Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(12):3022-3031. doi:10.7150/ijms.113676 This issue Cite
Research Paper
NSUN2 rs13181449 variant decreases the risk of oral cancer development
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. Department of Radiation Oncology, Changhua Christian Hospital, Changhua, Taiwan.
3. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
4. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan.
5. Department of Otolaryngology, St. Martin De Porres Hospital, Chiayi, Taiwan.
6. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
7. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan.
8. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan.
9. Division of Hematology and Oncology, Department of Internal Medicine, Chung Shan Medical University Hospital, Taichung, Taiwan.
Received 2025-3-12; Accepted 2025-6-9; Published 2025-6-20
Abstract
NOP2/Sun RNA methyltransferase 2 (NSUN2), encoded by the NSUN2 gene, is a nuclear RNA methyltransferase that catalyzes the methylation of cytosine to 5-methylcytosine (m5C). Although RNA modification has been widely discussed in cancer development and prognosis, the role of the NSUN2 gene in oral cavity squamous cell carcinoma (OCSCC) is unclear. This was a retrospective, case-control study. A total of 2514 participants were enrolled, including 52.4% (1318/2514) diagnosed with OCSCC and others as health control. The impact of NSUN2 rs4702373, rs166049, rs13181449, and rs8192120 on cancer development and prognosis were analyzed. Our results revealed that NSUN2 rs13181449 allele TT was significantly associated with lower OCSCC risk, with an adjusted odd ratio (AOR) [95% confidence index (CI)] of 0.757[0.575-0.997]. For cigarette smokers, the impact of rs13181449 was more obvious that AOR [95% CI] of allele CT, TT, and CT+TT were 0.760 [0.583-0.991], 0.699 [0.493-0.990], 0.746 [0.580-0.960], respectively. For OCSCC patients, rs4702373 allele CT was independently associated with advanced histological grade. Expression levels between different allele mutations were various in the GTE database. Mutant rs4702373 and rs166049 were associated with higher expression than the wild type, conversely mutant rs13181449 had lower expression. In the TCGA database, the trend that patients with higher expression had worse survival than those with lower expression was shown. In conclusion, NSUN2 rs13181449 was associated with lower cancer risk, especially for cigarette smokers. Unlikely other allele mutations, rs13181449 was correlated to lower expression. Patients with higher expression had worsened clinical outcomes.
Keywords: NSUN2, oral cavity squamous cell carcinoma, single-nucleotide polymorphism, susceptibility, clinicopathologic progression
Introduction
Oral cavity squamous cell carcinoma (OCSCC) is one of the largest subpopulations of head and neck squamous cell carcinoma (HNSCC), the eighth most common cancer globally and the fourth most common cancer of males in Taiwan [1, 2]. Although previous studies reported personal health habits impacted the risk of OCSCC development [3], genetic risks of OCSCC development were rarely discussed [4-8]. Once the disease progresses to recurrent metastatic status, a poor prognostic situation [9, 10], genetic roles in prognosis also need to be clear. In the genomics era, genetic profiling associated with developmental risk and prognosis for OCSCC is significant.
Developing next-generation sequencing technologies improves genomic studies and treatment decisions in the modern era [11, 12]. From the aspect of prevention medicine, some genetic variants also have been reported as significantly associated with cancer development, such as BRCA1/BRCA2 mutations [13]. The roles of single nucleotide polymorphisms were also discussed in the relationships of cancer risks across various cancer types [14, 15]. Additionally, RNA modification is another critical issue in the occurrence and progression of cancer [16, 17]. The mutation and regulation of long-noncoding RNA have been proven major role in cancer development for various cancer types [18-21]. In previous study, the role of RNA modification, such as RNA methylation mediated by the NSUN2 gene, was discussed, especially in cancer development and prognosis [22].
