Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(12):3084-3100. doi:10.7150/ijms.110640 This issue Cite
Review
Research Progress of Epigenetic Modifications in Myopia
1. Shandong University of Traditional Chinese Medicine, Jinan 250002, China.
2. Affiliated Eye Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250002, China.
3. Medical College of Optometry and Ophthalmology, Shandong University of Traditional Chinese Medicine, Jinan 250002, China.
4. Shandong Provincial Key Laboratory of Integrated Traditional Chinese and Western Medicine for Prevention and Therapy of Ocular Diseases; Shandong Academy of Eye Disease Prevention and Therapy, Jinan 250002, China.
Received 2025-1-17; Accepted 2025-6-9; Published 2025-6-23
Abstract
Myopia, also known as nearsightedness, refers to a refractive error of the eye that causes parallel rays of light to focus in front of the retina, affecting distance vision. High myopia significantly increases the risk of pathological myopia, leading to severe complications and an increased likelihood of myopia-related eye diseases. In recent decades, the incidence of myopia has continued to rise, posing significant social and human health issues. The complex interplay between genetic and environmental variables affects the development of myopia. Gene control depends to a large extent on epigenetic changes, which are reversible, inheritable, and sensitive to ecological shifts. Therefore, the pathophysiology and development of myopia are tightly linked to gene regulation mediated by epigenetic changes. To explore epigenetic modifications related to myopia, a PubMed search was conducted using keywords such as epigenetic modification, epigenetics, DNA methylation, RNA methylation, non-coding RNA, long non-coding RNA, short interfering RNA, microRNA, ribosomal RNA, circular RNA, transfer RNA, histone modification, histone methylation, and histone acetylation. This review presents the current understanding of these epigenetic modifications in myopia to provide new insights for advancing myopia research.
Keywords: myopia, epigenetic modification, DNA methylation, RNA methylation, non-coding RNA, histone modification
1. Introduction
Myopia is a spherical equivalent ≤ -0.5 diopter refractive error [1]. Myopia is usually caused by an excessively long eyeball, especially the long vitreous cavity [2]. Patients with high myopia (HM) are at a higher risk of various secondary complications, including glaucoma, cataracts, choroidal neovascularization (CNV), optic neuropathy, uveitis, retinal detachment, and myopia-related macular degeneration [3].
In the twenty-first century, myopia has become a severe public health problem [4]. Myopia has become far more common than it was in earlier decades. By 2050, 938 million people (9.8% of the world's population) will suffer from HM, while 4.758 billion individuals (49.8% of the world's population) will have myopia. This growing demand will require significant healthcare resources to manage nearly 1 billion HM patients worldwide [5]. The incidence of myopia continues to rise, which has a substantial impact on human eye health.
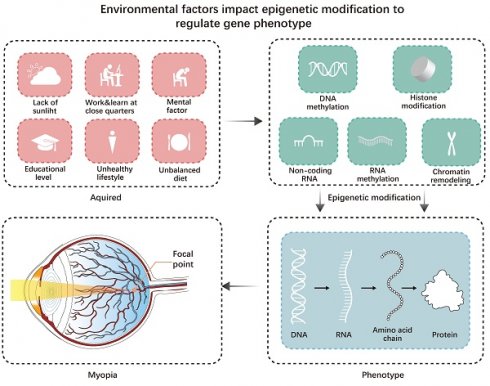
For most myopic patients, modern lifestyle factors may be the main factor contributing to myopia [1]. For instance, increased educational pressure and decreased outdoor time for children are the main lifestyle factors contributing to myopia. However, Inuit communities have much lower educational pressure than developed East and Southeast Asian countries. The lack of outdoor light may have a more significant impact on them [6]. Hereditary factors influence an individual's susceptibility to way-of-life risk factors for myopia (Figure 1).
Environmental factors impact epigenetic modification to regulate gene-phenotype. Acquired factors can all affect epigenetic modifications, leading to changes in the genetic phenotype and ultimately triggering myopia.
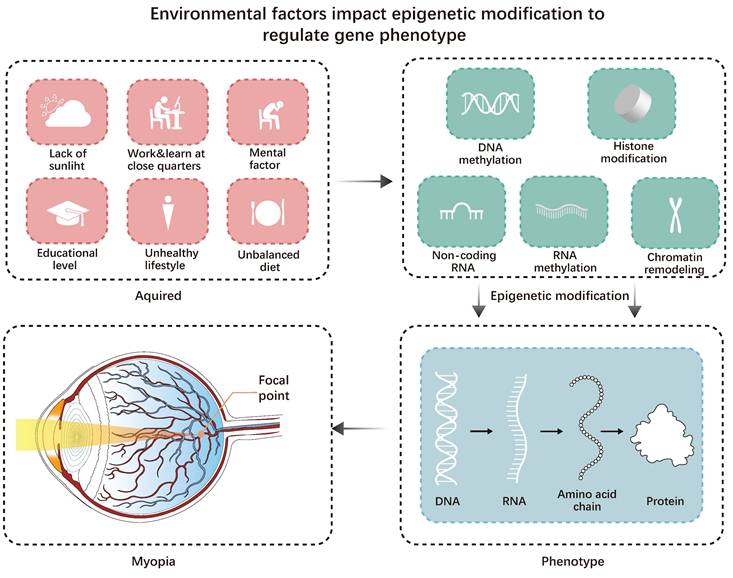
Epigenetics affects many diseases. Some diseases, including autoimmune diseases and cancer, may result from disturbances in the transcription patterns controlled by epigenetic processes [7]. Inheritance and external factors may influence disease progression through epigenetic changes.
Diseases related to the eyes are significantly influenced by epigenetic changes, like glaucoma, cataracts, corneal diseases, proliferative vitreoretinopathy (PVR), age-related macular degeneration (AMD), eye tumors, and certain systemic diseases like endocrine and immune disorders (uveitis, diabetic retinopathy, Sjögren's syndrome). For example, Wang et al. found that the most significantly associated demethylated region in people with Sjögren's syndrome is located in a gene controlled by type I interferon (IFN1), which is associated with B cell invasion and disease progression disease. Sjögren's syndrome is linked to MX dynamin-like GTPase 1 (MX1), RUNX family transcription factor 2 (RUNX1), and lymphotoxin alpha (LTA) [8]. It has also been found that the methylation level of the IL17RC promoter is significantly reduced in the retina, choroid, and peripheral blood of patients with AMD, leading to elevated IL17RC expression and that IL17RC may be a biomarker for AMD diagnosis [9]. Myopia is one of the most common eye diseases, and its occurrence and development are significantly affected by epigenetic modifications.
2. Molecular Mechanisms of Myopia
2.1 Myopia pathogenesis
The combination of environmental and genetic factors causes myopia [10]. External factors complete the information in the genome and continuously produce molecular-level information [11]. Zhang et al. calculated an environmental and genetic index value of 0.125 through a survey of Chinese children aged 6 to 9, suggesting that genetic variables may have an impact of 12.5% on the development of myopia. 87.5% of myopia progress may be influenced by external factors [12].
2.2 Heredity and myopia
The appearance of myopia is significantly influenced by genetic markers associated with myopia. Myopia has multiple inheritance mechanisms, including autosomal dominant, autosomal recessive, and X-linked. Autosomal dominant inheritance means that the defective gene is expressed dominantly. As long as one allele of the gene is abnormal, it can lead to autosomal dominant inheritance, and 50% of the children have the possibility of developing the disease. A pathogenic gene inherited in an autosomal recessive manner is located on an autosome, and its trait is recessive, meaning that the disease manifests only in homozygous individuals. Offspring have a 25% probability of inheriting the disease, with an equal likelihood for both male and female children. X-linked inheritance refers to the inheritance pattern of genes located on the X chromosome and can be classified into X-linked recessive and X-linked dominant inheritance. A few rare cases of pathological myopia occur exclusively in males, with female carriers transmitting the condition to the next generation. Li et al. discovered a novel SNP (rs1064261). They found that polymorphisms in the mechanistic target of rapamycin kinase (MTOR) and platelet-derived growth factor receptor alpha (PDGFRA) genes were associated with different degrees of myopia severity [13]. Zhao et al. discovered that the eyes of photoreceptor-deficient mice had lower phosphodiesterase 4B (PDE4B) levels and that PDE4B knockout mice had higher myopic dioptre shift. These findings suggest that PDE4B inhibition and down-regulation cause myopia [14].
3. The State of Research on Epigenetic Changes in Myopia
3.1 Methylation of DNA
DNA methylation is the most characteristic epigenetic mechanism [15]. It refers to the covalent addition of a methyl group to the 5th carbon of cytosine in CpG dinucleotides under the action of DNA methyltransferases (DNMTs), which is essential for the growth of mammals [16,17].
Some studies have found that single-point methylation alterations in NGFI-A binding protein 2 (NAB2), AP-1 transcription factor subunit (FOS), and early growth response 1 (EGR1) are associated with changes in in the speed of optically induced eye development rate, but not with large-scale changes [18]. Through DNA methylation, external environmental changes can affect eye growth and are associated with the onset and incidence of myopia.
DNA methylation generally silences genes, while DNA demethylation promotes gene expression. The delicate balance between continuous DNA methylation and demethylation forms the cell's final methylation pattern.
3.1.1 DNA demethylation of myopia-related genes
The gene that promotes myopia can increase its expression by hypomethylating its promoter region. In the guinea pig form deprivation myopia (FDM), Ding et al. discovered that hypomethylation of four CpG sites in the promoter region of the insulin-like growth factor 1 (IGF1) gene promotes an increase in IGF1 transcriptional levels in the sclera, which leads to myopia [19]. A team in Singapore found that in myopic children, the methylation level of the upstream homoeobox A9 (HOXA9) gene promoter region of the retinal pigment epithelium (RPE) is low, resulting in increased HOXA9 expression, which in turn causes increased expression of insulin-like growth factor 1 receptor (IGF1R), matrix metalloproteinase 2 (MMP2), fibroblast growth factor 2 (FGF2) and transforming growth factor beta (TGFB). These factors are responsible for the growth of the RPE, elongation of the eyeball, and the deepening of myopia [20]. Swierkowska's team found that the cytosine-phosphate-guanine sites (CpG islands) in the promoter region of the protocadherin alpha 10 (PCDHA10) gene cluster at the 5q31 locus was most significantly hypomethylated, overlapping with the intron of PCDHA19, which may be connected to the early onset of HM [21].
3.1.2 High levels of DNA methylation in genes linked to myopia
When the methylation level in the promoter region of the myopia suppressor gene is too high, the expression of the gene is inhibited. A study found that the methylation level of long interspersed nucleotide element 1 (LINE-1) in myopic individuals was significantly higher than in the normal group. FDM mice also showed higher methylation of LINE-1 in leukocytes, retina, and sclera. Dopamine (DA) can reduce LINE-1 methylation and DNA methyltransferases (DNMTs), revealing part of the mechanism of DA treatment of myopia [22]. Zhu et al. found that HM eyeballs undergo early vitreous liquefaction due to excessive axial length (AL), which increases the oxygen content around the lens and subjects myopic eyes to more oxidative stress. Increased expression of DNMT1 induces hypermethylation of antioxidant genes (thioredoxin reductase 2 (TXNRD2) and glutathione S-transferase pi 1 (GSTP1)), inhibiting the expression of antioxidant genes, promoting the development of HM [23]. One study found that the high methylation of the CpG islands in the promoter of the collagen type I alpha 1 chain (COL1A1) in the sclera of FDM mice hindered the transcription of COL1A1, thereby inhibiting the synthesis of scleral collagen and accelerating the onset of myopia [24]. Compared with individuals with age-related cataracts (ARC), nuclear cataracts and HM patients have more hypermethylation in the CpG regions in the crystallin alpha A (CRYAA) promoter. Furthermore, there is less CRYAA in the lenses of HM patients. The reduced CRYAA expression may make lens proteins more susceptible to oxidative stress associated with aging, which could explain how nuclear cataracts worsen the pathophysiology of HM [25].
