Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(12):3142-3153. doi:10.7150/ijms.112922 This issue Cite
Research Paper
Causal Relationship between Gut Microbiota and Endometrial Cancer: A Two-Sample Mendelian Randomization Study
1. Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, No. 36, Sanhao Street, Heping District, Shenyang, 110004, China.
2. Key Laboratory of Maternal-Fetal Medicine of Liaoning Province, Key Laboratory of Obstetrics and Gynecology of Higher Education of Liaoning Province, Shenyang, China.
3. Center of Reproductive Medicine, Shengjing Hospital of China Medical University, No. 39 Huaxiang Road, Shenyang 110004, China.
4. Shenyang Reproductive Health Clinical Medicine Research Center, Shenyang 110004, China.
Received 2025-2-27; Accepted 2025-5-29; Published 2025-6-23
Abstract
Several relevant reports have shown that changes in the composition of the gut microbiota are related to the pathogenesis of endometrial cancer (EC). However, the causal effect of the gut microbiota on EC remains unknown. A two-sample Mendelian randomization (MR) study was used to assess the causal effects of the gut microbiota on EC, EC with endometrioid histologies and EC with non-endometrioid histologies. The genetic statistics of the gut microbiota, including 18,340 participants, were acquired from the MiBioGen database. The summary statistics of EC, EC with endometrioid histologies and EC with non-endometrioid histologies were obtained from the publicly available Genome-wide Association Study (GWAS) database. Suitable single nucleotide polymorphisms (SNPs) were selected as instrumental variables (IVs) (P < 5×10-8, r2 < 0.001). The causal effects were evaluated via the MR-Egger regression method, the inverse-variance weighted (IVW) method, the weighted median test, the weighted mode test, and the simple mode test. The IVW analysis suggested that Ruminococcusgnavusgroup (OR=0.82, 95%CI=0.78-0.85, P=1.29×10-17), Euryarchaeota (OR=0.90, 95%CI=0.87-0.94, P=3.78×10-6), and CandidatusSoleaferrea (OR=0.92, 95%CI=0.87-0.98, P=0.01) had protective effects on EC and its subtypes. Gammaproteobacteria (OR=1.29, 95%CI=1.19-1.39, P=2.32×10-10) served as a risk factor for EC, and Intestinimonas (OR=1.33, 95%CI=1.10-1.62, P=3.68×10-3) had detrimental effects on EC with non-endometrioid histologies. The causal relationship between the gut microbiota and EC was explored through two-sample MR analysis, which is helpful for further understanding the gut microbiota-mediated development mechanism underlying EC.
Keywords: Mendelian randomization, gut microbiota, endometrial cancer, causality, genetics
Introduction
Endometrial cancer (EC) is one of the most common gynecological cancers worldwide, and is an epithelial malignancy that occurs in the endometrium. Recently, the incidence of EC has been increasing globally [1]. The most common symptom of EC is postmenopausal bleeding. Although the treatment of EC is still challenging, understanding various pathogenic state drivers of this disease and genetic diversity has led to the development of different treatment approaches to improve the precision of this complex malignancy [2]. Histologically, two ECs with morphological and molecular differences and therapeutic implications have been identified. The occurrence of type I EC is directly related to the continuous stimulation of estrogen without progesterone antagonism. In the absence of progesterone antagonism, the endometrium is in a state of hyperplasia for a long period of time and further develops into EC. Type II EC shows non-endometrioid differentiation and follows an estrogen-free pathway, and the mechanism is not fully understood [3]. The main risk factors for EC are obesity, diabetes mellitus, hypertension, and reproductive endocrine disorders, such as polycystic ovarian syndrome (PCOS) [2]. The prevention of risk factors can reduce the occurrence and development of EC to a certain extent.
The gut microbiota is a complex microbial population in the human gastrointestinal tract with important effects on homeostasis and disease in the host. The gut microbiota plays a crucial role in protecting against pathogens and maintaining immune and metabolic homeostasis. An increasing number of studies suggest that the gut microbiota is not only a central regulator of various inflammatory and proliferative diseases but also essential for physiological gastrointestinal function. Changes in the gut bacterial composition are implicated in the pathogenesis of many inflammatory diseases and infections [4]. Current evidence suggests that the composition of the gut microbiota is significantly different between EC patients and controls. Compared with those in the normal group, the abundances of Bacteroidota and Verrucomicrobiota increased in the EC group, whereas that of Proteobacteria decreased in the EC group [5]. The gut microbiota affects the metabolism of estrogen by secreting beta-glucuronidase (GUS), an enzyme that deconjugates estrogen to estrogen binding receptors, which indirectly influences the occurrence and development of EC [6]. However, most previous studies on the gut microbiota and cancer are case control studies, whose results are difficult to determine. Furthermore, the relationships between the gut microbiota and disease are susceptible to the environment, lifestyle, age, dietary patterns and other confounding factors, which cannot be avoided in observational studies. The causal inference of the gut microbiota and EC is limited by these conditions.
Mendelian randomization (MR) is a new epidemiological method based on whole-genome sequencing data, and single nucleotide polymorphisms (SNPs) were chosen as instrumental variables (IVs) to reveal causality [7]. Compared with observational studies such as cohort studies, MR is less affected by reverse causality and confounding factors, thus effectively reducing bias [8]. MR requires the satisfaction of three important hypotheses: (1) In the relevance hypothesis, IVs should be closely related to exposure factors. (2) In the independence hypothesis, the IV should be independent of any confounders and should be independent of whether these confounding factors can be observed. (3) Exclusivity hypothesis, IVs mediate outcomes only via exposure factors, not other pathways. At present, MR analysis is used for the causal exploration of gut microbiota and a variety of diseases, such as autoimmune diseases [9], heart failure [10], and primary liver cancer [11]. However, there are no published MR studies of the gut microbiota and EC.
This study conducted a two-sample MR analysis between the gut microbiota and EC, aiming to explore this association, to provide new theoretical support for further understanding the occurrence and development of EC.
Materials and Methods
Study design
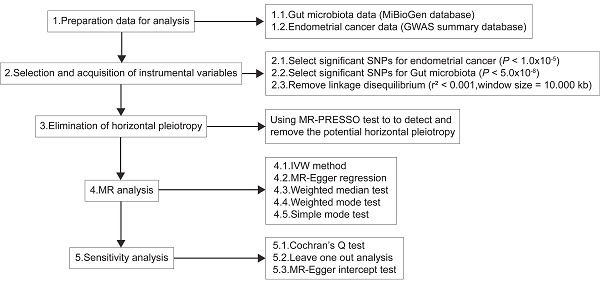
The causal relationship between the gut microbiota and EC was evaluated on the basis of statistics from two MR datasets. Figure 1 shows the flowchart of the study design.