NOP2/Sun RNA methyltransferase 2 (NSUN2), a nuclear RNA methyltransferase that catalyzes the methylation of cytosine to 5-methylcytosine (m5C) at position 34 of intron-containing tRNA precursors, is a protein encoded by the NSUN2 gene [23, 24]. The physiologic function of NSUN2 is to regulate RNA modification, which is necessary to stabilize the anticodon-codon pairing and correctly translate the mRNA [24]. NSUN2 directly plays an important role in tissue homeostasis, especially mitotic spindle stability. Other functions including cellular proliferation, migration, and differentiation were also presented [25]. For malignancies, NSUN2 was discovered as a target of MYC and the depletion of NSUN2 wound impaired MYC-dependent proliferation [26]. Prognostic roles of NSUN2 were also presented in various cancers, including gastric cancer, breast cancer, and others [27-29]. In addition to the studies discussing the function of NSUN2 in malignancies, NSUN2 polymorphisms have been reported to reduce cancer development in neuroblastoma [22]. However, the roles of NSUN2 polymorphisms as well as methyltransferase NSUN2 in OCSCC are limited.
To identify the impact of NSUN2 polymorphisms on cancer development and prognosis, our study retrospectively analyzed their roles between patients with OCSCC and healthy people. Published databases, such as the Genotype-Tissue Expression (GTEx) Portal and TCGA, were used to validate our results.
Materials and methods
Study subjects
This retrospective case-control study included patients diagnosed with OCSCC, selected from the BioBank database of Chung Shan Medical University Hospital. The control group comprised healthy individuals aged 30 to 70 years, with normal cognitive function and no history of cancer, selected from the Taiwan Biobank. For the case group, individuals without a pathological diagnosis or those with second primary malignancies were excluded. Female participants were excluded from both the case and control groups, as nearly 90% of OCSCC patients were male [30]. The study was approved by the Institutional Review Board of Chung Shan Medical University Hospital (CSMUH No: CS1-21151).
Clinical data collection
Basic characteristics, including age, cigarette smoking, alcohol drinking, and betel quid chewing, were collected from the BioBank databases. Pathologic factors of the case group were also supported in our previous studies [31, 32]. The American Joint Committee on Cancer's staging system (seventh edition) was used in this study [33]. However, due to delinking and anonymity, cancer-specific clinical outcomes of the case group were limited and difficult to available.
DNA extraction and genotyping
Genomic DNA was extracted from peripheral blood leukocytes by using QIAamp DNA blood mini kits (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Because NSUN2 polymorphisms, including rs4702373, rs166049, rs13181449, and rs8192120, were associated with cancer risk in previous studies [22], these polymorphisms were studied for OCSCC in our study. The results were further analyzed using SDS version 3.0. The details of DNA extraction and genotyping were published in our previous study [34].
Published databases for validation
Several published databases were used to verify our results, including the Genotype-Tissue Expression (GTEx) portal and cBioPortal. The GTEx portal is a comprehensive public resource to study human gene expression and regulation, and its relationship to genetic variation across multiple diverse tissues and individuals, including whole-genome sequencing (WGS), whole-exome sequencing, and RNA-seq (https://gtexportal.org/home/) [35]. The cBioPortal is an open-access, open-source resource for interactive exploration of multidimensional cancer genomics data sets, including the TCGA database (https://www.cbioportal.org/) [36].
Statistical analysis
Clinicopathological parameters were compared using the χ2 and Fisher exact tests. Independent risks of cancer development were analyzed by using univariate and multivariate logistic regressions with the odd ratios (ORs) [95% confidence index (CI)]. Additionally, the impact of NSUN2 polymorphisms in the OCSCC development was adjusted by age and personal health habits, with the so-called adjusted odd ratios (AOR). Significant association between genotypes and NSUN2 expression levels in the GTEx portal was detected with the linear regression model. Survival outcomes from the TCGA database were analyzed using Kaplan-Meier plots and the log-rank test. A two-sided P value of <0.05 was statistically significant. SPSS (version 21.0, SPSS Inc., Chicago, IL, USA) was used for all statistical analyses.
Results
Baseline characteristics
A total of 2514 participants were enrolled in the study, including 52.4% (1318/2514) in the case group and others (47.6%, 1196/2514) in the control group. The case group, diagnosed with OCSCC, were elderly, with more cigarette smoking, alcohol drinking, and betel quid chewing than the control group (all P < 0.001). Basic characteristics are shown in Table 1.