Early onset of myopia is also directly linked to high DNA methylation. Seow's team found that the higher frequency of early-stage myopia in 3-year-olds may be related to DNA site-specific CpG methylation in neonatal umbilical cord tissue [26]. One type of transcription factor involved in developing the central nervous system and the eye is paired box 6 (PAX6). It is the dominant gene for eye development [27]. A study found that grandmothers who smoked during pregnancy could reduce the prevalence of myopia in their grandchildren before seven. Whole genome methylation analysis is consistent with this view, and an association near the PAX6 gene indicates a clear association between methylation and early-onset myopia [28]. A study comparing HM cases with typical cases (4 to 12 years old) revealed that myopia-related genes such as suppressor of cytokine signaling 1 (SOCS1), zinc and ring finger 3 (ZNRF3), presenilin 1 (PSEN1), PAX6, adaptor related protein complex 1 subunit beta 1 (AP1B1), adenylate cyclase 3 (ADCY3), the regulator of G protein signaling 5 (RGS5), serum response factor (SRF) and growth factor receptor binding protein 2 (GRB2), are highly methylated, promoting the development of myopia [29]. A study found 1541 highly methylated CpGs, including PAX6, which interact with one another and are associated with the growth and maintenance of retinal ganglion cells (RGCs), changes in axon length, synaptic connectivity, and dysfunction of corneal and sclera [30]. The methylation level of the PAX6 gene in the myopic group of junior high school students was slightly higher than that in the non-myopic team, and the more severe the myopia, the lower the level of the methylation PAX6 gene. This proves that early myopia is caused by hypermethylation of the PAX6 gene promoter [31]. Multiple genes interact to cause myopia, not a single gene.
3.1.3 DNA methylation interacts with non-coding RNAs to promote myopia progression
The genes encoding miR-1178, miR-LET-7A2, miR-885, miR-548-I3, miR-6854, miR-675, miR-LET-7C, and miR-99A have hypomethylated CpG islands in their promoter regions, while the genes encoding miR-3621, miR-34C, and miR-423 have highly methylated CpG islands in their promoter regions. These differences in DNA methylation levels affect the transcriptional regulation of target genes and alter the pathways, leading to HM [32]. This study shows that epigenetic modifications of DNA methylation and micro RNAs (miRNAs) do not exist in isolation but are interconnected and interact (Figure 2).
DNA methylation is associated with myopia. 1. DNA methylation related to myopia in the lens increases crystallin's susceptibility to oxidative stress. 2. DNA methylation related to myopia in the retina, ultimately causing damage to RPE. 3. DNA methylation in the sclera associated with myopia, leading to reduced collagen synthesis by scleral fibroblasts.
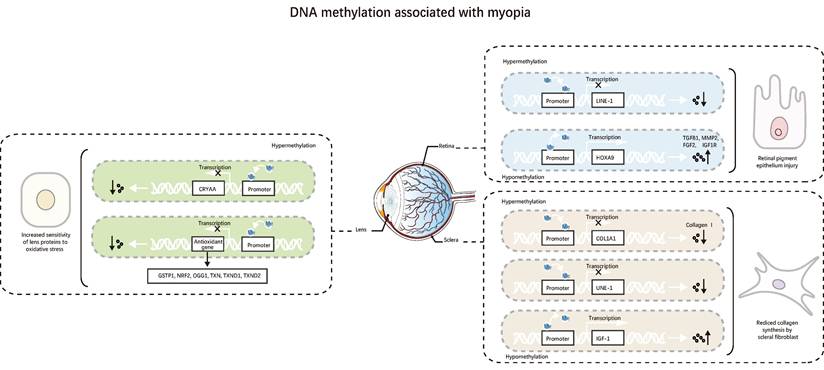
In short, methylation markers show how myopia genes are linked to epigenetic targets, how DNA methylation and non-coding RNA interactions affect eye growth, and how the environment affects gene expression. Methylation targets can prevent myopia from developing, while methylation markers can be used to detect early-onset myopia. However, individual variability and the high cost and complexity of methylation detection make their implementation more challenging. More research is needed to determine the precise dynamic equilibrium mechanism between methylation and demethylation.
3.2 Non-coding RNAs
Although non-coding RNAs (ncRNAs) were once considered “junk,” advances in molecular biology have revealed that they control gene expression in various cellular networks and processes. These ncRNAs include regulatory and housekeeping ncRNAs [33,34].
3.2.1 microRNAs
miRNA is a short (less than 22 nt) endogenous ncRNA that regulates gene expression post-transcriptionally through translation inhibition or messenger RNA (mRNA) degradation [35,36]. By controlling autophagy, gene expression, and scleral remodeling, choroidal fibrosis, among other processes, miRNAs are known to contribute to the development of myopia [37].
miRNAs associated with myopia in aqueous humor
In both HM and normal samples' aqueous humor (AH), Li et al. found 37 miRNAs that were differently expressed. Among these is hsa-miR-142-3p, which targets and inhibits TGFB1, suppresses type I collagen expression in human scleral fibroblasts (HSFs), and positively correlates with ocular AL [38]. The myopic group's total RNA content was 2.78 times higher than the cataract groups. Fifteen miRNAs are specific to myopia, while four were missing. It has been discovered that six well-known myopia-related genes are targeted by five myopia-specific miRNAs (has-miR-582-3p, has-miR-885-3p, has-miR-19b-3p, has-miR-450b-5p, and has-miR-17-5p): cyclic nucleotide-gated channel subunit beta 3 (CNGB3), lumican (LUM), vascular endothelial growth factor A (VEGFA), cholinergic receptor muscarinic 2 (CHRM2), IGF1, and adenosine A2a receptor (ADORA2A). The CHRM2 may be a target of the missing miRNA in myopia (has-miR-378a-5p) [39]. Compared to cataract patients, HM patients have higher expression of LET-7b/c/e, LET-7i, miR-98, miR-103, miR-214, and miR-29b in their AH. These abnormally produced miRNAs may regulate some signaling pathways, including phosphatidylinositol 3 kinase/protein kinase B (PI3K/AKT), hypoxia-inducible factor 1 subunit alpha (HIF1A), mitogen-activated protein kinase (MAPK), and tumor necrosis factor (TNF) [40]. Zhu et al. discovered that HM patients have noticeably increased levels of miR-29a in AH compared to cataract patients. Although miR-29a has no significant effect on cell proliferation, it can increase cell migration and suppress the synthesis of type I collagen in HSFs [41].
Myopic aqueous humor has variations in miRNA expression, and these molecules regulate ocular signaling pathways.
miRNAs associated with myopia in the vitreous humor
Jiang et al. discovered a significant decrease in miR-204-5p in vitreous humor (VH) from HM patients. Since the miR-204-5p-TXNIP axis directly targets a thioredoxin-interacting protein (TXNIP) to prevent proliferation, migration, invasion, and apoptosis and reduces reactive oxygen species (ROS) accumulation, it may be involved in regulating retinal cell activity and oxidative damage [42]. You et al. recovered high-purity exosomes from VH tissues from people with pathological myopia (PM) and a non-myopic group. They discovered that inhibiting miR-145 and miR-143 may increase IGF1R levels, trigger the insulin-resistant pathway, and lead to AL growth. miR-145-5p and miR-143-3p may be markers of myopic maculopathy progression [43]. Ando et al. discovered that the group of macular holes with HM had significantly higher levels of LET-7c and lower levels of miR-200a compared to the group of macular holes alone. LET-7c targets genes, including MAPK and several inflammatory signaling pathways. miR-200a targets genes, among which PI3K/AKT is the most abundant. In the macular holes with high myopia, the levels of the inflammatory cytokines C-C motif chemokine ligand 2 (CCL2), C-X-C motif chemokine ligand 10 (CXCL10), and interferon-gamma (IFNG) in the vitreous are significantly elevated. This suggests that myopia development, tissue reshaping, inflammation, and AL elongation are linked to LET-7c and miR-200a [44].
Changes in miRNA expression in the vitreous of eyes with macular degeneration and high myopia are linked to pathways involved in tissue remodeling and inflammation. The miRNA-based signal pathway axis controls retinal cell behavior and oxidative stress.
miRNAs associated with myopia in the retina
Patients with myopic macular degeneration have elevated levels of hsa-miR-328-3p in the blood, and the RPE layer's optical density decreased [45]. The C allele rs662702 of the PAX6 gene increases the risk of myopia, and the expression of miR-328 in the blood cells of myopic patients is higher than that of the average group [46]. Chen et al. also discovered that the dangerous allele C of the 3'UTR SNP rs644242 did strongly respond to miR-328 and downregulate PAX6 expression. When PAX6 transcription is downregulated, RPE proliferation increases, and HSF proliferation decreases. Type I collagen and integrin subunit beta 1 (ITGB1) are reduced in the sclera, whereas TGFB1 and MMP2 increase. Retinoic acid (RA) increases miRNA-328 expression and inhibits PAX6 expression [47]. According to Liang et al., miR-328-3p decreased both protein expression and mRNA dose-dependent manner. Fibromodulin (FMOD) raises p38-MAPK and MAPK8 phosphorylation levels and stimulates TGFB1 expression. Anti-miR-328-3p also inhibits AL elongation [48]. Patients with myopia exhibit peripheral blood and retinal upregulation of miR-328, indicating that miR-328 may develop into a biomarker for myopia (Figure 3).
Mei et al. detected that miR-466c-5p, miR-466h-5p, miR-466j, miR-468, miR-669e, miR-15a, miR-16-1, and miR-294 were significantly elevated in FDM retinas and whole eye samples. Seven were enriched considerably in transcriptional regulation, axon guidance, and the TGFB1 signaling pathway [49]. In addition, a study found that seven elevated miRNAs (miR-101a, miR-6690, miR-466f, miR-291a, miR-465b, miR-696, and miR-18b) were expressed five times greater in the retina of FDM mice than in the sclera. This implies they might play a role in controlling processes specific to the retina. A small group of retinal cells may exhibit miR-145 downregulation, as its expression in the retina is 25.4 times higher than in the sclera [50].
Liu et al. found that miR-92b-3p transcription was downregulated in lens-induced myopia (LIM) guinea pigs, resulting in elevated levels of p53 and BTG anti-proliferation factor 2 (BTG2). Activated BTG2 can cause retinal tissue apoptosis, induce DNA damage in guinea pig retinal tissue, promote BCL2 apoptosis regulator (BCL2) expression and apoptosis, and increase the levels of cyclin-dependent kinase 2 (CDK2) and BCL2-associated X (BAX). These effects can lead to decreased retinal thickness, electrophysiological dysfunction, and impaired visual function [51]. Myopic retina has higher levels of miR-182-5p, miR-181a-5p, miR-183-5p, miR-9-5p, and miR-96-5p. Myopia and myopia-related retinopathy are caused by overexpression of miR-181a-5p, which can also trigger autophagy, promote RPE cell proliferation, and target N-Sulfoglucosamine Sulfohydrolase (SGSH) in ARPE19 [52]. In FDM and LIM animal models of the retina, Cui et al. discovered that mutations in TEA domain family member 1 (TEAD1) may downregulate miR-671-5p, leading to modifications to cyclic adenosine monophosphate (cAMP) response element binding protein 1 (CREB1) and MAPK1 hub genes that are enriched in extracellular estrogen signaling and visual learning. Additionally, atropine targets MAPK1 and CREB1, key signals in the retinal choroid sclera cascade [53]. Liu et al. discovered elevated levels of mmu-miR-1936, mmu-miR-673-3p, and mmu-miR-338-5p in the retinas of FDM mice. Their target genes, such as Pax6 and SMAD family member 3 (SMAD3), are primarily enriched in retinal tissue morphogenesis and developmental growth. Through post-transcriptional gene regulation, these three miRNAs may contribute to myopia progression [54].