Data source
The Genome-wide Association Study (GWAS) statistics of EC (12,906 cases and 108,979 controls, 9,470,555 SNPs), EC with endometrioid histologies (8,758 cases and 46,126 controls, 9,464,330 SNPs) and EC with non-endometrioid histologies (1,230 cases and 35,447 controls, 8,974,630 SNPs) were downloaded as outcomes from the Open GWAS database. According to pathology reports, the histological subtype of EC was confirmed [12, 13]. The genetic statistics of the gut microbiota were obtained from the MiBioGen database, a large-scale multi-ethnic GWAS for 16S rRNA gene sequencing data collected from 24 cohorts including approximately 18,340 individuals [14]. A total of 211 taxa (9 phyla, 16 classes, 20 orders, 35 families and 131 genera) identified via microbiota quantitative trait locus (mbQTL) mapping analysis were included. After excluding 12 unknown genera, 119 taxa were included for analysis.
Acquisition of instrumental variables (IVs)
According to the GWAS data and IVs screened out from the previous step, the outcome-related IVs were removed at the locus-wide significant threshold (P < 1.0×10-5). The IVs (SNPs) that were sensibly associated with exposure factors were identified via the extract instrument function of the R package TwoSampleMR (version 0.5.6) [15] By setting P < 5×10-8, IVs with linkage disequilibrium (LD) were removed via clump = TRUE. On the basis of European ancestry reference data from the 1000 Genomes Project, the SNPs with LD (r2 < 0.001 and clumping window size = 10,000 kb) were excluded. Potential horizontal pleiotropy was detected using the MR pleiotropy residual sum and outlier (MR-PRESSO) test, and the effect of pleiotropy was eliminated by removing outliers [16]. Eventually, the effect sizes and alleles of the SNPs on the exposure and outcome data were harmonized, and the incompatible alleles were excluded.
Diagrammatic description of MR analysis. IVW, inverse-variance weighted; PRESSO, Pleiotropy RESidual Sum and Outlier; MR, Mendelian randomization; SNP, single nucleotide polymorphism.
Statistical analysis
In this study, the “TwoSampleMR” R package [15] was used for two-sample MR analysis between exposure and outcome, and five common MR methods were used for features that contained more than one IV: simple mode test, weighted mode test, weighted median test, MR-Egger regression, and inverse-variance weighted (IVW) method. The IVW test is the main method for studying the causal relationship between the gut microbiota and EC. The IVW method was first proposed by Burgess et al. [17] for MR studies with multiple IVs. This method was used to calculate the Wald ratio for each IV and to combine the outcome with the use of IVW analysis. Each IV was weighted according to the inverse of the effect variance. Thus, large studies with smaller standard errors can gain more weight than small studies with larger standard errors. This weight choice can minimize imprecision in the pooled effect estimates. The slope of the IVW method can be interpreted as the causal effect of exposure on the outcome. The fixed or multiplicative random effects model can be used to estimate the variance of the effect. Bowden et al. [18] proposed the MR-Egger method , which calculates the Wald ratio for each IV and combines the outcomes via Egger regression. Egger regression can be used to test for pleiotropy bias, and its slope coefficient reflects the size of the causal effect. It can provide a valid test for the null hypothesis of causality and unbalanced directional pleiotropy. The weighted median test calculates the Wald ratio for each instrument, and the median can be selected as the causal estimate variable. This method does not rely on Instrument Strength Independent of Direct Effect (InSIDE) assumptions and provides reliable causal estimates despite having fewer than 50% valid SNPs [19]. The calculation process of the simple model is roughly the same as that of a weighted model method, that is, the causal effect estimates of individual IVs are clustered. Then, the causal effect estimate is calculated for the largest set of IVs [20], but the weighted model method assigns the weight to each IV. To ensure the accuracy of the MR results, sensitivity analysis was performed. The presence of heterogeneity was tested via Cochran's Q test, and there was no significant heterogeneity at a P value greater than 0.05. Whether the pooled estimation was biased by single SNPs was appraised via the leave-one-out analysis. This method calculates the overall effect of the remaining IVs by gradually eliminating each IV and observing whether the outcome changes after eliminating each IV. If the outcome changed significantly after excluding a variable, it indicated that the variable might be an invalid SNP or have a special effect on the outcome. Horizontal pleiotropy was detected via the MR-Egger intercept test. Under the InSIDE hypothesis, the intercept of the line fitted by the MR-Egger regression intercept reflects horizontal pleiotropy, and there is horizontal pleiotropy of MR if the intercept is not equal to zero.
Results
Two-sample MR analysis
After a series of quality selective steps, 270 SNPs related to six microbiota features for EC, including genus CandidatusSoleaferrea, genus Ruminococcusgnavusgroup, phylum Euryarchaeota, class. Gammaproteobacteria, genus Eubacteriumeligensgroup and genus Intestinimonas were identified via at least one MR method. The screening results of specific SNPs are shown in Supplementary Tables 1-3.
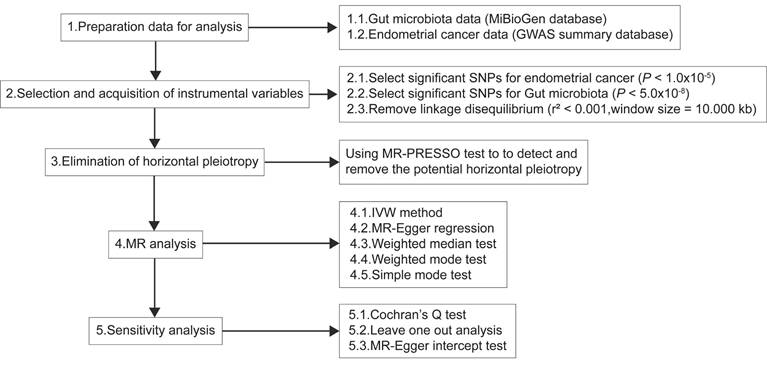
On the basis of the IVW results (Table 1), Ruminococcusgnavusgroup (OR=0.82, 95%CI=0.78-0.85, P=1.29×10-17), Euryarchaeota (OR=0.90, 95%CI=0.87-0.94, P=3.78×10-6), CandidatusSoleaferrea (OR=0.92, 95%CI=0.87-0.98, P=0.01) and Gammaproteobacteria (OR=1.29, 95%CI=1.19-1.39, P=2.32×10-10) had significant causal relationships with EC, while Eubacteriumeligensgroup and Intestinimonas had no causal effect with EC. These findings suggested that Ruminococcusgnavusgroup, Euryarchaeota and CandidatusSoleaferrea were related to the decreased risk of EC, and Gammaproteobacteria was related to the increased risk of EC. In the secondary analysis, Ruminococcusgnavusgroup, Euryarchaeota and CandidatusSoleaferrea had preventive effects in EC with endometrioid histologies and EC with non-endometrioid histologies, while Intestinimonas (OR=1.33, 95%CI=1.10-1.62, P=3.68×10-3) had a positive effect on the risk of EC with non-endometrioid histologies. As shown in Supplementary Figures 1-3, the forest plot showed that the IVW results for Ruminococcusgnavusgroup, Euryarchaeota, CandidatusSoleaferrea were less than 0, indicating that these three microorganisms were preventive factors for EC, EC with endometrioid histologies and EC with non-endometrioid histologies. Gammaproteobacteria was a risk factor for EC and Intestinimonas was positively related to the risk of EC with non-endometrioid histologies. Moreover, four other methods were also used to test the MR, and the results were consistent with the results estimated by the IVW model in terms of magnitude and direction (Table 2). Figure 2 presented a scatter plot of the causal relationship between the gut microbiota and EC, the results of which further confirmed that all methods have a stable direction without the occurrence of outliers.