NSUN2 rs13181449 prevents the development of OCSCC
The distributions of all allele mutations were shown. Among all participants, the allele mutant frequencies of NSUN2 rs4702373, rs166049, rs13181449, and rs8192120 were 32.4% (815/2514), 35.6% (895/2514), 67.3% (1692/2514), and 50.9% (1280/2514), respectively. The distributions of the allele mutations between the case and control groups were similar, with P values of 0.312, 0.872, 0.348, and 0.360, respectively. To know the impacts of NSUN2 polymorphisms on the development of OCSCC, logistic regression analysis was done to identify the odd ratio (95% confidence index [CI]) of each allele. After adjusting to age, cigarette smoking, alcohol drinking, and betel quid chewing, the participants with rs13181449 allele TT significantly had lower OCSCC development than those without (AOR [95% CI], 0.757[0.575-0.997]) (Table 2).
For cigarette smokers, one of the most common subgroups representing 65.4% (1643/2514) of all participants, rs13181449 allele mutations were also significantly associated with low OCSCC development. AORs [95% CI] of rs13181449 allele CT, TT, and CT+TT were 0.760 [0.583-0.991], 0.699 [0.493-0.990], 0.746 [0.580-0.960], respectively. NSUN2 rs13181449 decreasing OCSCC risk was advanced for cigarette smokers (Table 2).
Basic characteristics of the patients with oral cancer and healthy controls
| Variable | Patients (N=1318) | Controls (N=1196) | P value |
|---|---|---|---|
| Age (yrs) | 0.002 | ||
| ≥ 55 | 771 (58.5%) | 630 (52.7%) | |
| <55 | 547 (41.5%) | 566 (47.3%) | |
| Cigarette smoking | <0.001 | ||
| Yes | 1008 (76.5%) | 635 (53.1%) | |
| No | 310 (23.5%) | 561 (46.9%) | |
| Alcohol drinking | <0.001 | ||
| Yes | 497 (37.7%) | 236 (19.7%) | |
| No | 821 (62.3%) | 960 (80.3%) | |
| Betel quid chewing | <0.001 | ||
| Yes | 878 (66.6%) | 198 (16.6%) | |
| No | 440 (33.4%) | 998 (83.4%) | |
| Pathologic staging | NA | ||
| I+II | 603 (45.8%) | ||
| III+IV | 715 (54.2%) | ||
| Pathologic T staging | NA | ||
| T1+T2 | 634 (48.1%) | ||
| T3+T4 | 684 (51.9%) | ||
| Pathologic N staging | NA | ||
| N0 | 900 (68.3%) | ||
| N+ | 418 (31.7%) | ||
| Pathologic M staging | NA | ||
| M0 | 1312 (99.5%) | ||
| M1 | 6 (0.5%) | ||
| Histological differentiation | NA | ||
| Well | 204 (15.5%) | ||
| Moderate to poor | 1114 (84.5%) |
NSUN2 mRNA expression among different tissues
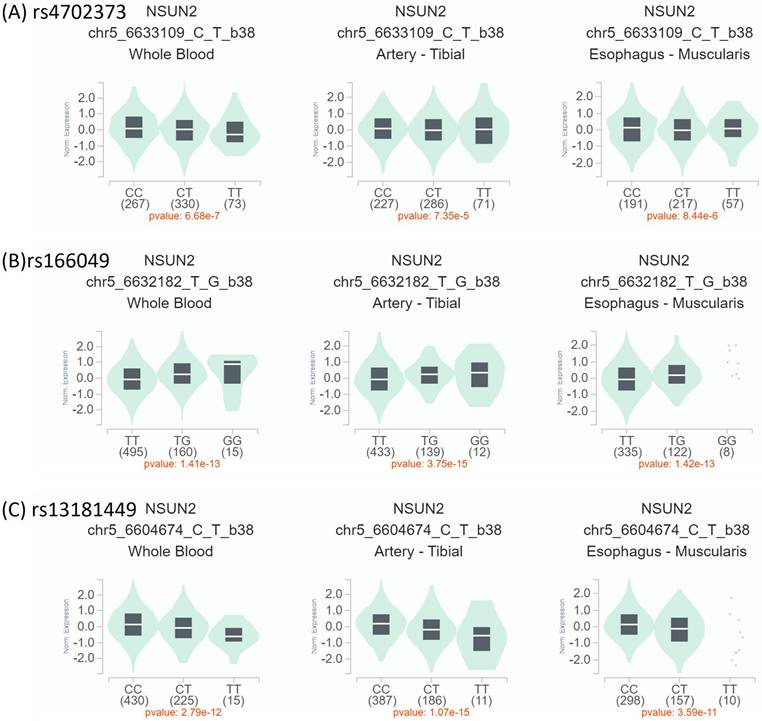
To verify our finding, the expressions of NSUN2 allele mutations among different tissues were compared based on the GTEx database. The participants with rs4702373 and rs166049 mutations were significantly associated with higher NSUN2 expression in whole blood, artery, and upper aerodigestive soft tissue (esophageal muscularis) than those without (all P values < 0.05). However, the participants with rs13181449 mutation had lower expression than others (P < 0.05) (Figure 1). It might indirectly support that the participants with lower NSUN2 expression levels were related to lower OCSCC development. Future warranted studies were needed.