The mechanism of miR-328 in myopia. Elevated expression of miR-328 leads to 1. A decrease in PAX6 results in (1) a decrease in the proliferation of scleral fibroblast, an increase in MMP2, and a decrease in collagen I and integrinB1, and (2) an increase in TGFB3, which causes an increase in the proliferation of retinal pigment epithelium cells. 2. A decrese in FMOD and COL1A1 leads to a decrease in p38-MAPK and MAPK8.
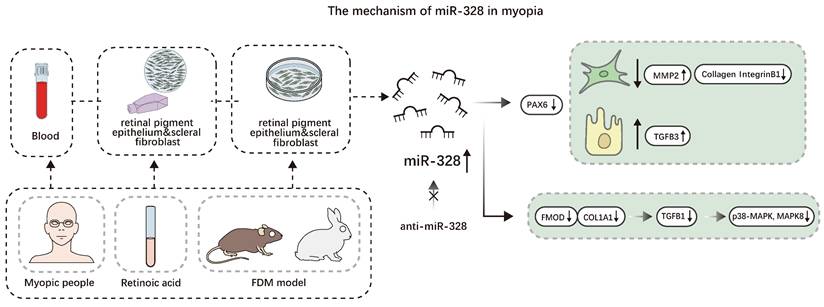
Overexpression of miR-328 might help in the early detection of myopia. Specific miRNAs are involved in tissue repair and the inflammatory response and are associated with retinal proliferation, autophagy, and apoptosis.
miRNAs associated with myopia in the choroid
Li et al. discovered that miR-138-5p suppresses the expression of α-smooth muscle actin (α-SMA), hydroxyproline (HYP), interleukin 1 beta (IL1B), TNF, TGFB1, and type I collagen by blocking the HIF1A signaling pathway. It successfully reduced the experimental myopic guinea pigs' axial length and refractive power, improved their choroidal fibrosis, and inhibited the progression of myopia [55]. According to Liu et al., peak expression of miR-21 in laser-induced CNV in FMD guinea pigs occurs on day 7, earlier than the expression of HIF1A and VEGF (day 14) and the peak development of CNV (day 21). miR-21 can promote the development of HM guinea pigs' CNV and is well correlated with the HIF1A-VEGF signaling pathway [56]. Current research on choroidal miRNAs mainly focuses on choroidal fibrosis and CNV formation.
In summary, miR-138-5p significantly suppresses choroidal fibrosis, while miR-21 can interfere with the establishment of CNV.
miRNAs associated with myopia in the sclera
Guo et al. found 27 differently expressed miRNAs in the sclerotic membranes of LIM guinea pigs. The primary signaling pathways were closely related to peroxisome proliferator-activated receptor alpha (PPARA), TGFB1, pyruvate, and propionic acid metabolism [57].
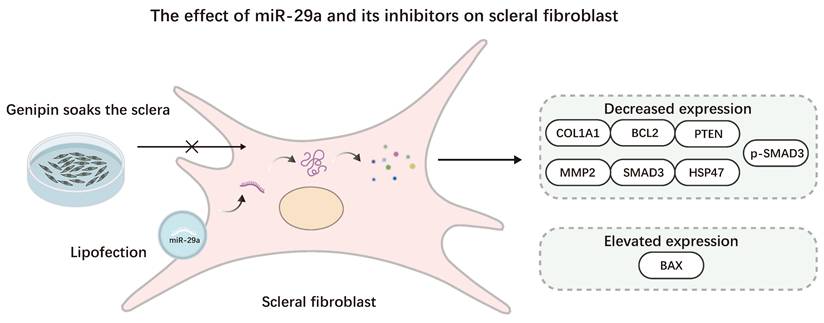
Zhang et al. found that transfection with miR-29a analogs dramatically increased MMP2 mRNA levels in RPE and scleral fibroblasts (SF), according to Matrix metalloproteinases (MMPs) hydrolyze proteins in the extracellular matrix (ECM). Targeting miR-29a may be beneficial for preventing and treating myopia [58]. Yang et al. discovered that in SF, elevated miR-29a expression suppresses the expression of type I collagen, MMP2, phosphoinositide, phosphatase, and tensin homolog (PTEN). Although MMP2 downregulation leads to increased type I collagen, miR-29a may more effectively inhibit type I collagen under the action of the gene expression regulatory complex, ultimately leading to a decrease in type I collagen [59]. Tang et al. found that miR-29a in HSFs inhibits the expression of serpin family H member 1 (SERPINH1), the SMAD3 pathway, COL1A1, and BCL2, upregulates the expression of BAX, reduces the expression level of collagen in HSFs, inhibits the proliferation of HSFs, and increases the apoptosis rate of HSFs [60]. In guinea pig myopia models' sclera, Wang et al. found an increase in the miR-29 cluster (a, b, and c) in the sclera of a guinea pig myopia model, leading to downregulation of COL1A1 expression and overexpression of MMP2. Genipin can counteract this process by lowering miR-29 expression [61].
Similarly, the G allele at the miR-29a SNP rs157907 locus reduces the incidence of HM, according to a clinical study by Xie et al. [62]. Jiang et al. collected the expression profiles of myopia-related miRNAs (miR-328, miR-184, miR-29a, and miR-7i) and fetal sclera miRNAs (miR-328, miR-103, miR-98, miR-107, miR-29b, miR-LET-7, and miR-214) to provide more accurate prognosis, diagnosis, and response prediction for possible myopia treatment [63].
SF, AH, and blood of myopic patients all contain miR-29a, whose primary function is to prevent the formation of type I collagen from forming in SF, thereby promoting scleral remodeling. This implies that miR-29a may be a potential biomarker for myopia and a target for future myopia treatments. However, the expression trajectory of MMP2 in HSF transfected with miR-29 was different from that in myopic guinea pigs. It is necessary to consider whether species differences exist or whether the cells in the in vitro cell experiment are from myopic eyes (Figure 4).
A study found significant variations in the levels of miRNAs (miR-16-2, LET-7a) and mRNAs (peripherin 2 (PRPH2), G protein subunit alpha transducin 1 (GNAT1), and sperm motility kinase 4A (SMOK4A)) in the sclera of FDM mice. G proteins, phototransduction, calcium ions, cytoskeletal protein binding, intermediate filament organization, and stimulus detection are important ontologies of gene overexpression. This implies that myopia is linked to differences in the expression of sclera miRNAs, which controls eye growth [64]. Before going to bed at night, atropine eye drops can decrease melatonin breakdown in the SF, down-regulate miR-2682-5p, and target the voltage-gated potassium channel J subfamily member 5 (KCNJ5) and prolactin receptor (PRLR), which reduces eye growth and scleral remodeling [65]. Zhang et al. reported that in LIM guinea pigs, the miR-15b-5p/miR-379-3p/IGF1R/PTEN/forkhead box (FOXO)/cyclin-dependent kinase inhibitor 1B (CDKN1B) axis can block the G1 cell cycle, induce apoptosis, affect the process of scleral fibrosis, lead to scleral reshaping, and aggravate the development of myopia [66].
Ren et al. discovered that the serum of PM patients had high laminin subunit alpha 4 (LAMA4) and low expression of miR-150-5p. LAMA4 activates the p38-MAPK signaling pathway, and miR-150-5p expression was downregulated due to the hypoxia-cultured HSF increasing HIF1A in the promoter region of miR-150-5p. As a result, MMP2 decreased, COL1A1, TIMP metallopeptidase inhibitor 2 (TIMP2), and α-SMA increased, which inhibit HSF ECM degradation and induce PM [67]. In the FMD animal model, MMP2 expression is elevated, TIMP and type I collagen expression are reduced [68,69], and sclera ECM degradation is accelerated, leading to ECM remodeling. The results of Ren's study run counter to common sense. Factors such as species differences, differences between in vitro and in vivo experiments (HSFs are not derived from myopic eyes), and tissue differences should be considered.
Bioinformatics can also be used to search for differentially expressed miRNAs. Tanaka et al. found that individual ocular tissues of LIM mice, including the sclera, cornea, retina, iris, lens, and choroid, showed significant overlapping changes in miRNA expression [70]. Hsa-miR-17-5p is one of the miRNAs with the most crucial number of gene links, according to Xiao et al. Myopia-specific miRNAs control eye development and DA biological processes [71]. miR-411-5p, miR-376c-3p, miR-155-5p, miR-132-3p, and miR-543 were downregulated in animal myopia samples and upregulated in human myopia samples. To find biomarkers of human myopia, animal models alone are not enough [72]. miRNA expression in human and animal models of myopia is different, and animal experiments combined with cell experiments or clinical trials may be necessary.
Lastly, miRNAs are unrelated to myopia. Two new heterozygous miR-184 alternative variants were found in two cases of isolated keratoconus. The miR-184 stem-loop rs41280052 was not substantially linked to keratoconus, and the axial myopia group had no miR-184 mutations [73].
The sclera of myopic eyes contains differentially expressed miRNAs, including miR-29 and miR-328, which control collagen, ECM proteins, and associated signal pathways in various tissues. These miRNAs are also involved in the remodeling and degradation of the scleral ECM and affect eye growth. They expected to be a targete for myopia treatment. However, studies using FDM and LIM models show that the expression patterns of myopia-related miRNAs in human and animal models are different. This suggests that caution should be exercised when comparing studies conducted in other species.
In conclusion, miRNAs are predicted to develop into myopia biomarkers since they exhibit differential expression in different tissues of myopic individuals. Numerous miRNAs control cell migration, proliferation, and apoptosis and are associated with eye development, tissue remodeling, and fibrosis. By influencing the metabolism of collagen and ECM, some miRNAs can control scleral remodeling and the length of the ocular axis. They may be used as therapeutic targets for future myopia treatment. Most miRNA research, however, has been conducted in animal models (such as FDM and LIM animal models), and the patterns of miRNA expression in these models differ from those in humans. It takes time and effort to extrapolate animal research accurately results to humans. Moreover, miRNAs play distinct roles in various tissues and experimental paradigms. More accurate experimental design and analysis are needed to investigate the tissue specificity of miRNAs and their mechanism of action in myopia. Lastly, one miRNA can affect multiple target genes and signal pathways due to its multi-target effect. Future studies must identify more precise targets, as this widespread regulatory action may have negative implications.
The effect of miR-29a and its antagonists on Scleral fibroblast. Myopia was promoted using the lipofection method to transfect SFs with miR-29a, resulting in elevated BAX expression and reduced COL1A1, BCL2, PTEN, MMP2, SMAD3, HSP47, and p-SMAD3 expression. This process can be reversed by using Genipin.
3.2.2. Long non-coding RNAs
Long non-coding RNAs (lncRNAs) are generally considered to be ncRNA molecules longer than 200 nucleotides with a 3' poly(A) tail and a 5' 7-methylguanosine cap [74]. Many aspects of cell development, differentiation, and other physiological processes are regulated by lncRNAs [75].