Heterogeneity and sensitivity analysis
Funnel plot is a subjective visualization method to assess heterogeneity based on the distribution of individual SNPs, which provides the distribution and strength of each genetic variant in the association between the gut microbiota and EC. The funnel diagram (Figure 3) showed that the SNPs were randomly distributed on both sides of the IVW line, indicating that Mendel's second law was followed. Meanwhile, the results of the heterogeneity test showed that all P values were greater than 0.05 (Table 3), indicating that there was no significant heterogeneity in this study. The magnitude of assessed level pleiotropy was tested by MR Egger intercept analysis, and the results of the pleiotropy test are shown in Table 4. The MR Egger intercepts of the MR pleiotropy tests of EC and CandidatusSoleaferrea (P=0.7598), Ruminococcusgnavusgroup (P=0.7342), Euryarchaeota (P =0.7304) and Gammaproteobacteria (P=0.9204) were -0.0072, -0.0051, -0.0047 and 0.0015, respectively. In addition, there was no evidence of pleiotropy detected by the MR Egger analysis between the two subtypes of EC (endometrioid EC and non-endometrioid EC) and the gut microbiota (all P > 0.05). Sensitivity analyses were then performed with the use of the Leave-one-out method. Single SNP was eliminated one by one, and the remaining SNPS were re-analyzed by IVW method. Leave-one-out sensitivity tests demonstrated that the overall results were consistent and there were no SNPs with extremely high sensitivity. Oversensitivity to individual SNP loci according to the of MR results was not exhibited (Supplementary Figures 4-6). In summary, the MR results were reliable.
Results of MR analysis using IVW model.
| Bacterial taxa (exposure) | No. SNP | EC | ECEH | ECNEH | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | ||
| CandidatusSoleaferrea | 41 | 0.92 (0.87-0.98) | 0.01 | 0.90 (0.84-0.97) | 3.63E-03 | 0.77 (0.65-0.90) | 1.77E-03 |
| Ruminococcusgnavusgroup | 63 | 0.82 (0.78-0.85) | 1.29E-17 | 0.81 (0.76-0.85) | 1.93E-14 | 0.50 (0.44-0.57) | 7.46E-24 |
| Euryarchaeota | 44 | 0.90 (0.87-0.94) | 3.78E-06 | 0.91 (0.87-0.96) | 5.57E-04 | 0.76 (0.67-0.86) | 1.30E-05 |
| Gammaproteobacteria | 58 | 1.29 (1.19-1.39) | 2.32E-10 | 1.07 (0.97-1.17) | 0.16 | 1.93 (1.54-2.41) | 0.76 |
| Eubacteriumeligensgroup | 12 | 1.10 (0.93-1.29) | 0.26 | 1.22 (1.00-1.48) | 0.05 | 1.17 (0.62-2.19) | 0.63 |
| Intestinimonas | 52 | 1.05 (0.98-1.12) | 0.20 | 0.99 (0.91-1.07) | 0.76 | 1.33 (1.10-1.62) | 3.68E-03 |
No. SNP, the number of SNPs as instrumental variables; IVW, inverse-variance weighted; MR, Mendelian randomization; EC, endometrial cancer; ECEH, endometrial cancer with endometrioid histologies; ECNEH, endometrial cancer with non-endometrioid histologies; OR, odds ratio; CI, confidence interval.
Mendelian randomization analysis results using the MR-Egger regression, the weighted median test, the weighted mode test and the simple mode test.
| Traits (outcome) | Bacterial taxa (exposure) | MR methods | No. SNP | OR (95% CI) | P value |
|---|---|---|---|---|---|
| EC | CandidatusSoleaferrea | MR Egger | 41 | 0.98 (0.64-1.52) | 0.94 |
| Weighted median | 0.91 (0.84-0.98) | 0.01 | |||
| Simple mode | 0.92 (0.78-1.07) | 0.29 | |||
| Weighted mode | 0.92 (0.78-1.07) | 0.27 | |||
| Ruminococcusgnavusgroup | MR Egger | 63 | 0.86 (0.63-1.17) | 0.35 | |
| Weighted median | 0.77 (0.72-0.82) | 1.06E-15 | |||
| Simple mode | 0.75 (0.65-0.86) | 1.74E-04 | |||
| Weighted mode | 0.75 (0.64-0.88) | 6.16E-04 | |||
| Euryarchaeota | MR Egger | 44 | 0.93 (0.79-1.10) | 0.41 | |
| Weighted median | 0.86 (0.81-0.91) | 7.27E-07 | |||
| Simple mode | 0.81 (0.69-0.94) | 0.01 | |||
| Weighted mode | 0.81 (0.69-0.95) | 0.01 | |||
| Gammaproteobacteria | MR Egger | 58 | 1.25 (0.77-2.04) | 0.36 | |
| Weighted median | 1.31 (1.18-1.46) | 6.58E-07 | |||
| Simple mode | 1.36 (1.05-1.76) | 0.02 | |||
| Weighted mode | 1.35 (1.07-1.71) | 0.02 | |||
| Eubacteriumeligensgroup | MR Egger | 12 | 1.60 (0.88-2.91) | 0.15 | |
| Weighted median | 1.10 (0.89-1.37) | 0.37 | |||
| Simple mode | 1.28 (0.87-1.89) | 0.24 | |||
| Weighted mode | 1.29 (0.89-1.88) | 0.21 | |||
| Intestinimonas | MR Egger | 52 | 1.21 (0.95-1.54) | 0.13 | |
| Weighted median | 1.07 (0.98-1.18) | 0.14 | |||
| Simple mode | 1.08 (0.89-1.30) | 0.45 | |||
| Weighted mode | 1.08 (0.90-1.29) | 0.44 | |||
| ECEH | CandidatusSoleaferrea | MR Egger | 41 | 0.95 (0.57-1.57) | 0.84 |