Prognostic role of NSUN2 polymorphisms in OCSCC
The prognostic roles of NSUN2 allele mutations in OCSCC were also studied. Among the participants diagnosed with OCSCC, allele mutations of rs4702373, rs166049, rs13181449, and rs8192120 were 33.5% (442/1318), 35.9% (473/1318), 66.5% (877/1318), and 52.2% (668/1318), respectively. Basic characteristics between the OCSCC patients with and without allele mutations were insignificant differences, except for the patients with rs4702373 allele mutation who were associated with advanced histological grade than those without (P = 0.038) (Table 3). However, for the patients with cigarette smoking, the patients with rs4702373 mutation also had a trend of advanced histological grade than those without but insignificant in statistic (advanced histologic grade, patients with and without rs4702373 mutation, 85.5% vs. 81.3%, respectively, P = 0.058) (Table 4).
Independent roles of NSUN2 alleles for histologic grades were analyzed by using univariant and multivariate logistic regression. In Table 5, cigarette smoking, betel quid chewing, pathologic N staging, and rs4702373 allele CT were significantly associated with moderate-to-poor histological grade in univariant analysis. But only rs4702373 allele CT was independent impacting histological grade (OR [95% CI], 1.451[1.017-2.069]). Other allele mutations were not associated with the impact of histological grade formation in analyses.
NSUN2 expression by Genotype-Tissue Expression (GTEx) Portal (https://www.gtexportal.org/home/). The expression levels of individual NSUN2 polymorphisms. Expression quantitative trait loci (eQTL) violin plots between mutant- and wild-type polymorphisms were compared in whole blood, artery-tibial, and esophagus muscularis. (A, B) The participants with mutant rs4702373 and rs166049 were associated with higher expression than those with wild type. (C) Conversely, those with mutant rs13181449 had lower expression than others with wild type.
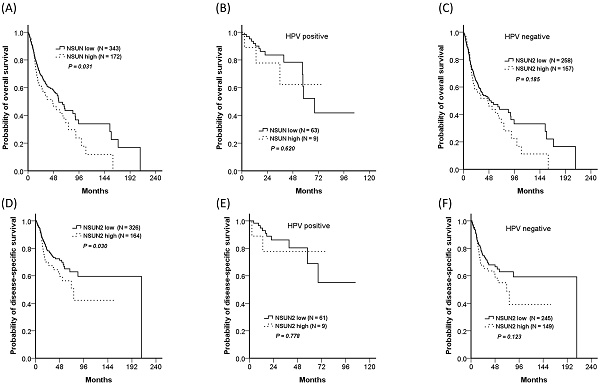
Relationship between NSUN2 expression and clinical outcomes
In addition, the TCGA database was also used for validation. Because nearly one-third of OCSCC patients in our cohort had rs4702373 allele mutation and the mutant allele might be associated with higher NSUN2 expression, a total of 515 HNSCC patients from the TCGA database were divided into the two groups of high- (33.4%, 172/515) and low- (66.6%, 343/515) NSUN2 expression. The high-NSUN2 group significantly worsened in overall survival and disease-free survival, with P values of 0.031 and 0.030, respectively. The impact of NSUN2 expression was also presented in both patients with and without human papillomavirus (HPV) associated. Although there were insignificant differences in overall survival and disease-free survival between the patients with high- and low- NSUN2 expression, either with or without HPV associated, the trend showed that the patients with high expression seemed worse survivals than those with low expression.