Wang et al. suggested that the lncRNA-XR_002792574.1/miR-760-3p/adenylate cyclase 1 (ADCY1) axis may negatively affect RGCs in FDM guinea pigs by inhibiting the Apelin and cyclic guanosine monophosphate (cGMP)/protein kinase cGMP-dependent 1 (PKG) signaling pathways. The ERK-MMP2 pathway may also contribute to myopic scleral remodeling [76]. Wu et al. discovered that the lncRNAs in the ciliary body of LIM guinea pigs are mainly enriched in the complement, cytokine-cytokine receptor interactions, coagulation cascade, and RAP1 GTPase activating protein (RAP1GAP) signaling pathways. Three hub genes—phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), ITGB1, and catenin beta 1 (CTNNB1)—are involved in the RAP1GAP signaling pathway. Retinal photoreceptors primarily express two key genes: G protein-coupled receptor kinase 1 (GRK1) and phosphodiesterase 6A (PDE6A). They improve the ciliary body's visual perception [77]. Li et al. identified 655 lncRNAs differently expressed in FDM retinas. These lncRNAs were primarily enriched in retinol metabolism, cytokine-cytokine receptor interactions, cellular activities, and rhythmic processes. lncRNA Gm35369 was mainly expressed in horizontal cells and RGCs [78]. Geng et al. found aberrant expression of the posterior pole lncRNA in the eyes of FDM and LIM guinea pigs compared to the healthy group. This expression is primarily enriched in cellular components (structural components of ECM), molecular functions (kinase activity, metabolism, and growth), and pathways (such as ECM receptor interaction, glycosaminoglycan degradation, and mucin-type O-glycan biosynthesis) [79]. According to the idea of molecular structural effects proposed by Wang et al., lncRNAs fold into complex spatial patterns that attract transcription factor (TF) targets for scaffold binding. Base mutations affecting key components and variable splicing regions have been discovered in transcription factor binding sites (TFBSs) of myopia-associated lncRNA transcripts. These mutations caused substantial changes in molecular conformation and mediate the development and progression of myopia [80]. The research on myopia-related lncRNAs mainly involves sequencing animal models' eye tissues (guinea pigs and mice) to find target genes, enriched networks, and specifically expressed lncRNAs.
In short, lncRNAs affect signal transduction, cell proliferation, and remodeling in different tissues in myopia. Their complex spatial structure allows them to participate in multiple physiological pathways. In addition to providing new molecular indicators for myopia development, lncRNAs may also become targets for intervention in tissue damage and structural remodeling linked to myopia. However, target identification is challenging due to the complicated three-dimensional structure of lncRNA and their multi-target characteristics, which may lead to adverse reactions. Furthermore, the stability and distribution system of lncRNAs are still problematic. Future studies must expand the structural and functional analysis of lncRNAs to support their clinical application and the development of precise regulatory techniques.
3.2.3. Circular RNAs
Exon circular RNAs (circRNAs) are covalently closed endogenous RNAs produced by the anti-splicing process. Their functions include protein recruiters, decoys, sponges, scaffolds, and miRNA. They can also encode various proteins, from tiny peptides to proteins, and act as translation templates in pathological processes [81,82].
Zhang et al. found that has-circ-Nbea and has-circ-Pank1 promote AL growth in HM patients; the target genes are mainly concentrated in MTOR, insulin, cAMP, vascular endothelial growth factor, and other signaling pathways. The sclera of the FDM mouse model contains the circNbea/miR-204-5p/ inositol 1,4,5-trisphosphate receptor type 1 (ITPR1) and circPank1/miR-145-5p/ NRAS proto-oncogene (NRAS) axes. These axes could be targets for myopia treatment or biomarkers for myopia progression [83]. Ma et al. discovered that compared with ARC patients, high myopia cataract (HMC) patients have increased tropomyosin 1 (TPM1) expression by circ-AFF1 through sponge miRNA760. It also controls apoptosis, proliferation, and migration of lens epithelial cells (LECs) [84]. Current research on circRNAs focuses on RNA sequencing of VH and lens epithelium samples from myopic patients to map circRNA-related networks and profiles.
Through particular signaling pathways, circRNAs can control the sclera and promote the growth of AL. They may also be involved in pathological changes of the eye's lens and can prevent the growth and death of LECs. By interfering with the pathway regulated by circRNA, it should be possible to regulate the myopic lesion tissue precisely. However, due to the complex regulation mechanism of circRNAs, learning more about their specific roles as protein and miRNA sponges is essential.
3.2.4. Short interfering RNAs
Dicer enzymes cleave non-coding double-stranded RNA (dsRNA) into short interfering RNAs (siRNAs) [85]. RNA interference (RNAi) is an evolutionarily conserved mechanism for post-transcriptional gene silencing [86]. siRNAs control gene expression via RNAi. They can also be used as a possible therapeutic agent. The local distribution of naked siRNA in two tissues, the lung and the eye, shows promise [87].
siRNAs can be used to knockout genes in the sclera that are associated with myopia. When CHRM2 in the sclera of mice is knocked out using siRNA, the levels of type I collagen increase, and type V collagen decreases, the growth of SF is reduced, and mice without CHRM2 were less likely to develop LIM than wild-type mice [88]. The use of transglutaminase 2 (TGM2) specific siRNA in HSFs slowed the prolongation of AL. Drugs that block CHRM2 also slowed myopia progression in mice, and this effect was accompanied by a decrease in TGM2 expression [89].
Furthermore, Zhao et al. discovered that COL1A1 is downregulated in HSF when PDE4B is knocked down by siRNA, indicating that PDE4B may be a new gene susceptible to HM [90]. Myofibroblast development is controlled by the ras homolog family member A (RHOA) / rho-associated coiled-coil containing protein kinase 2 (ROCK2), a critical mechanotransduction pathway. Elevated α-SMA expression is due to upregulated RhoA expression leading to activation of ROCK2. Mechanical strain cannot enhance the production of ROCK2, myocardin-related transcription factor A (MRTFA), and SRF if siRNA is used to block RhoA [91]. Li et al. found that the LIM group transfected with hepatocyte growth factor (HGF) siRNA had significantly lower MMP2 protein expression in SF than the normal group, indicating that HGF is an upstream mediator of MMP2 in guinea pigs SF [92].
siRNAs can also knock out myopia-related genes in the retina and lens. Dopamine receptor D2 (DRD2) protein expression increased in human RPE when ADORA2A was knocked down using siRNA, suggesting that ADORA2A and DRD2 heterodimers are present. To limit myopia development, 7-methylxanthine (7-MX) can increase the expression of DRD2, pERK1/2, and ERK1/2 while inhibiting the expression of ADORA2A [93]. HM is associated with endoplasmic reticulum stress-induced apoptosis of LECs, which causes cataracts. Apoptosis of human LECs is induced by exposing human LEC cells to endoplasmic reticulum stress under UV irradiation through activation of the activating transcription factor 4 (ATF4)-ATF3 -DNA damage-inducible transcript 3 (DDIT3) pathway. Downregulating ATF4, ATF3, and DDIT3 by siRNA can block KLF transcription factor 6 (KLF6)-induced apoptosis [94].
In other words, siRNA can knock out particular genes to confirm their function. It is a precise treatment technique that can silence genes linked to myopia. However, enhancing siRNA's specificity and targeting while minimizing off-target effects is a key therapeutic issue. The stability of siRNA in clinical applications needs to be further optimized.
3.2.5. Ribosomal RNAs
Ribosomal RNA (rRNA) accounts for about 80% of cellular RNA [95]. In all known organisms, rRNA is essential for protein synthesis and maintaining cellular function [96]. In bacteria, the structure of 16S rRNA has evolved into a highly conserved and diverse part with a medium-length gene sequence. This biomarker sequence is ideal [97].
In myopia, 16s rRNA is mainly used to reveal the potential relationship between the microbial community (primarily intestinal microorganisms) and myopia and confirm the existence of the gut-eye axis.
Fecal samples from adolescents sequenced with 16S RNA showed that the microbial communities in the myopia group were different from those in the control group in composition and abundance. The Firmicutes and Actinobacteria phyla had different relative abundances and absolute quantities, suggesting that the characteristics of the intestinal flora can significantly distinguish myopia from healthy controls [98]. According to 16S rRNA analysis, the intestinal microbiota of LIM mice—myopic mice with impaired intestinal barrier function—showed a much lower relative abundance of the Firmicutes phylum and a much higher relative abundance of the Actinobacteria phylum [99]. Wu et al. found through 16S rRNA gene sequencing that Ruminococcus_albus is a dominant and abundant bacterium in the intestines of FDM guinea pigs and is positively correlated with the levels of ten metabolites such as L-glutamic acid, as well as with vasoactive intestinal peptide (VIP) and lipopolysaccharide (LPS) [100]. The amount of the intestinal microbial metabolite indole-3-acetic acid (3-IAA) in the plasma of HM patients and the amount of beneficial intestinal bacteria were significantly reduced using 16S rRNA gene sequencing. When HM mice received a fecal microbiota transplant from a healthy donor, 3-IAA plasma levels increased, and HM progression slowed. The expression of COL1A1 in the sclera is maintained through Sp1 transcription factor (SP1)-dependent transcriptional activation [101].
The researchers also performed 16s rRNA sequencing on conjunctival sac swab specimens. They found that patients with HM had higher scores on the Ocular Surface Disease Index (OSDI), an imbalance in the bacterial microbiome in the conjunctival sac. Proteus, Actinomyces, and Acinetobacter may be associated with HM-related ocular surface inflammation [102].
In conclusion, 16S rRNA sequencing showed that the intestinal and ocular surface microbiomes of myopic eyes are significantly different from those of healthy eyes, thus confirming the existence of the gut-eye axis. Therefore, 16S rRNA sequencing can help with the early myopia diagnosis. By controlling the intestinal microbiota, fecal microbiota transplantation (FMT) may slow the progression of HM. Yet, the microbiomes of myopic patients vary greatly, and the long-term effectiveness and safety of FMT need to be confirmed by further research.
3.2.6. Transfer RNAs
A small ribonucleic acid molecule known as transfer RNA (tRNA) carries and transports amino acids. Under the direction of mRNA, its main job is to transport amino acids to the ribosome for protein synthesis.
Studies on the connection between tRNA and myopia are scarce. In the growing fetal brain, glutaminyl-tRNA synthetase (QARS) is extensively expressed. It is hypothesized that the catalytic domain of the QARS gene contains a homozygous missense mutation (V476I). It then causes glutamine-tRNA synthetase deficiency, which induces a novel recessive disease, including intellectual disability, microcephaly, severe linear growth retardation, HM, syndactyly, and distinctive facial features [103].
Briefly, QARS mutations can be used as genetic indicators of high myopia and provide clues for the early detection of rare myopia-related diseases. However, more research is still needed to determine their mode of action.
When ncRNA-related research is summarized, these RNA molecules have multiple functions in myopia's genetic control, signal regulation, and microbiomics. This information is crucial for understanding myopia's molecular mechanism and guiding the development of prospective therapeutic approaches. However, the clinical translation of the research results may be hampered by differences in RNA expression between human myopia and animal models. Determining the differences in RNA expression between humans and other species is crucial. Second, because ncRNAs are widely expressed in various tissues, it is challenging to ensure therapeutic specificity and prevent side effects.
3.3 Histone modification
Histones and their core octamer complexes form the basic structural unit of eukaryotic chromosomes [104]. The ribosomal core particles are formed by wrapping the DNA of each cell around the histone octamer. Numerous residues at these termini are susceptible to post-translational modification, which affects all DNA-based functions [105].
3.3.1 Histone methylation
Lysine methylation marks different sites on the tail and globular domains of histones, and a precise balance is achieved through the action of methyltransferases (“writers”) and demethylases (“erasers”); in addition, how different effector proteins (“readers”) recognize specific methyl lysines depends on the adjacent amino acid sequence and methylation status. H3K27me3 is a classic histone modification site [106,107].