| Weighted median | 0.85 (0.78-0.93) | 4.95E-04 | |||
| Simple mode | 0.85 (0.69-1.04) | 0.12 | |||
| Weighted mode | 0.85 (0.70-1.02) | 0.09 | |||
| Ruminococcusgnavusgroup | MR Egger | 63 | 0.84 (0.58-1.21) | 0.35 | |
| Weighted median | 0.80 (0.74-0.87) | 1.17E-07 | |||
| Simple mode | 0.79 (0.64-0.97) | 0.03 | |||
| Weighted mode | 0.80 (0.66-0.96) | 0.02 | |||
| Euryarchaeota | MR Egger | 44 | 0.82 (0.67-1.01) | 0.07 | |
| Weighted median | 0.86 (0.80-0.92) | 1.35E-05 | |||
| Simple mode | 0.82 (0.69-0.97) | 0.02 | |||
| Weighted mode | 0.82 (0.69-0.97) | 0.02 | |||
| Gammaproteobacteria | MR Egger | 58 | 1.27 (0.72-2.26) | 0.41 | |
| Weighted median | 1.12 (0.99-1.26) | 0.08 | |||
| Simple mode | 1.18 (0.87-1.59) | 0.28 | |||
| Weighted mode | 1.17 (0.87-1.55) | 0.30 | |||
| Eubacteriumeligensgroup | MR Egger | 12 | 1.72 (0.85-3.47) | 0.16 | |
| Weighted median | 1.36 (1.06-1.76) | 0.02 | |||
| Simple mode | 1.48 (1.00-2.19) | 0.08 | |||
| Weighted mode | 1.48 (1.01-2.16) | 0.07 | |||
| Intestinimonas | MR Egger | 52 | 1.28 (0.96-1.71) | 0.10 | |
| Weighted median | 0.93 (0.84-1.04) | 0.22 | |||
| Simple mode | 0.93 (0.75-1.16) | 0.53 | |||
| Weighted mode | 0.93 (0.75-1.15) | 0.53 | |||
| ECNEH | CandidatusSoleaferrea | MR Egger | 41 | 0.41 (0.12-1.40) | 0.16 |
| Weighted median | 0.73 (0.59-0.90) | 3.77E-03 | |||
| Simple mode | 0.73 (0.47-1.13) | 0.16 | |||
| Weighted mode | 0.73 (0.48-1.10) | 0.14 | |||
| Ruminococcusgnavusgroup | MR Egger | 63 | 0.57 (0.23-1.38) | 0.22 | |
| Weighted median | 0.43 (0.36-0.51) | 4.44E-20 | |||
| Simple mode | 0.41 (0.27-0.62) | 1.08E-04 | |||
| Weighted mode | 0.41 (0.27-0.62) | 9.24E-05 | |||
| Euryarchaeota | MR Egger | 44 | 0.58 (0.35-0.95) | 0.04 | |
| Weighted median | 0.76 (0.64-0.90) | 1.77E-03 | |||
| Simple mode | 0.59 (0.40-0.86) | 0.01 | |||
| Weighted mode | 0.59 (0.40-0.86) | 0.01 | |||
| Gammaproteobacteria | MR Egger | 58 | 0.14 (0.03-0.58) | 0.09 | |
| Weighted median | 2.57 (1.89-3.49) | 0.19 | |||
| Simple mode | 2.83 (1.42-5.66) | 0.46 | |||
| Weighted mode | 2.83 (1.42-5.67) | 0.47 | |||
| Eubacteriumeligensgroup | MR Egger | 12 | 13.34 (2.11-84.17) | 0.02 | |
| Weighted median | 1.49 (0.73-3.06) | 0.28 | |||
| Simple mode | 2.88 (0.55-14.96) | 0.23 | |||
| Weighted mode | 2.81 (0.58-13.60) | 0.23 | |||
| Intestinimonas | MR Egger | 52 | 1.01 (0.51-2.01) | 0.98 | |
| Weighted median | 1.94 (1.47-2.57) | 3.60E-06 | |||
| Simple mode | 1.96 (1.03-3.74) | 0.05 | |||
| Weighted mode | 1.96 (0.98-3.93) | 0.06 |
No. SNP, the number of SNPs being used as instrumental variables; MR, Mendelian randomization; EC, endometrial cancer; ECEH, endometrial cancer with endometrioid histologies; ECNEH, endometrial cancer with non-endometrioid histologies; OR, odds ratio; CI, confidence interval.
The heterogeneity results from Cochran's Q test
| Traits (outcome) | Bacterial taxa (exposure) | Q | P value |
|---|---|---|---|
| EC | CandidatusSoleaferrea | 8.38 | 1.00 |
| Ruminococcusgnavusgroup | 44.78 | 0.95 | |
| Euryarchaeota | 36.37 | 0.75 | |
| Gammaproteobacteria | 29.90 | 1.00 | |
| ECEH | CandidatusSoleaferrea | 14.61 | 1.00 |
| Ruminococcusgnavusgroup | 61.09 | 0.51 | |
| Euryarchaeota | 28.54 | 0.96 | |
| ECNEH | CandidatusSoleaferrea | 9.78 | 1.00 |
| Ruminococcusgnavusgroup | 23.04 | 1.00 | |
| Euryarchaeota | 28.61 | 0.95 | |
| Intestinimonas | 50.16 | 0.51 |
EC, endometrial cancer; ECEH, endometrial cancer with endometrioid histologies; ECNEH, endometrial cancer with non-endometrioid histologies.
Directional pleiotropy results from MR Egger intercept analysis
| Traits (outcome) | Bacterial taxa (exposure) | MR Egger_intercept | SE | P value |
|---|---|---|---|---|
| EC | CandidatusSoleaferrea | -0.0072 | 0.0234 | 0.7598 |
| Ruminococcusgnavusgroup | -0.0051 | 0.0150 | 0.7342 | |
| Euryarchaeota | -0.0047 | 0.0135 | 0.7304 | |
| Gammaproteobacteria | 0.0015 | 0.0151 | 0.9204 | |
| ECEH | CandidatusSoleaferrea | -0.0054 | 0.0274 | 0.8438 |
| Ruminococcusgnavusgroup | -0.0038 | 0.0177 | 0.8319 | |
| Euryarchaeota | 0.0167 | 0.0160 | 0.3027 | |
| ECNEH | CandidatusSoleaferrea | 0.0660 | 0.0661 | 0.3237 |
| Ruminococcusgnavusgroup | -0.0117 | 0.0432 | 0.7875 | |
| Euryarchaeota | 0.0431 | 0.0392 | 0.2772 | |
| Intestinimonas | 0.0240 | 0.0290 | 0.4126 |
EC, endometrial cancer; ECEH, endometrial cancer with endometrioid histologies; ECNEH, endometrial cancer with non-endometrioid histologies; MR, Mendelian randomization.
