-97
76. Kim D, Bae S, Park J, Kim E, Kim S, Yu HR. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nature methods. 2015;12:237-43
77. Lazzarotto CR, Malinin NL, Li Y, Zhang R, Yang Y, Lee G. et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Nature biotechnology. 2020;38:1317-27
78. Lazzarotto CR, Nguyen NT, Tang X, Malagon-Lopez J, Guo JA, Aryee MJ. et al. Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nature protocols. 2018;13:2615-42
79. Özden F, Minary P. Learning to quantify uncertainty in off-target activity for CRISPR guide RNAs. Nucleic Acids Research. 2024;52:e87-e
80. Mehta A, Merkel OM. Immunogenicity of Cas9 protein. Journal of pharmaceutical sciences. 2020;109:62-7
81. Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim Y-h. et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nature communications. 2018;9:3048
82. Li J, Wu P, Cao Z, Huang G, Lu Z, Yan J. et al. Machine learning-based prediction models to guide the selection of Cas9 variants for efficient gene editing. Cell Reports. 2024 43
83. Harrington LB, Paez-Espino D, Staahl BT, Chen JS, Ma E, Kyrpides NC. et al. A thermostable Cas9 with increased lifetime in human plasma. Nature communications. 2017;8:1424
84. Kim S, Kim D, Cho SW, Kim J, Kim J-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome research. 2014;24:1012-9
85. Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. Journal of controlled release. 2017;266:17-26
86. Sakurai T, Watanabe S, Kamiyoshi A, Sato M, Shindo T. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC biotechnology. 2014;14:1-11
87. Huang Z, Nair M. A CRISPR/Cas9 guidance RNA screen platform for HIV provirus disruption and HIV/AIDS gene therapy in astrocytes. Scientific reports. 2017;7:5955
88. Farsani AM, Mokhtari N, Nooraei S, Bahrulolum H, Akbari A, Farsani ZM. et al. Lipid nanoparticles: The game-changer in CRISPR-Cas9 genome editing. Heliyon. 2024 10
89. Ashok B, Peppas NA, Wechsler ME. Lipid-and polymer-based nanoparticle systems for the delivery of CRISPR/Cas9. Journal of drug delivery science and technology. 2021;65:102728
90. Kim D, Luk K, Wolfe SA, Kim J-S. Evaluating and enhancing target specificity of gene-editing nucleases and deaminases. Annual review of biochemistry. 2019;88:191-220
91. Liu P, Erez A, Nagamani SCS, Dhar SU, Kołodziejska KE, Dharmadhikari AV. et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889-903
92. Kloosterman WP, Cuppen E. Chromothripsis in congenital disorders and cancer: similarities and differences. Current Opinion in Cell Biology. 2013;25:341-8
93. Ly P, Brunner SF, Shoshani O, Kim DH, Lan W, Pyntikova T. et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nature genetics. 2019;51:705-15
94. Cortés-Ciriano I, Lee JJ-K, Xi R, Jain D, Jung YL, Yang L. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nature genetics. 2020;52:331-41
95. Wang L, Zheng W, Liu S, Li B, Jiang X. Delivery of CRISPR/Cas9 by novel strategies for gene therapy. ChemBioChem. 2019;20:634-43
96. Xu C-F, Chen G-J, Luo Y-L, Zhang Y, Zhao G, Lu Z-D. et al. Rational designs of in vivo CRISPR-Cas delivery systems. Advanced Drug Delivery Reviews. 2021;168:3-29
97. Mellott AJ, Forrest ML, Detamore MS. Physical non-viral gene delivery methods for tissue engineering. Annals of biomedical engineering. 2013;41:446-68
98. Berdien B, Mock U, Atanackovic D, Fehse B. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene therapy. 2014;21:539-48
99. Jordan ET, Collins M, Terefe J, Ugozzoli L, Rubio T. Optimizing electroporation conditions in primary and other difficult-to-transfect cells. Journal of biomolecular techniques: JBT. 2008;19:328
100. Heller LC, Heller R. In vivo electroporation for gene therapy. Human gene therapy. 2006;17:890-7
101. Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370-9
102. Mashel TV, Tarakanchikova YV, Muslimov AR, Zyuzin MV, Timin AS, Lepik KV. et al. Overcoming the delivery problem for therapeutic genome editing: Current status and perspective of non-viral methods. Biomaterials. 2020;258:120282
103. Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184-8
104. Wan T, Niu D, Wu C, Xu F-J, Church G, Ping Y. Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Materials Today. 2019;26:40-66
105. Wang H-X, Li M, Lee CM, Chakraborty S, Kim H-W, Bao G. et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chemical Reviews. 2017;117:9874-906