Cui's team found that heat shock transcription factor 4(HSF4) recruits the methyltransferase enhancer of zeste 2 polycomb repressive complex 2 subunits (EZH2), increased H3K27me3 levels in the proximal promoter region of p21cip1, inhibit p21cip1 expression in mouse embryonic fibroblasts (MEFs), reduced UVB-induced senescence of LECs, and suggested that HSF4 may have an anti-aging effect during lens development [108]. Methyltransferases that regulate G9a, H3K27me3, EZH2, and H3K9me2 in the mouse retina are enriched during active retinal development in the embryo. RGCs exhibit H3K9me2 during their early development. These histone marks and the related methyltransferase mediate the epigenetic regulation of essential cell lineage genes throughout adult retinal cell plasticity, axonal regeneration, retinal cell capacity, retinal development, retinal cell survival, and retinal cancer [109]. Sumiko et al. learned that the methylation levels of histones H3K27 and H3K4, which are essential for retinal differentiation and proliferation, as well as the acetylation levels of histone H3 at specific gene loci, undergo significant changes during retinal development. Progenitor cell-specific gene expression is attenuated by low amounts of H3K4me3 and high amounts of H3K27me3 [110].
Genetic regulation of ocular tissue throughout development can be better understood by considering the effects of histone methylation on retinal development and lens cell aging.
3.3.2 Histone acetylation
Acetylation is a common and degradable post-translational protein modification catalyzed by two types of enzymes: lysine deacetylases (KDACs) remove the acetyl group, and lysine acetyltransferases (KATs) transfer the acetyl group to the lysine residue [111].
One study found that elevated level of glycolytic activity in the optic nerve sheath region of mouse embryos. Eye development was inhibited by removing the activity of the lactate dehydrogenase A (LDHA) gene and the solute carrier family 2 member 1 (SLC2A1) in developing retinal progenitor cells. Histone H3 acetylation is epigenetically modified by histone deacetylase (HDAC) operation, which regulates the lactate-mediated key field-specific transcription factors [112]. According to Park et al., the expression levels of HDAC2 and HDAC3 in limbal conjunctiva (LC) and peripapillary sclera (PPS) ECM of Koreans are lower than Caucasians ECM, the expression levels of acetylated histone H3, the elastic fiber components (fibrillin 1 and elastin) and lysine oxidase-like protein (LOXL2) are higher than those in Caucasians, indicating that the histone acetylation status of elastic fiber component and LOXL2 promoter region differs in LC and PPS of other ethnic groups [113]. HM is associated with five non-coding potassium voltage-gated channel Q member 5 (KCNQ5). SNPs rs7744813 and rs9342979 are located at or near the DNase I hypersensitive sites and histone marks of H3K27ac and H3K4me1. This demonstrates that KCNQ5 is a risk factor for HM and that KCNQ5 gene polymorphisms contribute to genetic predisposition to HM [114]. Histone acetylation is associated with eye development and HM (Figure 5).
The reversibility of lysine acetylation makes it possible to intervene in myopia development. Racial differences in histone acetylation reveal its function in the ocular architecture of different races and help develop tailored treatment plans for specific groups.
Changes in histone make new targets for myopia treatment possible. They can be employed as tools to control gene expression in the eye because of their reversibility, which will help to understand the pathophysiology and genetic basis of eye disorders. Racial differences in histone modifications make treatment for specific populations possible. However, histone modification involves multiple enzymes and regulatory pathways, and the regulatory mechanism is complex. Accurately controlling these processes takes time and effort. Finding a more unified treatment is difficult since the effects of histone modification fluctuate widely between races and individuals.
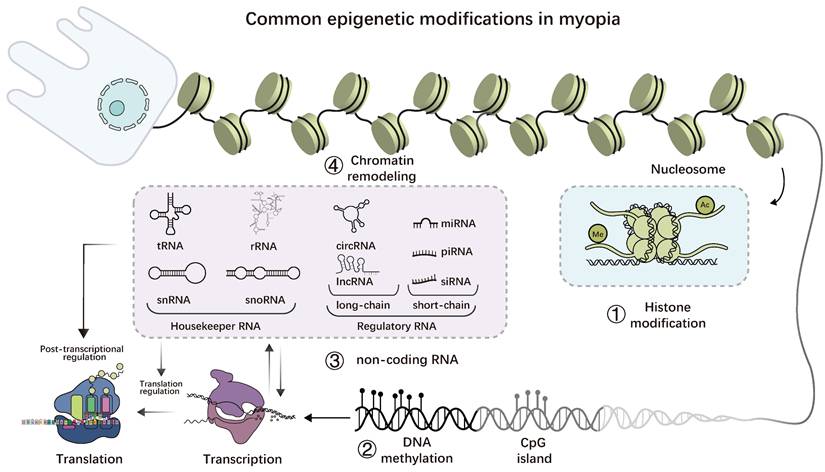
Common epigenetic modifications in myopia. 1. histone modification is a reversible covalent modification that mainly occurs at the tail of histones; 2. DNA methylation occurs on CpG islands, and the methyl group is acquired via covalent binding; 3. ncRNAs, including housekeeper and regulator types; 4. RNA methylation is a reversible post-transcriptional modification that primarily occurs on mRNA but is also found in non-coding RNAs, with N6-methyladenosine (m6A) being the most common form; 5. Numerous enzymes, including the SWI/SNF complex, are involved in chromatin remodeling. By interacting with one another, the four common epigenetic changes mentioned above can also impact gene expression.
3.4 RNA methylation
RNA methylation is a modification process catalyzed by RNA methyltransferases that incorporates methyl groups into RNA bases using s -adenosyl- l-methionine (SAMe) as a donor [115]. These include n6-methyladenosine (m6A), n1-methyladenosine (m1A), 5-methylcytosine (m5C), and 7-methylguanosine (m7G) [116].
Wen et al. found that upregulation of the m6a methylase methyltransferase-like 14 (METTL14) and downregulation of the demethylase obesity-associated protein (FTO), alkB homolog 5 (ALKBH5), catalyzed the hypermethylation of the m6a methylase-like protein 1 (CHI3L1) and its encoding protein, YKL-40, in the anterior capsule of the lens in patients with nuclear cataracts with high myopia, and contributes to the pathological state of high myopia by regulating the composition of the extracellular matrix. The remaining target genes also include collagen alpha-3(VI) chain (COL6A3) and peroxidasin homolog (PXDN) [117]. Xue et al. found that forkhead box M1 (FOXM1) increased the m6A methylation level of apolipoprotein A1 (APOA1) by inhibiting methyltransferase-like 3 (METTL3) transcription. APOA1 mRNA stability and transcription were enhanced by decreasing YTHN6- methyladenosine RNA binding protein F2 (YTHDF2)-recognized m6A-methylated transcripts [118]. Shan's team found that elevated levels of FTO in neovascularized corneas and endothelial cells of CNV decreased m6A methylation levels of focal adhesion kinase (FAK), leading to reduced RNA stability and increased RNA decay via YTHDF2, resulting in abnormal endothelial cell function and pathological corneal angiogenesis [119]. Liu et al. found that tRF-22 of the transfer RNA-derived fragments (tRFs) family was decreased in the choroid of C57BL/6 J mice with FDM and interacted with METTL3 to block Axin1 and AT-rich interaction domain 1B (ARID1B) methylation of m6A in mRNA transcripts leads to increased expression of Axin1 and Arid1b, resulting in their attenuated inhibition of Wnt signaling and promotion of choroidal neovascularization [120].
m6A plays a key role in myopia and ocular diseases and could provide new molecular targets for future therapies, especially in regulating gene expression and neovascularization. Although studies have revealed the role of m6A in myopia, the molecular mechanisms involved are still not completely clear, and m6A modifications and their roles still need to be studied. These studies are based on animal models and cellular experiments, and how to effectively translate them into clinical therapeutic approaches still faces excellent technical and practical challenges.
To date, we have summarized the research results on epigenetic modifications in myopia. Current research mainly focuses on ncRNAs, and there are few studies on histone modification, RNA methylation, and chromatin remodeling. In addition, relatively few studies have been conducted on the interactions between different types of epigenetic modifications. Myopia involves multiple epigenetic changes, and other types do not exist in isolation; they form a complex gene regulation network. More critical discoveries may come from connections between various epigenetic changes rather than changes in epigenetic modifications themselves.
4. Conclusion
Recent advancements in the study of epigenetic modifications have clarified the complex interactions between genetic susceptibility and environmental factors in myopia development. It is clear that epigenetic processes, including non-coding RNAs (ncRNAs), DNA methylation, histone modifications, and RNA methylation, play a crucial role in regulating gene expression patterns associated with refractive error and ocular development. Gene mutations are not the only factors determining myopia; they involve heritable but reversible epigenetic changes influenced by environmental factors.
DNA methylation patterns of key genes, such as those involved in eye growth and signaling pathways, are associated with myopia development. Furthermore, histone modifications can impact chromatin structure and gene accessibility, thereby affecting the regulation of ocular growth. In myopia, ncRNAs, particularly microRNAs, act as essential regulators of gene expression, providing a possible molecular mechanism for further control of eye elongation and growth. m6A RNA methylation affects myopia development by regulating gene expression and neovascularization and may be involved in key processes such as visual axis lengthening.
Future research should elucidate how to reverse or prevent these epigenetic changes to develop potential therapeutic interventions. Specifically, studies could explore epigenetic therapies, such as DNA demethylation agents or miRNA-based treatments, to avoid or mitigate myopia progression. Additionally, combining genomic, transcriptomic, and epigenetics data will provide a more comprehensive understanding of the molecular causes of myopia and enable the development of personalized approaches to prevention and treatment.
Overall, studying epigenetic modifications provides a promising approach to addressing the rising global prevalence of myopia. Understanding how environmental factors affect gene expression through epigenetic mechanisms can help develop effective strategies for early diagnosis, prevention, and individualized treatment of myopia.