Discussion
Although traditional observational studies are trying to investigate the role of the gut microbiota in diseases, the defects of the method itself are that it is easily affected by many confounding factors. Therefore, the direct and exact causal relationships between the gut microbiota and diseases cannot be elucidated. On the basis of the GWAS data, a two-sample MR analysis was conducted to obtain instrumental variable information on exposure and outcome, and its sensitivity was tested, with the goal of identifying evidence of the causal relationship between gut microbiota and EC. Since IVs are not affected by the reverse causality of traditional epidemiological studies and confounding factors, reliable evidence can be provided to support the correlation between gut microbiota and EC. These results indicated that Gammaproteobacteria were associated with the risk of EC. Among the histological subtypes of EC (EC with endometrioid histologies and EC with non-endometrioid histologies), the direction of risk estimation for Gammaproteobacteria was consistent with that for EC (although the results of IVW were not significant). Euryarchaeota, CandidatusSoleaferrea and Ruminococcusgnavusgroup may reduce the risk of EC.
The causality of gut microbiota and EC. (A) The causal effect of Ruminococcusgnavusgroup on EC. (B) The causal effect of Euryarchaeota on EC. (C) The causal effect of CandidatusSoleaferrea on EC. (D) The causal effect of Gammaproteobacteria on EC. (E) The causal effect of Ruminococcusgnavusgroup on ECEH. (F) The causal effect of Euryarchaeota on ECEH. (G) The causal effect of CandidatusSoleaferrea on ECEH. (H) The causal effect of Ruminococcusgnavusgroup on ECNEH. (I) The causal effect of Euryarchaeota on ECNEH. (J) The causal effect of CandidatusSoleaferrea on ECNEH. (K) The causal effect of Intestinimonas on ECNEH. EC, endometrial cancer; ECEH, endometrial cancer with endometrioid histologies; ECNEH, endometrial cancer with non-endometrioid histologies.
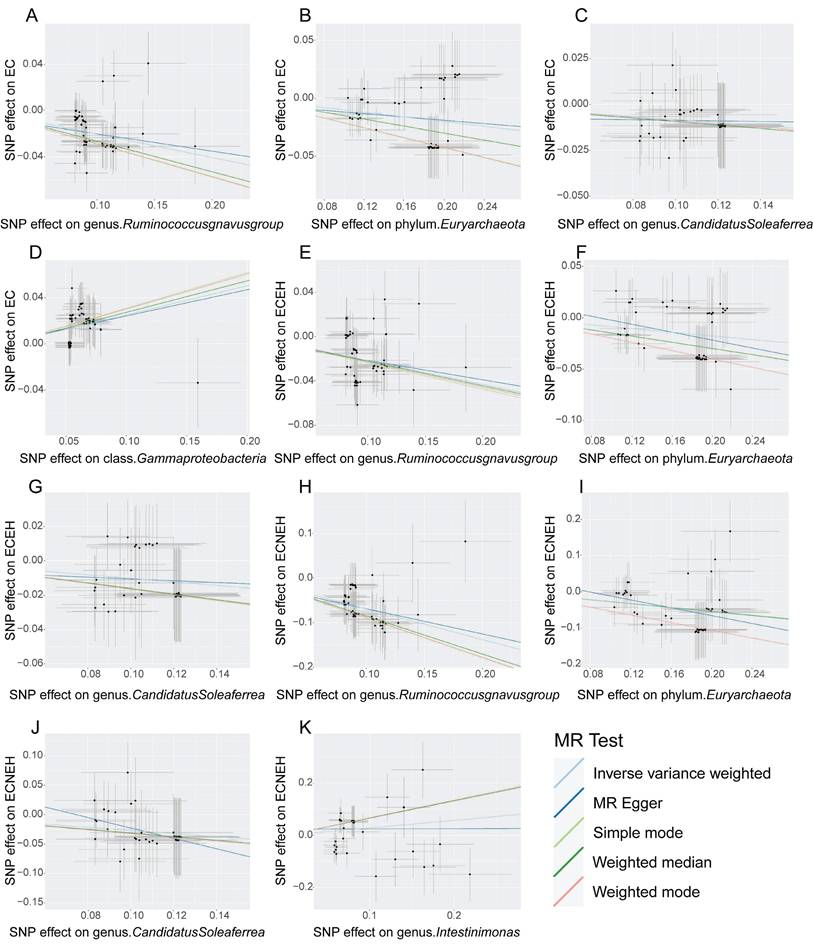
The judgment of randomness of Mendel's second Law. (A) Random judgment of MR analysis for SNPs of Ruminococcusgnavusgroup and EC. (B) Random judgment of MR analysis for SNPs of Euryarchaeota and EC. (C) Random judgment of MR analysis for SNPs of CandidatusSoleaferrea and EC. (D) Random judgment of MR analysis for SNPs of Gammaproteobacteria and EC. (E) Random judgment of MR analysis for SNPs of Ruminococcusgnavusgroup and ECEH. (F) Random judgment of MR analysis for SNPs of Euryarchaeota and ECEH. (G) Random judgment of MR analysis for SNPs of CandidatusSoleaferrea and ECEH. (H) Random judgment of MR analysis for SNPs of Ruminococcusgnavusgroup and ECNEH. (I) Random judgment of MR analysis for SNPs of Euryarchaeota and ECNEH. (J) Random judgment of MR analysis for SNPs of CandidatusSoleaferrea and ECNEH. (K) Random judgment of MR analysis for SNPs of Intestinimonas and ECNEH. MR, Mendelian randomization; SNP, single nucleotide polymorphism; EC, endometrial cancer; ECEH, endometrial cancer with endometrioid histologies; ECNEH, endometrial cancer with non-endometrioid histologies.