106. Glass Z, Lee M, Li Y, Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends in biotechnology. 2018;36:173-85
107. Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nature Reviews Drug Discovery. 2017;16:387-99
108. Böttger R, Pauli G, Chao P-H, Fayez NA, Hohenwarter L, Li S-D. Lipid-based nanoparticle technologies for liver targeting. Advanced Drug Delivery Reviews. 2020;154:79-101
109. Chang J, Chen X, Glass Z, Gao F, Mao L, Wang M. et al. Integrating combinatorial lipid nanoparticle and chemically modified protein for intracellular delivery and genome editing. Accounts of Chemical Research. 2018;52:665-75
110. Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nature nanotechnology. 2020;15:313-20
111. van den Berg AI, Yun C-O, Schiffelers RM, Hennink WE. Polymeric delivery systems for nucleic acid therapeutics: Approaching the clinic. Journal of Controlled Release. 2021;331:121-41
112. Wang H, Jin Y, Chen Y, Luo Y, Lv S, Li M. et al. Multifunctional hybrid sponge for in situ postoperative management to inhibit tumor recurrence. Biomaterials Science. 2021;9:4066-75
113. Zhang Z, Shen W, Ling J, Yan Y, Hu J, Cheng Y. The fluorination effect of fluoroamphiphiles in cytosolic protein delivery. Nature Communications. 2018;9:1377
114. Zhang L, Wang L, Xie Y, Wang P, Deng S, Qin A. et al. Triple-targeting delivery of CRISPR/Cas9 To reduce the risk of cardiovascular diseases. Angewandte Chemie International Edition. 2019;58:12404-8
115. Tao Y, Chan HF, Shi B, Li M, Leong KW. Light: a magical tool for controlled drug delivery. Advanced Functional Materials. 2020;30:2005029
116. Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA base-editing and prime-editing. International Journal of Molecular Sciences. 2020;21:6240
117. Koo T, Popplewell L, Athanasopoulos T, Dickson G. Triple trans-splicing adeno-associated virus vectors capable of transferring the coding sequence for full-length dystrophin protein into dystrophic mice. Human Gene Therapy. 2014;25:98-108
118. Maddalena A, Tornabene P, Tiberi P, Minopoli R, Manfredi A, Mutarelli M. et al. Triple vectors expand AAV transfer capacity in the retina. Molecular Therapy. 2018;26:524-41
119. Uddin F, Rudin CM, Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Frontiers in Oncology. 2020;10:1387
120. Fang H, Bygrave AM, Roth RH, Johnson RC, Huganir RL. An optimized CRISPR/Cas9 approach for precise genome editing in neurons. Elife. 2021;10:e65202
121. Hayashi H, Kubo Y, Izumida M, Matsuyama T. Efficient viral delivery of Cas9 into human safe harbor. Scientific Reports. 2020;10:1-14
122. Le Rhun A, Escalera-Maurer A, Bratovič M, Charpentier E. CRISPR-Cas in Streptococcus pyogenes. RNA Biology. 2019;16:380-9
123. Park SH, Bao G. CRISPR/Cas9 gene editing for curing sickle cell disease. Transfusion and Apheresis Science. 2021;60:103060
124. Yin S, Ma L, Shao T, Zhang M, Guan Y, Wang L. et al. Enhanced genome editing to ameliorate a genetic metabolic liver disease through co-delivery of adeno-associated virus receptor. Science China Life Sciences. 2020:1-13
125. Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E. et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400-3
126. Naso MF, Tomkowicz B, Perry III WL, Strohl WR. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs. 2017;31:317-34
127. Dong W, Kantor B. Lentiviral vectors for delivery of gene-editing systems based on CRISPR/Cas: current state and perspectives. Viruses. 2021;13:1288
128. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. Journal of Cell Biology. 2013;200:373-83
129. Gee P, Lung MS, Okuzaki Y, Sasakawa N, Iguchi T, Makita Y. et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nature Communications. 2020;11:1334
130. Duan L, Xu L, Xu X, Qin Z, Zhou X, Xiao Y. et al. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale. 2021;13:1387-97
131. Qiao L, Hu S, Huang K, Su T, Li Z, Vandergriff A. et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics. 2020;10:3474
132. Yong T, Zhang X, Bie N, Zhang H, Zhang X, Li F. et al. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nature Communications. 2019;10:3838
133. Kim SM, Yang Y, Oh SJ, Hong Y, Seo M, Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. Journal of Controlled Release. 2017;266:8-16
134. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology. 2011;29:341-5
135. Liang Y, Duan L, Lu J, Xia J. Engineering exosomes for targeted drug delivery. Theranostics. 2021;11:3183
136. Liang Y, Xu X, Li X, Xiong J, Li B, Duan L. et al. Chondrocyte-targeted microRNA delivery by engineered exosomes toward a cell-free osteoarthritis therapy. ACS Applied Materials & Interfaces. 2020;12:36938-47
137. Sun H, Zu Y. Aptamers and their applications in nanomedicine. Small. 2015;11:2352-64
138. Li J, Fan C, Pei H, Shi J, Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Advanced Materials. 2013;25:4386-96
139. Zhuang J, Tan J, Wu C, Zhang J, Liu T, Fan C. et al. Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy. Nucleic acids research. 2020;48:8870-82
140. Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Molecular Pharmaceutics. 2015;12:3650-7
141. Emam SE, Lila ASA, Elsadek NE, Ando H, Shimizu T, Okuhira K. et al. Cancer cell-type tropism is one of crucial determinants for the efficient systemic delivery of cancer cell-derived exosomes to tumor tissues. European Journal of Pharmaceutics and Biopharmaceutics. 2019;145:27-34
142. Lin Y, Wu J, Gu W, Huang Y, Tong Z, Huang L. et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Advanced Science. 2018;5:1700611
143. Wang D, Zhang F, Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136-50
144. Boucher P, Cui X, Curiel DT. Adenoviral vectors for in vivo delivery of CRISPR-Cas gene editors. Journal of Controlled Release. 2020;327:788-800
145. Asmamaw M, Zawdie B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics: Targets and Therapy. 2021:353-61
146. Verdera HC, Kuranda K, Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Molecular Therapy. 2020;28:723-46
147. Wilson RC, Gilbert LA. The promise and challenge of in vivo delivery for genome therapeutics. ACS Chemical Biology. 2018;13:376-82
148. Flotte T. Gene therapy progress and prospects: recombinant adeno-associated virus (rAAV) vectors. Gene Therapy. 2004;11:805-10
149. Vogel R, Seyffert M, de Andrade Pereira B, Fraefel C. Viral and cellular components of AAV2 replication compartments. The Open Virology Journal. 2013;7:98
150. Conlon TJ, Flotte TR. Recombinant adeno-associated virus vectors for gene therapy. Expert Opinion on Biological Therapy. 2004;4:1093-101
151. Zhao H, Li Y, He L, Pu W, Yu W, Li Y. et al. In vivo AAV-CRISPR/Cas9-mediated gene editing ameliorates atherosclerosis in familial hypercholesterolemia. Circulation. 2020;141:67-79
152. Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, Goodspeed K, Gray SJ, Kay CN. et al. Current clinical applications of in vivo gene therapy with AAVs. Molecular Therapy. 2021;29:464-88
153. Tatsis N, Ertl HC. Adenoviruses as vaccine vectors. Molecular Therapy. 2004;10:616-29
154. Barnes C, Scheideler O, Schaffer D. Engineering the AAV capsid to evade immune responses. Current Opinion in Biotechnology. 2019;60:99-103
155. Armentano D, Sookdeo CC, Hehir KM, Gregory RJ, George JAS, Prince GA. et al. Characterization of an adenovirus gene transfer vector containing an E4 deletion. Human Gene Therapy. 1995;6:1343-53
156. Palmer D, Ng P. Improved system for helper-dependent adenoviral vector production. Molecular Therapy. 2003;8:846-52
157. Stephens CJ, Kashentseva E, Everett W, Kaliberova L, Curiel DT. Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Therapy. 2018;25:139-56
158. Tsukamoto T, Sakai E, Iizuka S, Taracena-Gándara M, Sakurai F, Mizuguchi H. Generation of the adenovirus vector-mediated CRISPR/Cpf1 system and the application for primary human hepatocytes prepared from humanized mice with chimeric liver. Biological and Pharmaceutical Bulletin. 2018;41:1089-95
159. Srivastava A, Mallela KM, Deorkar N, Brophy G. Manufacturing challenges and rational formulation development for AAV viral vectors. Journal of Pharmaceutical Sciences. 2021;110:2609-24
160. Jacob-Dolan C, Barouch DH. COVID-19 vaccines: adenoviral vectors. Annual Review of Medicine. 2022;73:41-54
161. Ghaleh HEG, Bolandian M, Dorostkar R, Jafari A, Pour MF. Concise review on optimized methods in production and transduction of lentiviral vectors in order to facilitate immunotherapy and gene therapy. Biomedicine & Pharmacotherapy. 2020;128:110276
162. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84-7