Odds ratios (OR) and 95% confidence interval (CI) of oral cancer associated with NSUN2 genotypic frequencies.
| Variable | Patients (N, %) | Controls (N, %) | OR (95% CI) | AOR (95% CI) a | |
|---|---|---|---|---|---|
| All participants | |||||
| N = 1318 | N = 1196 | P value | |||
| rs4702373 | 0.312 | ||||
| CC | 876(66.5%) | 823(68.8%) | 1 | 1 | |
| CT | 391(29.7%) | 337(28.2%) | 1.090(0.916-1.297) | 1.111(0.907-1.361) | |
| TT | 51(3.9%) | 36(3.0%) | 1.331(0.860-2.061) | 1.359(0.825-2.238) | |
| CT+TT | 442(33.5%) | 373(31.2%) | 1.113(0.942-1.316) | 1.136(0.935-1.381) | |
| rs166049 | 0.872 | ||||
| TT | 845(64.1%) | 774(64.7%) | 1 | 1 | |
| TG | 419(31.8%) | 370(30.9%) | 1.037(0.875-1.230) | 1.041(0.854-1.269) | |
| GG | 54(4.1%) | 52(4.3%) | 0.951(0.642-1.409) | 0.922(0.579-1.467) | |
| TG+GG | 473(35.9%) | 422(35.3%) | 1.027(0.872-1.209) | 1.027(0.849-1.242) | |
| rs13181449 | 0.348 | ||||
| CC | 441(33.5%) | 381(31.9%) | 1 | 1 | |
| CT | 659(50.0%) | 592(49.5%) | 0.962(0.806-1.147) | 0.908(0.740-1.115) | |
| TT | 218(16.5%) | 223(18.6%) | 0.845(0.670-1.065) | 0.757(0.575-0.997) | |
| CT+TT | 877(66.5%) | 815(68.1%) | 0.930(0.787-1.099) | 0.870(0.716-1.057) | |
| rs8192120 | 0.360 | ||||
| CC | 630(47.8%) | 604(50.5%) | 1 | 1 | |
| CA | 584(44.3%) | 497(41.6%) | 1.127(0.957-1.327) | 1.153(0.953-1.395) | |
| AA | 104(7.9%) | 95(7.9%) | 1.050(0.778-1.416) | 1.047(0.736-1.489) | |
| CA+AA | 688(52.2%) | 592(49.5%) | 1.114(0.953-1.303) | 1.135(0.946-1.362) | |
| Cigarette smoking | |||||
| N = 1008 | N = 635 | ||||
| rs4702373 | 0.448 | ||||
| CC | 664(65.9%) | 437(68.8%) | 1 | 1 | |
| CT | 303(30.1%) | 176(27.7%) | 1.133(0.908-1.414) | 1.138(0.878-1.474) | |
| TT | 41(4.1%) | 22(3.5%) | 1.227(0.721-2.088) | 1.295(0.705-2.380) | |
| CT+TT | 344 (34.1%) | 198 (31.2%) | 1.143(0.925-1.414) | 1.157(0.902-1.483) | |
| rs166049 | 0.760 | ||||
| TT | 647(64.2%) | 406(63.9%) | 1 | 1 | |
| TG | 318(31.5%) | 197(31.0%) | 1.013(0.816-1.258) | 0.984(0.764-1.267) | |
| GG | 43(4.3%) | 32(5.0%) | 0.843(0.525-1.355) | 0.861(0.490-1.512) | |
| TG+GG | 361 (35.8%) | 229 (36.1%) | 0.989(0.804-1.217) | 0.965(0.758-1.230) | |
| rs13181449 | 0.154 | ||||
| CC | 341(33.8%) | 186(29.3%) | 1 | 1 | |
| CT | 499(49.5%) | 333(52.4%) | 0.817(0.652-1.025) | 0.760(0.583-0.991) | |
| TT | 168(16.7%) | 116(18.3%) | 0.790(0.587-1.063) | 0.699(0.493-0.990) | |
| CT+TT | 667 (66.2%) | 449 (70.7%) | 0.810(0.654-1.005) | 0.746(0.580-0.960) | |
| rs8192120 | 0.960 | ||||
| CC | 486(48.2%) | 310(48.8%) | 1 | 1 | |
| CA | 438(43.5%) | 274(43.1%) | 1.020(0.829-1.255) | 1.041(0.817-1.328) | |
| AA | 84(8.3%) | 51(8.0%) | 1.051(0.721-1.530) | 1.098(0.707-1.704) | |