Abbreviations
3-IAA: intestinal microbial metabolite indole-3-acetic acid; 7-MX: 7-methylxanthine; ADCY1: adenylate cyclase 1; ADCY3: adenylate cyclase 3; ADORA2A: adenosine A2a receptor; AH: aqueous humor; AL: axial length; AP1B1: adaptor-related protein complex 1 subunit beta 1; ARC: age-related cataracts; ATF4: activating transcription factor 4; BAX: BCL2-associated X; BCL2: BCL2 apoptosis regulator; BTG2: BTG anti-proliferation factor 2; cAMP: cyclic adenosine monophosphate; CCL2: C-C motif chemokine ligand 2; CDK2: cyclin-dependent kinase 2; CDKN1B: cyclin-dependent kinase inhibitor 1B; cGMP: cyclic guanosine monophosphate; CHRM2: cholinergic receptor muscarinic 2; circRNAs: circular RNAs; CNGB3: cyclic nucleotide-gated channel subunit beta 3; CNV: choroidal neovascularization; COL1A1: collagen type I alpha 1 chain; CpG islands: cytosine-phosphate-guanine sites; CREB1: cAMP response element binding protein 1; CRYAA: crystallin alpha A; CTNNB1: catenin beta 1; CXCL10: C-X-C motif chemokine ligand 10; DA: Dopamine; DDIT3: DNA damage-inducible transcript 3; DNMTs: DNA methyltransferases; DRD2: dopamine receptor D2; dsRNA: double-stranded RNA; ECM: extracellular matrix; EGR1: early growth response 1; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunits; FDM: form-deprivation myopia; FGF2: fibroblast growth factor 2; FMOD: fibromodulin; FOS: AP-1 transcription factor subunit; FOXO: forkhead box; GNAT1: G protein subunit alpha transducin 1; GRB2: growth factor receptor binding protein 2; GRK1: G protein-coupled receptor kinase 1; GSTP1: glutathione S-transferase pi 1; HDAC: histone deacetylase; HGF: hepatocyte growth factor; HIF1A: hypoxia-inducible factor 1 subunit alpha; HM: high myopia; HMC: high myopia cataract; HOXA9: homeobox A9; HSF4: heat shock transcription factor 4; HSFs: human scleral fibroblasts; HYP: hydroxyproline; IFN1: type I interferon; IFNG: interferon-gamma; IGF1: insulin-like growth factor 1; IGF1R: insulin-like growth factor 1 receptor; IL1B: interleukin 1 beta; ITGB1: integrin subunit beta 1; ITPR1: inositol 1,4,5-trisphosphate receptor type 1; KATs: lysine acetyltransferases; KCNJ5: voltage-gated potassium channel J subfamily member 5; KCNQ5: potassium voltage-gated channel subfamily Q member 5; KDACs: lysine deacetylases; KLF6: KLF transcription factor 6; LAMA4: laminin subunit alpha 4; LC: limbal conjunctiva; LDHA: lactate dehydrogenase A; LECs: lens epithelial cells; LIM: lens-induced myopia; LINE-1: long interspersed nucleotide element 1; lncRNAs: long non-coding RNAs; LOXL2: lysine oxidase-like protein; LPS: lipopolysaccharide; LTA: lymphotoxin alpha; LUM: lumican; MAPK: mitogen-activated protein kinase; MEFs: mouse embryonic fibroblasts; miRNAs: micro RNAs; MMP2: matrix metalloproteinase 2; MMPs: matrix metalloproteinases; mRNA: messenger RNA; MRTFA: myocardin-related transcription factor A; MTOR: mechanistic target of rapamycin kinase; MX1: MX dynamin-like GTPase 1; NAB2: NGFI-A binding protein 2; ncRNAs: non-coding RNAs; NRAS: NRAS proto-oncogene; OSDI: Ocular Surface Disease Index; PAX6: paired box 6; PCDHA10: protocadherin alpha 10; PDE4B: phosphodiesterase 4B; PDE6A: phosphodiesterase 6A; PDGFRA: platelet-derived growth factor receptor alpha; PI3K/AKT: phosphatidylinositol 3 kinase/protein kinase B; PIK3R1: phosphoinositide-3-kinase regulatory subunit 1; piRNA: piwi-associated RNA; PKG: protein kinase cGMP-dependent 1; PM: pathological myopia; PPARA: peroxisome proliferator-activated receptor alpha; PPS: peripapillary sclera; PRLR: prolactin receptor; PRPH2: peripherin 2; PSEN1: presenilin 1; PTEN: phosphatase and tensin homolog; QARS: glutaminyl-tRNA synthetase; RA: Retinoic acid; RAP1GAP: RAP1 GTPase activating protein; RGCs: retinal ganglion cells; RGS5: regulator of G protein signaling 5; RHOA: ras homolog family member A; RISC: RNA-induced silencing complex; RNAi: RNA interference; ROCK2: rho-associated coiled-coil containing protein kinase 2; ROS: reactive oxygen species; RPE: retinal pigment epithelial cells; rRNA: ribosomal RNA; RUNX1: RUNX family transcription factor 2; SERPINH1: serpin family H member 1; SF: scleral fibroblasts; SGSH: N-sulfoglucosamine sulfohydrolase; siRNAs: short interfering RNAs; SLC2A1: solute carrier family 2 member 1; SMAD3: SMAD family member 3; SMOK4A: sperm motility kinase 4A; snoRNA: small nucleolar RNA; snRNA: small nuclear RNA; SOCS1: suppressor of cytokine signaling 1; SP1: Sp1 transcription factor; SRF: serum response factor; TEAD1: TEA domain family member 1; TF: transcription factor; TFBSs: transcription factor binding sites; TGFB: transforming growth factor beta; TGM2: transglutaminase 2; TIMP2: TIMP metallopeptidase inhibitor 2; TNF: tumor necrosis factor; TPM1: tropomyosin 1; tRNA: transfer RNA; TXNIP: thioredoxin-interacting protein; TXNRD2: thioredoxin reductase 2; V476I: homozygous missense mutation; VEGFA: vascular endothelial growth factor A; VH: vitreous humor; VIP: vasoactive intestinal peptide; ZNRF3: zinc and ring finger 3; α-SMA: α-smooth muscle actin.
Acknowledgements
Acknowledgment is given to Chenxi Zhang (Shandong Jianzhu University) for optimizing and visualizing the image drafts of this manuscript, which significantly supported the illustration presentation.
Funding
This study was supported by the Natural Science Foundation of Shandong Province (ZR2024MH057), the National Key R&D Program of China (2021YFC2702103), the Science & Technology Project of Medicine and Health of Shandong Province (202307021591), and the “Taishan Scholar” Project Special Fund (tsqnz20231252).
Author contributions
Conceptualization & Supervision: Dadong Guo; Writing Original Draft&Image Drafts: Yinqiao Zhang; Visualization: Chenxi Zhang; Writing Review & Editing: Dadong Guo, Zhaohui Yang, Miao Zhang, Zhongyu Ma; Investigation: Yuanting Yang, Mengke Wu.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Baird PN, Saw S-M, Lanca C, Guggenheim JA, Smith Iii EL, Zhou X. et al. Myopia. Nat Rev Dis Primers. 2020;6:99 https://doi.org/10.1038/s41572-020-00231-4
2. Morgan IG, Ohno-Matsui K, Saw S-M. Myopia. The Lancet. 2012;379:1739-48 https://doi.org/10.1016/S0140-6736(12)60272-4
3. Modjtahedi BS, Ferris FL, Hunter DG, Fong DS. Public health burden and potential interventions for myopia. Ophthalmology. 2018;125:628-30 https://doi.org/10.1016/j.ophtha.2018.01.033
4. Resnikoff S, Jonas JB, Friedman D, He M, Jong M, Nichols JJ. et al. Myopia - a 21st century public health issue. Invest Ophthalmol Vis Sci. 2019;60:Mi https://doi.org/10.1167/iovs.18-25983
5. Holden BA, Fricke TR, Wilson DA, Jong M, Naidoo KS, Sankaridurg P. et al. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology. 2016;123:1036-42 https://doi.org/10.1016/j.ophtha.2016.01.006
6. Morgan IG, French AN, Ashby RS, Guo X, Ding X, He M. et al. The epidemics of myopia: Aetiology and prevention. Progress in Retinal and Eye Research. 2018;62:134-49 https://doi.org/10.1016/j.preteyeres.2017.09.004
7. Chang C, Lu Q, editors. Epigenetics in allergy and autoimmunity. vol. 1253. Singapore: Springer Singapore. 2020 https://doi.org/10.1007/978-981-15-3449-2
8. Wang Y, Riaz F, Wang W, Pu J, Liang Y, Wu Z. et al. Functional significance of DNA methylation: Epigenetic insights into sjögren's syndrome. Front Immunol. 2024;15:1289492 https://doi.org/10.3389/fimmu.2024.1289492
9. Wei L, Liu B, Tuo J, Shen D, Chen P, Li Z. et al. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Reports. 2012;2:1151-8 https://doi.org/10.1016/j.celrep.2012.10.013
10. Hao J, Yang Z, Zhang R, Ma Z, Liu J, Bi H. et al. Crosstalk between heredity and environment in myopia: An overview. Heliyon. 2024;10:e29715 https://doi.org/10.1016/j.heliyon.2024.e29715
11. Kaliman P. Epigenetics and meditation. Current Opinion in Psychology. 2019;28:76-80 https://doi.org/10.1016/j.copsyc.2018.11.010
12. Zhang X, Wang Y, Pan C, Yang W, Xiang Y, Yang J. et al. Effect of genetic-environmental interaction on Chinese childhood myopia. Journal of Ophthalmology. 2020;2020:1-6 https://doi.org/10.1155/2020/6308289
13. Li X, Long J, Liu Y, Cai Q, Zhao Y, Jin L. et al. Association of MTOR and PDGFRA gene polymorphisms with different degrees of myopia severity. Exp Eye Res. 2022;217:108962 https://doi.org/10.1016/j.exer.2022.108962
14. Zhao F, Zhou H, Chen W, Zhao C, Zheng Y, Tao Y. et al. Declines in PDE4B activity promote myopia progression through downregulation of scleral collagen expression. Experimental Eye Research. 2021;212:108758 https://doi.org/10.1016/j.exer.2021.108758
15. Esteve-Puig R, Bueno-Costa A, Esteller M. Writers, readers and erasers of RNA modifications in cancer. Cancer Letters. 2020;474:127-37 https://doi.org/10.1016/j.canlet.2020.01.021
16. Li S, Tollefsbol TO. DNA methylation methods: Global DNA methylation and methylomic analyses. Methods. 2021;187:28-43 https://doi.org/10.1016/j.ymeth.2020.10.002
17. Chen Z, Zhang Y. Role of mammalian DNA methyltransferases in development. Annu Rev Biochem. 2020;89:135-58 https://doi.org/10.1146/annurev-biochem-103019-102815
18. Thomson K, Game J, Karouta C, Morgan IG, Ashby R. Correlation between small-scale methylation changes and gene expression during the development of myopia. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2022;36:e22129 https://doi.org/10.1096/fj.202101487R
19. Ding X, Fu D, Ge S, Guan Q, Chen M, Yu Z. DNA methylation and mRNA expression of IGF-1 and MMP-2 after form-deprivation myopia in guinea pigs. Ophthalmic & Physiological Optics: The Journal of the British College of Ophthalmic Opticians (Optometrists). 2020;40:491-501 https://doi.org/10.1111/opo.12696
20. Liang C-L, Hsu P-Y, Ngo CS, Seow WJ, Karnani N, Pan H. et al. HOXA9 is a novel myopia risk gene. BMC Ophthalmology. 2019;19:28 https://doi.org/10.1186/s12886-019-1038-9
21. Swierkowska J, Karolak JA, Vishweswaraiah S, Mrugacz M, Radhakrishna U, Gajecka M. Decreased levels of DNA methylation in the PCDHA gene cluster as a risk factor for early-onset high myopia in young children. Invest Ophth Vis Sci. 2022;63:31 https://doi.org/10.1167/iovs.63.9.31
22. Hsi E, Wang Y-S, Huang C-W, Yu M-L, Juo S-HH, Liang C-L. Genome-wide DNA hypermethylation and homocysteine increase a risk for myopia. Int J Ophthalmol-Chi. 2019;12:38-45 https://doi.org/10.18240/ijo.2019.01.06