163. Ryu S-M, Koo T, Kim K, Lim K, Baek G, Kim S-T. et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nature Biotechnology. 2018;36:536-9
164. Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME. et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nature Biotechnology. 2014;32:941-6
165. Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192-7
166. Choi J, Dang Y, Abraham S, Ma H, Zhang J, Guo H. et al. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Therapy. 2016;23:627-33
167. Joglekar AV, Hollis RP, Kuftinec G, Senadheera S, Chan R, Kohn DB. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Molecular Therapy. 2013;21:1705-17
168. Ortinski PI, O'Donovan B, Dong X, Kantor B. Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-mediated gene editing. Molecular Therapy-Methods & Clinical Development. 2017;5:153-64
169. Liu Q, Fan D, Adah D, Wu Z, Liu R, Yan QT. et al. CRISPR/Cas9-mediated hypoxia inducible factor-1α knockout enhances the antitumor effect of transarterial embolization in hepatocellular carcinoma. Oncology Reports. 2018;40:2547-57
170. Ghalesardi OK, Khosravi A, Azizi E, Ahmadi SE, Hajifathali A, Bonakchi H. et al. The prognostic importance of BCR-ABL transcripts in Chronic Myeloid Leukemia: A systematic review and meta-analysis. Leukemia Research. 2021;101:106512
171. Volpe G, Panuzzo C, Ulisciani S, Cilloni D. Imatinib resistance in CML. Cancer Letters. 2009;274:1-9
172. Martinez-Lage M, Torres-Ruiz R, Puig-Serra P, Moreno-Gaona P, Martin M, Moya F. et al. In vivo CRISPR/Cas9 targeting of fusion oncogenes for selective elimination of cancer cells. Nature Communications. 2020;11:5060
173. Firquet S, Beaujard S, Lobert P-E, Sané F, Caloone D, Izard D. et al. Survival of enveloped and non-enveloped viruses on inanimate surfaces. Microbes and Environments. 2015;30:140-4
174. Saari H, Turunen T, Lõhmus A, Turunen M, Jalasvuori M, Butcher SJ. et al. Extracellular vesicles provide a capsid-free vector for oncolytic adenoviral DNA delivery. Journal of Extracellular Vesicles. 2020;9:1747206
175. Wood MJ, O'Loughlin AJ, Lakhal S. Exosomes and the blood-brain barrier: implications for neurological diseases. Therapeutic Delivery. 2011;2:1095-9
176. Hudry E, Andres-Mateos E, Lerner EP, Volak A, Cohen O, Hyman BT. et al. Efficient gene transfer to the central nervous system by single-stranded Anc80L65. Molecular Therapy-Methods & Clinical Development. 2018;10:197-209
177. Orefice NS, Souchet B, Braudeau J, Alves S, Piguet F, Collaud F. et al. Real-time monitoring of exosome enveloped-AAV spreading by endomicroscopy approach: a new tool for gene delivery in the brain. Molecular Therapy-Methods & Clinical Development. 2019;14:237-51
178. Meliani A, Boisgerault F, Fitzpatrick Z, Marmier S, Leborgne C, Collaud F. et al. Enhanced liver gene transfer and evasion of preexisting humoral immunity with exosome-enveloped AAV vectors. Blood Advances. 2017;1:2019-31
179. Zhang D, Lee H, Jin Y. Delivery of functional small RNAs via extracellular vesicles in vitro and in vivo. RNA Interference and CRISPR Technologies: Technical Advances and New Therapeutic Opportunities. 2020:107-17
180. Zhao L, Gu C, Gan Y, Shao L, Chen H, Zhu H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. Journal of Controlled Release. 2020;318:1-15
181. Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. New England Journal of Medicine. 2021;385:493-502
182. Frangoul H, Altshuler D, Cappellini MD, Chen Y-S, Domm J, Eustace BK. et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. New England Journal of Medicine. 2021;384:252-60
183. Konishi CT, Long C. Progress and challenges in CRISPR-mediated therapeutic genome editing for monogenic diseases. Journal of Biomedical Research. 2021;35:148
184. You L, Tong R, Li M, Liu Y, Xue J, Lu Y. Advancements and obstacles of CRISPR-Cas9 technology in translational research. Molecular Therapy-Methods & Clinical Development. 2019;13:359-70
185. Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E. et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365
186. Wang Z, Li N, Feng K, Chen M, Zhang Y, Liu Y. et al. Phase I study of CAR-T cells with PD-1 and TCR disruption in mesothelin-positive solid tumors. Cellular & molecular immunology. 2021;18:2188-98
187. Liao Y, Chen L, Feng Y, Shen J, Gao Y, Cote G. et al. Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget. 2017;8:30276