| CA+AA | 522 (51.8%) | 325 (51.2%) | 1.025(0.840-1.250) | 1.050(0.832-1.324) | |
a Adjusted for the effects of age, cigarette smoking, alcohol drinking, and betel quid chewing
The distributions of demographical characteristics of NSUN2 allele mutation in all OCSCC patients (N = 1318)
| rs4702373 | rs166049 | rs13181449 | rs8192120 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | CT+TT N = 442 (%) | CC N = 876 (%) | P value | TG+GG N = 476 (%) | TT N = 845 (%) | P value | CT+TT N = 887 (%) | CC N = 441 (%) | P value | CA+AA N = 688 (%) | CC N = 630 (%) | P value |
| Age >= 55 | 260 (58.8) | 551 (58.3) | 0.456 | 286(60.5) | 485(57.4) | 0.152 | 510(58.2) | 261(59.2) | 0.383 | 403(58.6) | 368(58.4) | 0.498 |
| Personal history | ||||||||||||
| Cigarette smoking | 344(77.8) | 664(75.8) | 0.227 | 361(76.3) | 647(76.6) | 0.485 | 667(76.1) | 341(77.3) | 0.330 | 522(75.9) | 486(77.1) | 0.316 |
| Alcohol drinking | 174(39.4) | 323(36.9) | 0.205 | 183(38.7) | 314(37.2) | 0.312 | 335(38.2) | 162(36.7) | 0.324 | 250(36.3) | 247(39.2) | 0.155 |
| Betel quid chewing | 299(67.6) | 579(66.1) | 0.308 | 308(65.1) | 570(67.5) | 0.211 | 587(66.9) | 291(66.0) | 0.388 | 454(66.0) | 424(67.3) | 0.328 |
| Pathologic staging | 0.513 | 0.450 | 0.396 | 0.057 | ||||||||
| Stage I+II | 202(45.7) | 401(45.8) | 218(46.1) | 385(45.6) | 404(46.1) | 199(45.1) | 300(43.6) | 303(48.1) | ||||
| Stage III+IV | 240(54.3) | 475(54.2) | 255(53.9) | 460(54.4) | 473(53.9) | 242(54.9) | 388(56.4) | 327(51.9) | ||||
| Pathologic T staging | 0.325 | 0.284 | 0.347 | 0.124 | ||||||||
| T1/2 | 217(49.1) | 417(47.6) | 233(49.3) | 401(47.5) | 418(47.7) | 216(49.0) | 320(46.5) | 314(49.8) | ||||
| T3/4 | 225(50.9) | 459(52.4) | 240(50.7) | 444(52.5) | 459(52.3) | 225(51.0) | 368(53.5) | 316(50.2) | ||||
| Pathologic N staging | 0.293 | 0.379 | 0.323 | 0.163 | ||||||||
| N0 | 297(67.2) | 603(68.8) | 326(68.9) | 574(67.9) | 603(68.8) | 297(67.3) | 461(67.0) | 439(69.7) | ||||
| N+ | 145(32.8) | 273(31.2) | 147(31.1) | 271(32.1) | 274(31.2) | 144(32.7) | 227(33.0) | 191(30.3) | ||||
| Metastasis | 0.677 | 0.371 | 0.678 | 0.614 | ||||||||
| M0 | 440(99.5) | 872(99.5) | 470(99.4) | 842(99.6) | 873(99.5) | 439(99.5) | 685(99.6) | 627(99.5) | ||||
| M1 | 2(0.5) | 4(0.5) | 3(0.6) | 3(0.4) | 4(0.5) | 2(0.5) | 3(0.4) | 3(0.5) | ||||
| Cell differentiation grade | 0.038 | 0.247 | 0.418 | 0.271 | ||||||||
| Well | 57(12.9) | 147(16.8) | 78(16.5) | 126(14.9) | 134(15.3) | 70(15.9) | 102(14.8) | 102(16.2) | ||||
| Moderate or poor | 385(87.1) | 729(83.2) | 395(83.5) | 719(85.1) | 743(84.7) | 371(84.1) | 586(85.2) | 528(83.8) | ||||