Approximately 10^14 microorganisms colonize the human intestinal tract; these microorganisms are diverse and have important functions such as promoting food digestion and absorption, maintaining the stability of the internal environment, protecting the intestinal mucosal barrier, and regulating metabolism and immunity. Disorders of the gut microbiota can cause pathological changes in the body, leading to the occurrence of chronic diseases such as obesity, diabetes, metabolic diseases and cardiovascular diseases. Moreover, the gut microbiota plays an irreplaceable role in the occurrence and development of tumors. Type I EC, representing 80-85% of the total incidence of EC, is mainly associated with monoestrogenic increase without progestogen opposition. Estrogen has a long-term effect on the endometrium, which can increase the proliferation, migration and invasion of endometrial cells, and then induce endometrial atypical hyperplasia and deterioration. Estrogen metabolism mainly occurs in the liver, where it is glucuronidated and sulfonated to form inactive glucuronide-estrogen conjugates, which are excreted into the intestine through bile and bound to GUS produced by intestinal bacteria. Estrogen exposure is caused by reabsorption into the circulation in the form of active free estrogen through the intestinal mucosa. It has been found that Bacteroides, Bifidobacterium, Collinella, Aliella, Edwardiella, Faecalibacterium, Lactobacillus and Rosberia can encode GUS and increase estrogen in the body [21]. Moreover, the gut microbiota is highly correlated with EC risk factors such as obesity, diabetes, and PCOS. Obesity further changes the abundance of intestinal microorganisms and affects the synthesis of estrogen. An increase of estrogen promotes the inflammatory response in the body. The increased inflammatory factors stimulate the synthesis of estrogen in the body by participating in the synthesis of aromatase and 17β-hydroxysteroid dehydrogenase, forming an interactive multi-pathway stimulation loop and increasing the risk of EC [3].
Yue et al. [22] demonstrated that the abundance of Gammaproteobacteria in EC patients was greater than that in the control group through 16S rRNA high-throughput gene sequencing. As an important group of Proteobacteria, Gammaproteobacteria is parasitic in the mucosa adjacent to the intestinal lumen and remains at a low level in the healthy intestine. The increase of its abundance is often associated with intestinal flora disorder, intestinal dysfunction and inflammation [23]. The intestinal barrier is disrupted by disorders of the gut microbiota and intestinal inflammation. Pathogenic factors in the intestinal lumen can enter the blood circulation via the damaged intestinal barrier reach distant organs, and thereby induce EC [24-27]. Lipopolysaccharide (LPS) and trimethylamine N-oxide (TMAO) are the main functional factors of Gammaproteobacteria. In addition to being a major component of the cell wall of Gram-negative bacteria, LPS is also a pathogen-associated molecular pattern (PAMP) and inflammatory inducer. Several studies have demonstrated that LPS can promote tumor progression via inflammatory and metabolic pathways. For instance, Jiang et al. [28] revealed that LPS affect cervical cancer cell proliferation and glucose metabolism by modulating the FRA1/MDM2/p53 pathway. In breast cancer, LPS has been shown to induce inflammation through the prostaglandin E2 (PGE2)-EP2 signaling pathway, thereby facilitating pulmonary metastasis [29]. TMAO, a gut microbiota-derived metabolite associated with increased cardiovascular risk, has also garnered attention for its potential role in tumorigenesis. Zhou et al. [30] demonstrated through in vivo and in vitro experiments that TMAO promotes hepatocellular carcinoma cell proliferation, migration, and epithelial-mesenchymal transition (EMT) by activating the MAPK pathway. Yang et al. [31] found that TMAO stimulates colorectal cancer cell proliferation and elevates vascular endothelial growth factor A (VEGFA) levels, further driving cancer progression. These findings suggest that LPS and TMAO may similarly influence EC progression through analogous mechanisms, though further experimental validation is required. Additionally, some studies have found that compared with normal control populations, EC-related metabolic diseases, including PCOS and type 2 diabetes mellitus (T2DM) patients, have intestinal barrier dysfunction and the content of Gammaproteobacteria in feces was significantly increased [32, 33], and hence it can be speculated that Gammaproteobacteria indirectly cause EC by promoting EC high-risk factors through its functional factors. Some studies have also shown that the peripheral LPS and TMAO levels are much higher in PCOS and T2DM patients than in control individuals, which further supports our speculation [34-37].
This study revealed that CandidatusSoleaferrea exerted preventive effects on EC the occurrence and development and that the reason may be related to glucagon-like peptide 2 (GLP-2). According to Cai et al. [38], the abundance of CandidatusSoleaferrea had a positive correlation with the level of GLP-2, a trophic hormone secreted by intestinal endocrine L cells in response to stimulation with short-chain fatty acids (SCFAs), a type of metabolite of the gut microbiota, that is involved in maintaining the morphology and function of the intestinal epithelium and improving the intestinal mucosal barrier. It can enhance the immune defense function, reduce the amount of LPS released from the intestinal barrier into the circulation, and exert the anti-tumor effect [39]. In addition, some studies have found that GLP-2 regulates hepatic glucose metabolism in mice through the activation of the GLP-2R-PI3K-Akt-FoxO1 signaling pathway, and mice that lack GLP-2 receptors show glucose intolerance and hepatic insulin resistance (IR), which means that GLP-2 contributes greatly to the control of glucose homeostasis and insulin sensitivity [40]. Therefore, it is speculated CandidatusSoleaferrea may exert a protection impact within EC by inhibiting inflammation and IR through GLP-2. By analyzing the symbiotic flora in the human normal colon, adjacent colorectal cancer and colon tumor tissues, Zhang et al. [41] found that normal colon tissues were rich in Ruminococcus gnavus. This inhibited the growth of colon tumors and promoted the immune surveillance function of CD8+T cells by degrading lysophospholipid in mice with normal immune function. Moreover, Ruminococcus gnavus can increase the number of regulatory T cells in mesenteric lymph nodes and the concentration of butyric acid in the cecum of mice [42]. As a beneficial metabolite produced by the gut microbiota, butyrate has been shown to inhibit tumor proliferation and promote the anticancer efficacy of chemotherapy. For example, in vitro experiments demonstrated that butyrate can strongly inhibit the proliferation of pancreatic cancer cells, enhance the sensitivity of pancreatic cancer cells to chemotherapeutic drugs, and promote gemcitabine-mediated tumor growth inhibition mainly by inducing apoptosis. An animal experiment revealed that butyrate can regulate the tumor microenvironment, reduce the levels of tumor extracellular matrix and macrophage markers in a mouse model of pancreatic ductal adenocarcinoma, and improve serum lipid metabolism, thus playing a tumor suppressive role [43]. However, studies have shown that the abundance of Ruminococcus gnavus is increased in the feces of patients with Crohn's disease, inflammatory bowel disease and metabolic diseases, including obesity, T2DM and gestational diabetes mellitus, which suggested that Ruminococcus gnavus is pathogenic [44]. Although functional metabolites of Ruminococcus gnavus have been identified, such as SCFAs, anti-inflammatory capsular polysaccharides and secondary bile acids, the contradictory reasons for the pathogenic or beneficial effects of Ruminococcus gnavus have not been elucidated. Additionally, the mechanism of the interaction between Ruminococcus gnavus and EC needs to be further explored. Contrary to the effects of Gammaproteobacteria mentioned above, Euryarchaeota in the human gut have the genetic potential to take hydrogen to reduce trimethylamine (TMA) and TMAO, and were found to be associated with lower fecal TMA concentrations [45]. Therefore, it is hypothesized that Euryarchaeota could prevent metabolic disorders and malignant tumors by eliminating TMA and TMAO before it enters the circulation.