188. Yang Y. PD-1 Knockout EBV-CTLs for Advanced Stage Epstein-Barr Virus (EBV) Associated Malignancies. 2020.
189. Feasibility A. Safety Study of Universal Dual Specificity CD19 and CD20 or CD22 CAR-T Cell Immunotherapy for Relapsed or Refractory Leukemia and Lymphoma 2018
190. Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Seminars in Cancer Biology: Elsevier. 2019 p. 106-19
191. Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA. et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. New England Journal of Medicine. 2017;377:2531-44
192. Lin H, Cheng J, Mu W, Zhou J, Zhu L. Advances in universal CAR-T cell therapy. Frontiers in Immunology. 2021: 4014.
193. Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM. et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113-7
194. Saayman S, Ali SA, Morris KV, Weinberg MS. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opinion on Biological Therapy. 2015;15:819-30
195. Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV-and HCV-associated hepatocellular carcinoma. Nature Reviews Cancer. 2013;13:123-35
196. Wang X, Huang X, Zhang Y. Involvement of human papillomaviruses in cervical cancer. Frontiers in Microbiology. 2018;9:2896
197. Kanda T, Yajima M, Ikuta K. Epstein-Barr virus strain variation and cancer. Cancer Science. 2019;110:1132-9
198. Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proceedings of the National Academy of Sciences. 2014;111:13157-62
199. Lopez-Gordo E, Podgorski II, Downes N, Alemany R. Circumventing antivector immunity: potential use of nonhuman adenoviral vectors. Human gene therapy. 2014;25:285-300
200. Meliani A, Boisgerault F, Hardet R, Marmier S, Collaud F, Ronzitti G. et al. Antigen-selective modulation of AAV immunogenicity with tolerogenic rapamycin nanoparticles enables successful vector re-administration. Nature communications. 2018;9:4098
201. Fournillier A, Frelin L, Jacquier E, Ahlén G, Brass A, Gerossier E. et al. A heterologous prime/boost vaccination strategy enhances the immunogenicity of therapeutic vaccines for hepatitis C virus. The Journal of infectious diseases. 2013;208:1008-19
202. Kaufmann JK, Nettelbeck DM. Virus chimeras for gene therapy, vaccination, and oncolysis: adenoviruses and beyond. Trends in molecular medicine. 2012;18:365-76
203. Ungerechts G, Springfeld C, Frenzke ME, Lampe J, Parker WB, Sorscher EJ. et al. An immunocompetent murine model for oncolysis with an armed and targeted measles virus. Molecular Therapy. 2007;15:1991-7
204. Parker AL, Waddington SN, Buckley SM, Custers J, Havenga MJ, van Rooijen N. et al. Effect of neutralizing sera on factor x-mediated adenovirus serotype 5 gene transfer. Journal of virology. 2009;83:479-83
205. Das SK, Menezes ME, Bhatia S, Wang XY, Emdad L, Sarkar D. et al. Gene therapies for cancer: strategies, challenges and successes. Journal of cellular physiology. 2015;230:259-71
206. Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P. et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. 2002; 195: 1303-16.
207. Shi Y, Zhang J, Mao Z, Jiang H, Liu W, Shi H. et al. Extracellular vesicles from gastric cancer cells induce PD-L1 expression on neutrophils to suppress T-cell immunity. 2020; 10: 629.
208. Jella KK, Nasti TH, Li Z, Malla SR, Buchwald ZS, Khan MKJV. Exosomes, their biogenesis and role in inter-cellular communication, tumor microenvironment and cancer immunotherapy. 2018; 6: 69.