23. Zhu X, Li D, Du Y, He W, Lu Y. DNA hypermethylation-mediated downregulation of antioxidant genes contributes to the early onset of cataracts in highly myopic eyes. Redox Biol. 2018;19:179-89 https://doi.org/10.1016/j.redox.2018.08.012
24. Zhou X, Ji F, An J, Zhao F, Shi F, Li Y. et al. Experimental murine myopia induces collagen type Iα1 (COL1A1) DNA methylation and altered COL1A1 messenger RNA expression in sclera. Mol Vis. 2012;18:1312-24
25. Zhu X-J, Zhou P, Zhang K-K, Yang J, Luo Y, Lu Y. Epigenetic regulation of αA-crystallin in high myopia-induced dark nuclear cataract. PloS One. 2013;8:e81900 https://doi.org/10.1371/journal.pone.0081900
26. Seow WJ, Ngo CS, Pan H, Barathi VA, Tompson SW, Whisenhunt KN. et al. In-utero epigenetic factors are associated with early-onset myopia in young children. PloS One. 2019;14:e0214791 https://doi.org/10.1371/journal.pone.0214791
27. Tsonis PA, Fuentes EJ. Focus on molecules: Pax-6, the eye master. Experimental Eye Research. 2006;83:233-4 https://doi.org/10.1016/j.exer.2005.11.019
28. Williams C, Suderman M, Guggenheim JA, Ellis G, Gregory S, Iles-Caven Y. et al. Grandmothers' smoking in pregnancy is associated with a reduced prevalence of early-onset myopia. Sci Rep. 2019;9:15413 https://doi.org/10.1038/s41598-019-51678-9
29. Vishweswaraiah S, Swierkowska J, Ratnamala U, Mishra NK, Guda C, Chettiar SS. et al. Epigenetically dysregulated genes and pathways implicated in the pathogenesis of non-syndromic high myopia. Sci Rep. 2019;9:4145 https://doi.org/10.1038/s41598-019-40299-x
30. He X, Li S-M. Genetic methylation in myopia. Ophthalmic Genetics. 2024;45:233-4 https://doi.org/10.1080/13816810.2024.2381123
31. Jiang D, Lin S, Gong Q, Hong J, Wang J, Gao H. et al. PAX6 gene promoter methylation is correlated with myopia in Chinese adolescents: A pilot sutdy. Ophthalmic Genet. 2024;45:219-25 https://doi.org/10.1080/13816810.2024.2315152
32. Swierkowska J, Vishweswaraiah S, Mrugacz M, Radhakrishna U, Gajecka M. Differential methylation of microRNA encoding genes may contribute to high myopia. Front Genet. 2022;13:1089784 https://doi.org/10.3389/fgene.2022.1089784
33. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18 https://doi.org/10.1038/nrc.2017.99
34. Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. In: Slaby O, Calin GA, editors. Non-coding RNAs in Colorectal Cancer, vol. 937, Cham: Springer International Publishing. 2016 p. 3-17. https://doi.org/10.1007/978-3-319-42059-2_1
35. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97 https://doi.org/10.1016/S0092-8674(04)00045-5
36. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147-54 https://doi.org/10.1016/S1672-0229(08)60044-3
37. Zhuang Z, Li L, Yu Y, Su X, Lin S, Hu J. Targeting MicroRNA in myopia: Current insights. Experimental Eye Research. 2024;243:109905 https://doi.org/10.1016/j.exer.2024.109905
38. Li Q, Zheng Q, He J, Li L, Xie X, Liang H. Hsa-miR-142-3p reduces collagen I in human scleral fibroblasts by targeting TGF-β1 in high myopia. Exp Eye Res. 2022;219:109023 https://doi.org/10.1016/j.exer.2022.109023
39. Chen C-F, Hua K, Woung L-C, Lin C-H, Chen C-T, Hsu C-H. et al. Expression profiling of exosomal miRNAs derived from the aqueous humor of myopia patients. Tohoku J Exp Med. 2019;249:213-21 https://doi.org/10.1620/tjem.249.213
40. Zhu Y, Li W, Zhu D, Zhou J. microRNA profiling in the aqueous humor of highly myopic eyes using next generation sequencing. Exp Eye Res. 2020;195:108034 https://doi.org/10.1016/j.exer.2020.108034
41. Zhu Y, Zhang Y, Jiang R, Zhao K, Zhou J. MicroRNA-29a may influence myopia development by regulating collagen I. Current Eye Research. 2022;47:468-76 https://doi.org/10.1080/02713683.2021.1998542
42. Jiang B, Hong N, Guo D, Shen J, Qian X, Dong F. MiR-204-5p may regulate oxidative stress in myopia. Sci Rep. 2024;14:9770 https://doi.org/10.1038/s41598-024-60688-1
43. You J, Wu Q, Xu G, Gu C, Allen E, Zhu T. et al. Exosomal MicroRNA profiling in vitreous humor derived from pathological myopia patients. Invest Ophthalmol Vis Sci. 2023;64:9 https://doi.org/10.1167/iovs.64.1.9
44. Ando Y, Keino H, Inoue M, Hirota K, Takahashi H, Sano K. et al. Circulating vitreous microRNA as possible biomarker in high myopic eyes with macular hole. Int J Mol Sci. 2022;23:3647 https://doi.org/10.3390/ijms23073647
45. Kunceviciene E, Budiene B, Smalinskiene A, Vilkeviciute A, Liutkeviciene R. Association of hsa-mir-328-3p expression in whole blood with optical density of retinal pigment epithelial cells. In Vivo. 2021;35:827-31 https://doi.org/10.21873/invivo.12323
46. Kunceviciene E, Liutkeviciene R, Budiene B, Sriubiene M, Smalinskiene A. Independent association of whole blood miR-328 expression and polymorphism at 3′UTR of the PAX6 gene with myopia. Gene. 2019;687:151-5 https://doi.org/10.1016/j.gene.2018.11.030
47. Chen K-C, Hsi E, Hu C-Y, Chou W-W, Liang C-L, Juo S-HH. MicroRNA-328 may influence myopia development by mediating the PAX6 gene. Invest Ophthalmol Vis Sci. 2012;53:2732-9 https://doi.org/10.1167/iovs.11-9272
48. Liang C-L, Chen K-C, Hsi E, Lin J-Y, Chen C-Y, Tseng J-K. et al. miR-328-3p affects axial length via multiple routes and anti-miR-328-3p possesses a potential to control myopia progression. Invest Ophthalmol Vis Sci. 2022;63:11 https://doi.org/10.1167/iovs.63.12.11
49. Mei F, Wang J, Chen Z, Yuan Z. Potentially important MicroRNAs in form-deprivation myopia revealed by bioinformatics analysis of MicroRNA profiling. Ophthalmic Res. 2017;57:186-93 https://doi.org/10.1159/000452421
50. Tkatchenko AV, Luo X, Tkatchenko TV, Vaz C, Tanavde VM, Maurer-Stroh S. et al. Large-scale microRNA expression profiling identifies putative retinal miRNA-mRNA signaling pathways underlying form-deprivation myopia in mice. PLoS ONE. 2016;11:e0162541 https://doi.org/10.1371/journal.pone.0162541
51. Liu J, Bao B, Li T, Yang Z, Du Y, Zhang R. et al. miR-92b-3p protects retinal tissues against DNA damage and apoptosis by targeting BTG2 in experimental myopia. J Transl Med. 2024;22:511 https://doi.org/10.1186/s12967-024-05288-3
52. Jiang B, Hong N, Zhang L, Xu B, He Q, Qian X. et al. MiR-181a-5p may regulate cell proliferation and autophagy in myopia and the associated retinopathy. Experimental Eye Research. 2024;241:109829 https://doi.org/10.1016/j.exer.2024.109829
53. Cui Z, Huang Y, Chen X, Chen T, Hou X, Yu N. et al. Identification of miR-671-5p and its related pathways as general mechanisms of both form-deprivation and lens-induced myopia in mice. Curr Issues Mol Biol. 2023;45:2060-72 https://doi.org/10.3390/cimb45030132
54. Liu S, Chen H, Ma W, Zhong Y, Liang Y, Gu L. et al. Non-coding RNAs and related molecules associated with form-deprivation myopia in mice. J Cell Mol Med. 2022;26:186-94 https://doi.org/10.1111/jcmm.17071
55. Li T, Li X, Hao Y, Liu J, Bao B, Yang Z. et al. Inhibitory effect of miR-138-5p on choroidal fibrosis in lens-induced myopia guinea pigs via suppressing the HIF-1α signaling pathway. Biochem Pharmacol. 2023;211:115517 https://doi.org/10.1016/j.bcp.2023.115517
56. Liu L, Zhu D, Ding W, Zhang T, Ma X, Zou J. MiRNA-21-HIF-1α-VEGF axis is associated with myopic choroidal neovascularization in guinea pigs. Ophthalmic Res. 2022;65:493-505 https://doi.org/10.1159/000522511
57. Guo D, Ding M, Song X, Sun Y, Li G, Li Z. et al. Regulatory roles of differentially expressed MicroRNAs in metabolic processes in negative lens-induced myopia guinea pigs. BMC Genomics. 2020;21:13 https://doi.org/10.1186/s12864-020-6447-x
58. Zhang Y, Hu D-N, Zhu Y, Sun H, Gu P, Zhu D. et al. Regulation of matrix metalloproteinase-2 secretion from scleral fibroblasts and retinal pigment epithelial cells by miR-29a. Biomed Res Int. 2017;2017:2647879 https://doi.org/10.1155/2017/2647879
59. Yang Q, Lv S, Zhu H, Zhang L, Li H, Song S. A potential research target for scleral remodeling: Effect of MiR-29a on scleral fibroblasts. Ophthalmic Res. 2022;65:566-74 https://doi.org/10.1159/000525189
60. Tang X, Liu L, Liu S, Song S, Li H. MicroRNA-29a inhibits collagen expression and induces apoptosis in human fetal scleral fibroblasts by targeting the Hsp47/Smad3 signaling pathway. Exp Eye Res. 2022;225:109275 https://doi.org/10.1016/j.exer.2022.109275
61. Wang M, Yang Z-K, Liu H, Li R-Q, Liu Y, Zhong W-J. Genipin inhibits the scleral expression of miR-29 and MMP2 and promotes COL1A1 expression in myopic eyes of guinea pigs. Graefes Arch Clin Exp Ophthalmol. 2020;258:1031-8 https://doi.org/10.1007/s00417-020-04634-7
62. Xie M, Li Y, Wu J, Wu J. Genetic variants in MiR-29a associated with high myopia. Ophthalmic Genetics. 2016;37:456-8 https://doi.org/10.3109/13816810.2015.1101776
63. Jiang B, Huo Y, Gu Y, Wang J. The role of microRNAs in myopia. Graefes Arch Clin Exp Ophthalmol. 2017;255:7-13 https://doi.org/10.1007/s00417-016-3532-6
64. Metlapally R, Park HN, Chakraborty R, Wang KK, Tan CC, Light JG. et al. Genome-wide scleral micro- and messenger-RNA regulation during myopia development in the mouse. Invest Ophthalmol Vis Sci. 2016;57:6089-97 https://doi.org/10.1167/iovs.16-19563
65. Hsiao Y-T, Chang W-A, Kuo M-T, Lo J, Lin H-C, Yen M-C. et al. Systematic analysis of transcriptomic profile of the effects of low dose atropine treatment on scleral fibroblasts using next-generation sequencing and bioinformatics. Int J Med Sci. 2019;16:1652-67 https://doi.org/10.7150/ijms.38571
66. Zhang R, Wen Y, Liu J, Hao J, Peng Y, Zhang M. et al. The miR-15b-5p/miR-379-3p-FOXO axis regulates cell cycle and apoptosis in scleral remodeling during experimental myopia. J Transl Med. 2024;22:710 https://doi.org/10.1186/s12967-024-05523-x
67. Ren Y, Yang X, Luo Z, Wu J, Lin H. HIF-1α aggravates pathologic myopia through the miR-150-5p/LAMA4/p38 MAPK signaling axis. Mol Cell Biochem. 2022;477:1065-74 https://doi.org/10.1007/s11010-021-04305-z
68. Rada JA, Brenza HL. Increased Latent Gelatinase Activity in the Sclera of Visually Deprived Chicks n.d
69. Jiang B, Shi C-S. Dynamic changes of periostin and collagen I in the sclera during progressive myopia in guinea pigs. ABO. 2020 83. https://doi.org/10.5935/0004-2749.20200034
70. Tanaka Y, Kurihara T, Hagiwara Y, Ikeda S-I, Mori K, Jiang X. et al. Ocular-component-specific miRNA expression in a murine model of lens-induced myopia. Int J Mol Sci. 2019;20:3629 https://doi.org/10.3390/ijms20153629
71. Xiao H, Lin S, Jiang D, Lin Y, Liu L, Zhang Q. et al. Association of extracellular signal-regulated kinase genes with myopia: A longitudinal study of Chinese children. Front Genet. 2021;12:654869 https://doi.org/10.3389/fgene.2021.654869