The research is among the first to explore the causal relationships between the gut microbiota and EC and its subtypes, which has the following important implications. First, SNPs obtained following the large-scale GWAS data are closely related to exposure factors, but not to outcomes, and therefore the impact of exposure factors on outcomes can be represented by the association effect between SNPs and outcomes. Due to the fact that alleles abide by the random assignment principle, this effect is not influenced by the confounding factors within the traditional observational research and can provide robust causal evidence. Second, given the paucity of data on the connection between the gut microbiota and EC, the research expands the current evidence on the causal relationship between the gut microbiota and EC. Third, the research assessed MR through multiple complementary sensitivity analyses to ensure the rigor of the study.
However, it is necessary to consider several limitations of this research. First of all, the GWAS database here was dominated by participants of European ancestry, and it remains uncertain whether the research results can be generalized to individuals of non-European ancestry because genetic differences exist between ethnic groups. In the future, we plan to actively collect multi-center, multi-regional clinical samples and data, incorporating diverse ethnic populations to reduce data heterogeneity and enhance the generalizability of findings. Second, since data on exposure factors were available only at the genus level, the connection between the gut microbiota and EC at the species level could not be analyzed. Metagenomic sequencing technologies will be employed to obtain species-level gut microbial profiles, enabling replication of MR analyses at the microbial species level. Third, although MR can reduce the influence of confounding factors, it cannot entirely eliminate the possibility that alterations in gut microbiota are a consequence rather than a cause of EC. Current basic research methodologies and techniques for the role of gut microbiota in disease pathogenesis are becoming increasingly improving. For instance, studies utilizing animal and cellular experiments have demonstrated the influence of specific gut bacterial taxa on tumorigenic behaviors [46, 47]. Furthermore, longitudinal cohort studies enable the tracking of temporal dynamics, thereby helping to determine whether the gut microbiota acts as a causal driver in the pathogenesis and progression of diseases rather than a consequence [48, 49]. To further substantiate the causal directionality of the associations discussed in this research, we will conduct foundational experiments (e.g., microbiota colonization experiments) and longitudinal cohort studies (e.g., monitoring microbial dynamics prior to EC onset) in future work, thereby providing more robust empirical evidence for our findings and enhancing the validity of this research.
Conclusions
In conclusion, this study demonstrates a potential causal association between the gut microbiota and EC. Future research will investigate the specific mechanism through which the gut microbiota (Gammaproteobacteria, CandidatusSoleaferrea, Ruminococcusgnavusgroup and Euryarchaeota) affects the occurrence and development of EC.
Abbreviations
EC: Endometrial cancer; PCOS: polycystic ovarian syndrome; GUS: glucuronidase; MR: Mendelian randomization; SNPs: single nucleotide polymorphisms; IVs: instrumental variables; GWAS: Genome-wide Association Study; mbQTL: microbiota quantitative trait locus; LD: linkage disequilibrium; MR-PRESSO: MR pleiotropy residual sum and outlier; IVW: inverse-variance weighted; InSIDE: Instrument Strength Independent of Direct Effect; LPS: Lipopolysaccharide; TMAO: trimethylamine N-oxide; PAMP: pathogen-associated molecular pattern; PGE2: prostaglandin E2; EMT: epithelial-mesenchymal transition; VEGFA: vascular endothelial growth factor A; T2DM: type 2 diabetes mellitus; GLP-2: glucagon-like peptide 2; SCFAs: short-chain fatty acids; IR: insulin resistance; TMA: trimethylamine.
Supplementary Material
Supplementary figures.
Supplementary table 1.
Supplementary table 2.
Supplementary table 3.
Acknowledgements
The authors sincerely thank the editors and reviewers for their helpful suggestions and comments. The authors also appreciate the MiBioGen consortium for providing the gut microbiota GWAS summary statistics.
Funding
This work was supported by the Liaoning Science and Technology Program (2022JH2/20200047), Shenyang Science and Technology Plan Project (22-315-6-16), National Natural Science Foundation of China (82303503), and Shenyang Science and Technology Plan Project (22-321-33-19).
Data availability statement
The datasets used in this research include the Open GWAS database (https://gwas.mrcieu.ac.uk/) and the MiBioGen database (https://mibiogen.gcc.rug.nl).
Author contributions
Bei Lin and Xinrui Yao were the major contributors in conceptualization. Sitong Dong, Xiao Li, Ouxuan Liu analyzed and verified the correctness of the data. Xinrui Yao, Yuxuan Wang and Xiangcheng Fan were major contributors in writing the manuscript. Bei Lin and Yuexin Hu reviewed and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Crosbie EJ, Kitson SJ, McAlpine JN, Mukhopadhyay A, Powell ME, Singh N. Endometrial cancer. Lancet. 2022;399:1412-28
2. Makker V, MacKay H, Ray-Coquard I, Levine DA, Westin SN, Aoki D. et al. Endometrial cancer. Nat Rev Dis Primers. 2021;7:88
3. Boutriq S, González-González A, Plaza-Andrades I, Laborda-Illanes A, Sánchez-Alcoholado L, Peralta-Linero J. et al. Gut and Endometrial Microbiome Dysbiosis: A New Emergent Risk Factor for Endometrial Cancer. J Pers Med. 2021;11:659
4. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823-36
5. Zhao SS, Chen L, Yang J, Wu ZH, Wang XY, Zhang Q. et al. Altered Gut Microbial Profile Accompanied by Abnormal Fatty Acid Metabolism Activity Exacerbates Endometrial Cancer Progression. Microbiology spectrum. 2022;10:e0261222
6. Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324-35
7. Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251-60
8. Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1-22
9. Xu Q, Ni JJ, Han BX, Yan SS, Wei XT, Feng GJ. et al. Causal Relationship Between Gut Microbiota and Autoimmune Diseases: A Two-Sample Mendelian Randomization Study. Front Immunol. 2021;12:746998
10. Luo Q, Hu Y, Chen X, Luo Y, Chen J, Wang H. Effects of Gut Microbiota and Metabolites on Heart Failure and Its Risk Factors: A Two-Sample Mendelian Randomization Study. Front Nutr. 2022;9:899746