209. Liu Y, Gu Y, Cao XJO. The exosomes in tumor immunity. 2015; 4: e1027472.
210. Fan Z, Jiang C, Wang Y, Wang K, Marsh J, Zhang D. et al. Engineered extracellular vesicles as intelligent nanosystems for next-generation nanomedicine. 2022; 7: 682-714.
211. Xie D, Ouyang SJFii. The role and mechanisms of macrophage polarization and hepatocyte pyroptosis in acute liver failure. 2023; 14: 1279264.
212. Lou J, Wang B, Li J, Ni P, Jin Y, Chen S. et al. The CRISPR-Cas system as a tool for diagnosing and treating infectious diseases. Molecular biology reports. 2022;49:11301-11
213. Madani A, Krause B, Greene ER, Subramanian S, Mohr BP, Holton JM. et al. Large language models generate functional protein sequences across diverse families. Nature biotechnology. 2023;41:1099-106
214. Shin H, Choi BH, Shim O, Kim J, Park Y, Cho SK. et al. Single test-based diagnosis of multiple cancer types using Exosome-SERS-AI for early stage cancers. Nature communications. 2023;14:1644
215. Lu D, Shangguan Z, Su Z, Lin C, Huang Z, Xie H. Artificial intelligence-based plasma exosome label-free SERS profiling strategy for early lung cancer detection. Analytical and Bioanalytical Chemistry. 2024;416:5089-96
216. Yin H, Xie J, Xing S, Lu X, Yu Y, Ren Y. et al. Machine learning-based analysis identifies and validates serum exosomal proteomic signatures for the diagnosis of colorectal cancer. Cell Reports Medicine. 2024 5
217. Lin X, Zhu J, Shen J, Zhang Y, Zhu J. Advances in exosome plasmonic sensing: device integration strategies and AI-aided diagnosis. Biosensors and Bioelectronics. 2024;266:116718
218. Li B, Kugeratski FG, Kalluri R. A novel machine learning algorithm selects proteome signature to specifically identify cancer exosomes. Elife. 2024;12:RP90390
219. Ghosh S, Rajendran RL, Mahajan AA, Chowdhury A, Bera A, Guha S. et al. Harnessing exosomes as cancer biomarkers in clinical oncology. Cancer Cell International. 2024;24:278
220. Haider S. Nanoparticles: the future of drug delivery. J Young Investig. 2020 38
221. Vargason AM, Anselmo AC, Mitragotri S. The evolution of commercial drug delivery technologies. Nature biomedical engineering. 2021;5:951-67
222. Adepu S, Ramakrishna S. Controlled drug delivery systems: current status and future directions. Molecules. 2021;26:5905
223. Leibowitz ML, Papathanasiou S, Doerfler PA, Blaine LJ, Sun L, Yao Y. et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nature Genetics. 2021;53:895-905
224. Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nature Medicine. 2018;24:1216-24
225. Ikeda A, Fujii W, Sugiura K, Naito K. High-fidelity endonuclease variant HypaCas9 facilitates accurate allele-specific gene modification in mouse zygotes. Communications Biology. 2019;2:371
226. Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK. et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nature Medicine. 2019;25:249-54
227. Wagner DL, Amini L, Wendering DJ, Burkhardt L-M, Akyüz L, Reinke P. et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nature Medicine. 2019;25:242-8
228. Ferdosi SR, Ewaisha R, Moghadam F, Krishna S, Park JG, Ebrahimkhani MR. et al. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nature Communications. 2019;10:1842
Author contact
![]() Corresponding authors: Dr. Xiangmin Tong, Department of Hematology, the Hangzhou First People's Hospital, No. 261 Huansha Road, Hangzhou 310006, China. Email: tongxiangmincom. Dr. Lifen Zhu, Department of Pathology, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, No. 158 Shangtang Road, Hangzhou 310014, China. Email: zhulifenedu.cn.
Corresponding authors: Dr. Xiangmin Tong, Department of Hematology, the Hangzhou First People's Hospital, No. 261 Huansha Road, Hangzhou 310006, China. Email: tongxiangmincom. Dr. Lifen Zhu, Department of Pathology, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, No. 158 Shangtang Road, Hangzhou 310014, China. Email: zhulifenedu.cn.

 Global reach, higher impact
Global reach, higher impact