72. Hong N, Jiang B, Gu L, Chen S-Y, Tong J-P. MicroRNA-expression profiling in myopia: A meta-analysis and systematic review. Ophthalmic Res. 2022;65:254-63 https://doi.org/10.1159/000521300
73. Lechner J, Bae HA, Guduric-Fuchs J, Rice A, Govindarajan G, Siddiqui S. et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci. 2013;54:5266-72 https://doi.org/10.1167/iovs.13-12035
74. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206 https://doi.org/10.1186/s13059-017-1348-2
75. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen L-L. et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24:430-47 https://doi.org/10.1038/s41580-022-00566-8
76. Wang X, Lin Q, Liu S, Li X, Kong X, Wang Y. et al. LncRNA-XR_002792574.1-mediated ceRNA network reveals potential biomarkers in myopia-induced retinal ganglion cell damage. J Transl Med. 2023;21:785 https://doi.org/10.1186/s12967-023-04662-x
77. Wu S, Hao J, Guo D, Ma Z, Wu Q, Zhang M. et al. Characterization of lncRNA and mRNA profiles in ciliary body in experimental myopia. Exp Eye Res. 2024;241:109849 https://doi.org/10.1016/j.exer.2024.109849
78. Li Y, Lu Y, Du K, Yin Y, Hu T, Fu Q. et al. RNA-sequencing analysis reveals the long noncoding RNA profile in the mouse myopic retina. Front Genet. 2022;13:1014031 https://doi.org/10.3389/fgene.2022.1014031
79. Geng C, Li Y, Guo F, Wang J, Yue Y, Zhou K. et al. RNA sequencing analysis of long non-coding RNA expression in ocular posterior poles of guinea pig myopia models. Mol Vis. 2020;26:117-34
80. Wang H, Li J, Wang S, Lu X, Zhang G, Zhuang Y. et al. Contribution of structural accessibility to the cooperative relationship of TF-lncRNA in myopia. Brief Bioinform. 2021;22:bbab082 https://doi.org/10.1093/bib/bbab082
81. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 2020;10:3503-17 https://doi.org/10.7150/thno.42174
82. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836 https://doi.org/10.15252/embj.2018100836
83. Zhang L, Yu X, Hong N, Xia Y, Zhang X, Wang L. et al. CircRNA expression profiles and regulatory networks in the vitreous humor of people with high myopia. Exp Eye Res. 2024;241:109827 https://doi.org/10.1016/j.exer.2024.109827
84. Ma S, Zhu X, Li D, Yang F, Meng J, Jiang Y. et al. The differential expression of circular RNAs and the role of circAFF1 in lens epithelial cells of high-myopic cataract. J Clin Med. 2023;12:813 https://doi.org/10.3390/jcm12030813
85. Lam JKW, Chow MYT, Zhang Y, Leung SWS. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252 https://doi.org/10.1038/mtna.2015.23
86. Park JH, Hong SW, Yun S, Lee D-K, Shin C. Effect of siRNA with an asymmetric RNA/dTdT overhang on RNA interference activity. Nucleic Acid Ther. 2014;24:364-71 https://doi.org/10.1089/nat.2014.0494
87. Dana H, Chalbatani GM, Mahmoodzadeh H, Karimloo R, Rezaiean O, Moradzadeh A. et al. Molecular mechanisms and biological functions of siRNA. Int J Biomed Sci. 2017;13:48-57
88. Barathi VA, Kwan JL, Tan QSW, Weon SR, Seet LF, Goh LK. et al. Muscarinic cholinergic receptor (M2) plays a crucial role in the development of myopia in mice. Dis Model Mech. 2013;6:1146-58 https://doi.org/10.1242/dmm.010967
89. Barathi VA, Ho CEH, Tong L. Molecular basis of transglutaminase-2 and muscarinic cholinergic receptors in experimental myopia: A target for myopia treatment. Biomolecules. 2023;13:1045 https://doi.org/10.3390/biom13071045
90. Zhao F, Chen W, Zhou H, Reinach PS, Wang Y, Juo S-HH. et al. PDE4B proposed as a high myopia susceptibility gene in Chinese population. Front Genet. 2021;12:775797 https://doi.org/10.3389/fgene.2021.775797
91. Yuan Y, Li M, To CH, Lam TC, Wang P, Yu Y. et al. The role of the RhoA/ROCK signaling pathway in mechanical strain-induced scleral myofibroblast differentiation. Invest Ophthalmol Vis Sci. 2018;59:3619-29 https://doi.org/10.1167/iovs.17-23580
92. Li X-J, Yang X-P, Wan G-M, Wang Y-Y, Zhang J-S. Effects of hepatocyte growth factor on MMP-2 expression in scleral fibroblasts from a guinea pig myopia model. Int J Ophthalmol. 2014;7:239-44 https://doi.org/10.3980/j.issn.2222-3959.2014.02.09
93. Fan Y, Li J, Huang L, Wang K, Zhao M. 7-methylxanthine influences the behavior of ADORA2A-DRD2 heterodimers in human retinal pigment epithelial cells. Ophthalmic Res. 2022;65:678-84 https://doi.org/10.1159/000525563
94. Tian F, Zhao J, Bu S, Teng H, Yang J, Zhang X. et al. KLF6 induces apoptosis in human lens epithelial cells through the ATF4-ATF3-CHOP axis. Drug Des Devel Ther. 2020;14:1041-55 https://doi.org/10.2147/DDDT.S218467
95. Denisenko O. Epigenetics of ribosomal RNA genes. Biochemistry Moscow. 2022;87:S103-10 https://doi.org/10.1134/S0006297922140097
96. Hori Y, Engel C, Kobayashi T. Regulation of ribosomal RNA gene copy number, transcription and nucleolus organization in eukaryotes. Nat Rev Mol Cell Biol. 2023;24:414-29 https://doi.org/10.1038/s41580-022-00573-9
97. Clarridge JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840-62 https://doi.org/10.1128/CMR.17.4.840-862.2004
98. Sun Y, Xie Y, Li J, Hou X, Sha Y, Bai S. et al. Study on the relationship between adolescent myopia and gut microbiota via 16S rRNA sequencing. Exp Eye Res. 2024;247:110067 https://doi.org/10.1016/j.exer.2024.110067
99. Li H, Liu S, Zhang K, Zhu X, Dai J, Lu Y. Gut microbiome and plasma metabolome alterations in myopic mice. Front Microbiol. 2023;14:1251243 https://doi.org/10.3389/fmicb.2023.1251243
100. Wu Y, Fan H, Feng Y, Yang J, Cen X, Li W. Unveiling the gut microbiota and metabolite profiles in guinea pigs with form deprivation myopia through 16S rRNA gene sequencing and untargeted metabolomics. Heliyon. 2024;10:e30491 https://doi.org/10.1016/j.heliyon.2024.e30491
101. Li H, Du Y, Cheng K, Chen Y, Wei L, Pei Y. et al. Gut microbiota-derived indole-3-acetic acid suppresses high myopia progression by promoting type I collagen synthesis. Cell Discov. 2024;10:89 https://doi.org/10.1038/s41421-024-00709-5
102. Xiao K, Chen Z, Long Q. Comparison of conjunctival sac microbiome between low and high myopic eyes. J Microbiol. 2023;61:571-8 https://doi.org/10.1007/s12275-023-00045-5
103. Leshinsky-Silver E, Ling J, Wu J, Vinkler C, Yosovich K, Bahar S. et al. Severe growth deficiency, microcephaly, intellectual disability, and characteristic facial features are due to a homozygous QARS mutation. Neurogenetics. 2017;18:141-6 https://doi.org/10.1007/s10048-017-0516-6
104. Smith MM. Histone structure and function. Current Opinion in Cell Biology. 1991;3:429-37 https://doi.org/10.1016/0955-0674(91)90070-F
105. Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends in Genetics. 2016;32:42-56 https://doi.org/10.1016/j.tig.2015.10.007
106. Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49:e324-e324 https://doi.org/10.1038/emm.2017.11
107. Yamagishi M, Kuze Y, Kobayashi S, Nakashima M, Morishima S, Kawamata T. et al. Mechanisms of action and resistance in histone methylation-targeted therapy. Nature. 2024;627:221-8 https://doi.org/10.1038/s41586-024-07103-x
108. Cui X, Du C, Wan S, Wu D, Yan L, Zhang J. et al. Deficiency of heat shock factor 4 promotes lens epithelial cell senescence through upregulating p21cip1 expression. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2021;1867:166233 https://doi.org/10.1016/j.bbadis.2021.166233
109. Rao RC, Tchedre KT, Malik MTA, Coleman N, Fang Y, Marquez VE. et al. Dynamic patterns of histone lysine methylation in the developing retina. Invest Ophthalmol Vis Sci. 2010;51:6784 https://doi.org/10.1167/iovs.09-4730
110. Watanabe S, Murakami A. Regulation of retinal development via the epigenetic modification of histone H3. In: Bowes Rickman C, LaVail MM, Anderson RE, Grimm C, Hollyfield J, Ash J, editors. Retinal Degenerative Diseases, vol. 854, Cham: Springer International Publishing. 2016 p. 635-41. https://doi.org/10.1007/978-3-319-17121-0_84
111. Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022;23:329-49 https://doi.org/10.1038/s41580-021-00441-y
112. Takata N, Miska JM, Morgan MA, Patel P, Billingham LK, Joshi N. et al. Lactate-dependent transcriptional regulation controls mammalian eye morphogenesis. Nat Commun. 2023;14:4129 https://doi.org/10.1038/s41467-023-39672-2
113. Park H-YL, Kim JH, Jung Y, Park CK. Racial differences in the extracellular matrix and histone acetylation of the lamina cribrosa and peripapillary sclera. Invest Ophthalmol Vis Sci. 2017;58:4143 https://doi.org/10.1167/iovs.17-21474
114. Liao X, Yap MKH, Leung KH, Kao PYP, Liu LQ, Yip SP. Genetic association study of KCNQ5 polymorphisms with high myopia. BioMed Research International. 2017;2017:1-7 https://doi.org/10.1155/2017/3024156
115. Liu S, Cao Y, Zhang Y. Regulatory roles of RNA methylation in vascular lesions in ocular and cardiopulmonary diseases. Critical Reviews in Clinical Laboratory Sciences. 2024;61:726-40 https://doi.org/10.1080/10408363.2024.2370267
116. Tang J, Zhou C, Ye F, Zuo S, Zhou M, Lu L. et al. RNA methylation homeostasis in ocular diseases: All eyes on Me. Progress in Retinal and Eye Research. 2025;105:101335 https://doi.org/10.1016/j.preteyeres.2025.101335
117. Wen K, Zhang Y, Li Y, Wang Q, Sun J. Comprehensive analysis of transcriptome-wide m6 A methylome in the anterior capsule of the lens of high myopia patients. Epigenetics. 2021;16:955-68 https://doi.org/10.1080/15592294.2020.1834917
118. Xue M, Li B, Lu Y, Zhang L, Yang B, Shi L. FOXM1 Participates in Scleral Remodeling in Myopia by Upregulating APOA1 Expression Through METTL3/YTHDF2. Invest Ophthalmol Vis Sci. 2024;65:19 https://doi.org/10.1167/iovs.65.1.19
119. Shan K, Zhou R, Xiang J, Sun Y, Liu C, Lv M. et al. FTO regulates ocular angiogenesis via m6A-YTHDF2-dependent mechanism. Experimental Eye Research. 2020;197:108107 https://doi.org/10.1016/j.exer.2020.108107
120. Liu C, Li M, Shen Y, Han X, Wei R, Wang Y. et al. Targeting choroidal vasculopathy via up-regulation of tRNA-derived fragment tRF-22 expression for controlling progression of myopia. J Transl Med. 2023;21:412 https://doi.org/10.1186/s12967-023-04274-5
Author contact
![]() Corresponding author: Dadong Guo, No. 48#, Yingxiongshan Road, Jinan 250002, P. R. China; Email: dadonggenecom.
Corresponding author: Dadong Guo, No. 48#, Yingxiongshan Road, Jinan 250002, P. R. China; Email: dadonggenecom.

 Global reach, higher impact
Global reach, higher impact