11. Ma J, Li J, Jin C, Yang J, Zheng C, Chen K. et al. Association of gut microbiome and primary liver cancer: A two-sample Mendelian randomization and case-control study. Liver Int. 2023;43:221-33
12. Cheng TH, Thompson DJ, O'Mara TA, Painter JN, Glubb DM, Flach S. et al. Five endometrial cancer risk loci identified through genome-wide association analysis. Nat Genet. 2016;48:667-74
13. O'Mara TA, Glubb DM, Amant F, Annibali D, Ashton K, Attia J. et al. Identification of nine new susceptibility loci for endometrial cancer. Nat Commun. 2018;9:3166
14. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53:156-65
15. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081
16. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-8
17. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-65
18. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-25
19. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-14
20. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985-98
21. Edwinson AL, Yang L, Peters S, Hanning N, Jeraldo P, Jagtap P. et al. Gut microbial β-glucuronidases regulate host luminal proteases and are depleted in irritable bowel syndrome. Nat Microbiol. 2022;7:680-94
22. Li Y, Liu G, Gong R, Xi Y. Gut Microbiome Dysbiosis in Patients with Endometrial Cancer vs. Healthy Controls Based on 16S rRNA Gene Sequencing. Curr Microbiol. 2023;80:239
23. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708-11
24. Martel J, Chang SH, Ko YF, Hwang TL, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends in endocrinology and metabolism: TEM. 2022;33:247-65
25. Shu LZ, Ding YD, Xue QM, Cai W, Deng H. Direct and indirect effects of pathogenic bacteria on the integrity of intestinal barrier. Therapeutic advances in gastroenterology. 2023;16:17562848231176427
26. Di Tommaso N, Gasbarrini A, Ponziani FR. Intestinal Barrier in Human Health and Disease. International journal of environmental research and public health. 2021;18:12836
27. Shen Y, Fan N, Ma SX, Cheng X, Yang X, Wang G. Gut Microbiota Dysbiosis: Pathogenesis, Diseases, Prevention, and Therapy. MedComm. 2025;6:e70168
28. Jiang X, Yuan J, Dou Y, Zeng D, Xiao S. Lipopolysaccharide Affects the Proliferation and Glucose Metabolism of Cervical Cancer Cells Through the FRA1/MDM2/p53 Pathway. International journal of medical sciences. 2021;18:1030-8
29. Li S, Xu X, Jiang M, Bi Y, Xu J, Han M. Lipopolysaccharide induces inflammation and facilitates lung metastasis in a breast cancer model via the prostaglandin E2-EP2 pathway. Molecular medicine reports. 2015;11:4454-62
30. Zhou C, Basnet R, Zhen C, Ma S, Guo X, Wang Z. et al. Trimethylamine N-oxide promotes the proliferation and migration of hepatocellular carcinoma cell through the MAPK pathway. Discover oncology. 2024;15:346
31. Yang S, Dai H, Lu Y, Li R, Gao C, Pan S. Trimethylamine N-Oxide Promotes Cell Proliferation and Angiogenesis in Colorectal Cancer. Journal of immunology research. 2022;2022:7043856
32. Mammadova G, Ozkul C, Yilmaz Isikhan S, Acikgoz A, Yildiz BO. Characterization of gut microbiota in polycystic ovary syndrome: Findings from a lean population. Eur J Clin Invest. 2021;51:e13417
33. Diviccaro S, Falvo E, Piazza R, Cioffi L, Herian M, Brivio P. et al. Gut microbiota composition is altered in a preclinical model of type 1 diabetes mellitus: Influence on gut steroids, permeability, and cognitive abilities. Neuropharmacology. 2023;226:109405
34. Banaszewska B, Siakowska M, Chudzicka-Strugala I, Chang RJ, Pawelczyk L, Zwozdziak B. et al. Elevation of markers of endotoxemia in women with polycystic ovary syndrome. Hum Reprod. 2020;35:2303-11
35. Jayashree B, Bibin YS, Prabhu D, Shanthirani CS, Gokulakrishnan K, Lakshmi BS. et al. Increased circulatory levels of lipopolysaccharide (LPS) and zonulin signify novel biomarkers of proinflammation in patients with type 2 diabetes. Mol Cell Biochem. 2014;388:203-10
36. Huang J, Liu L, Chen C, Gao Y. PCOS without hyperandrogenism is associated with higher plasma Trimethylamine N-oxide levels. BMC Endocr Disord. 2020;20:3
37. Shan Z, Sun T, Huang H, Chen S, Chen L, Luo C. et al. Association between microbiota-dependent metabolite trimethylamine-N-oxide and type 2 diabetes. Am J Clin Nutr. 2017;106:888-94
38. Cai J, Zhou L, Song X, Yin M, Liang G, Xu H. et al. Alteration of Intestinal Microbiota in 3-Deoxyglucosone-Induced Prediabetic Rats. Biomed Res Int. 2020;2020:8406846
39. Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O. et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091-103
40. Shi X, Zhou F, Li X, Chang B, Li D, Wang Y. et al. Central GLP-2 enhances hepatic insulin sensitivity via activating PI3K signaling in POMC neurons. Cell Metab. 2013;18:86-98
41. Zhang X, Yu D, Wu D, Gao X, Shao F, Zhao M. et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe. 2023;31:418-32.e8
42. Ahn JR, Lee SH, Kim B, Nam MH, Ahn YK, Park YM. et al. Ruminococcus gnavus ameliorates atopic dermatitis by enhancing Treg cell and metabolites in BALB/c mice. Pediatr Allergy Immunol. 2022;33:e13678
43. Panebianco C, Villani A, Pisati F, Orsenigo F, Ulaszewska M, Latiano TP. et al. Butyrate, a postbiotic of intestinal bacteria, affects pancreatic cancer and gemcitabine response in in vitro and in vivo models. Biomed Pharmacother. 2022;151:113163
44. Crost EH, Coletto E, Bell A, Juge N. Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol Rev. 2023;47:fuad014
45. Borrel G, McCann A, Deane J, Neto MC, Lynch DB, Brugère JF. et al. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. Isme j. 2017;11:2059-74
46. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A. et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer discovery. 2018;8:403-16
47. Zhang JW, Zhang D, Yin HS, Zhang H, Hong KQ, Yuan JP. et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression and chemoresistance by enhancing the secretion of chemotherapy-induced senescence-associated secretory phenotype via activation of DNA damage response pathway. Gut microbes. 2023;15:2197836
48. Chun Y, Grishin A, Rose R, Zhao W, Arditi Z, Zhang L. et al. Longitudinal dynamics of the gut microbiome and metabolome in peanut allergy development. The Journal of allergy and clinical immunology. 2023;152:1569-80
49. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY. et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698-706
Author contact
![]() Corresponding author: Bei Lin, email: linbei88com; Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, No. 36, Sanhao Street, Heping District, Shenyang, 110004, China.
Corresponding author: Bei Lin, email: linbei88com; Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, No. 36, Sanhao Street, Heping District, Shenyang, 110004, China.

 Global reach, higher impact
Global reach, higher impact