The distributions of demographical characteristics of NSUN2 allele mutation in OCSCC patients with Cigarette smoking (N = 1008)
| rs4702373 | rs166049 | rs13181449 | rs8192120 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | CT+TT N = 344 (%) | CC N = 664 (%) | P value | TG+GG N = 361 (%) | TT N = 647 (%) | P value | CT+TT N = 667 (%) | CC N = 341 (%) | P value | CA+AA N = 522 (%) | CC N = 486 (%) | P value |
| Age >= 55 | 191(55.5) | 370(55.7) | 0.502 | 212(58.7) | 349(53.9) | 0.081 | 368(55.2) | 193(56.6) | 0.358 | 290(55.6) | 27(55.8) | 0.499 |
| Personal history | ||||||||||||
| Cigarette smoking | NA | NA | NA | NA | NA | NA | NA | NA | ||||
| Alcohol drinking | 160(46.5) | 293(44.1) | 0.256 | 170(47.1) | 283(43.7) | 0.169 | 303(45.4) | 150(44.0) | 0.357 | 223(42.7) | 230(47.3) | 0.080 |
| Betel quid chewing | 283(82.3) | 531(80.0) | 0.214 | 286(79.2) | 528(81.6) | 0.201 | 544(81.6) | 270(79.2) | 0.205 | 421(80.7) | 393(80.9) | 0.498 |
| Pathologic staging | 0.288 | 0.256 | 0.405 | 0.035 | ||||||||
| Stage I+II | 154(44.8) | 311(46.8) | 172(47.6) | 293(45.3) | 310(46.5) | 155(45.5) | 226(43.3) | 239(49.2) | ||||
| Stage III+IV | 190(55.2) | 353(53.2) | 189(52.4) | 354(54.7) | 357(53.5) | 186(54.5) | 296(56.7) | 247(50.8) | ||||
| Pathologic T staging | 0.419 | 0.389 | 0.397 | 0.145 | ||||||||
| T1/2 | 172(50.0) | 326(49.1) | 181(50.1) | 317(49.0) | 332(49.8) | 166(48.7) | 249(47.7) | 249(51.2) | ||||
| T3/4 | 172(50.0) | 338(50.9) | 180(49.9) | 330(51.0) | 335(50.2) | 175(51.3) | 273(52.3) | 237(48.8) | ||||
| Pathologic N staging | 0.407 | 0.350 | 0.409 | 0.130 | ||||||||
| N0 | 233(67.7) | 456(68.7) | 250(69.3) | 439(67.9) | 458(68.7) | 231(67.7) | 348(66.7) | 341(70.2) | ||||
| N+ | 111(31.6) | 208(31.3) | 111(30.7) | 208(32.1) | 209(31.3) | 110(32.3) | 174(33.3) | 145(29.8) | ||||
| Metastasis | 0.445 | 0.588 | 0.451 | 0.466 | ||||||||
| M0 | 343(99.7) | 660(99.4) | 359(99.4) | 644(99.5) | 663(99.4) | 340(99.7) | 520(99.6) | 483(99.4) | ||||
| M1 | 1(0.3) | 4(0.6) | 2(0.6) | 3(0.5) | 4(0.6) | 1(0.3) | 2(0.4) | 3(0.6) | ||||
| Cell differentiated grade | 0.058 | 0.183 | 0.523 | 0.274 | ||||||||
| Well | 50(14.5) | 124(18.7) | 68(18.8) | 106(16.4) | 115(17.2) | 59(17.3) | 86(16.5) | 88(18.1) | ||||
| Moderate or poor | 294(85.5) | 540(81.3) | 293(81.2) | 541(83.6) | 554(82.8) | 282(82.7) | 436(83.5) | 398(81.9) |
Overall survivals of the high- and low-NSUN2 HNSCC patients from the TCGA database. The trend showed the high NSUN2 group had worsened survival than the low NSUN2 group. (A) For all patients, 5-year overall survivals were 44.0% and 49.5% for the high- and low-NSUN2 groups, respectively (P = 0.031), (B) 62.2% and 52.2% for HPV positive population (P = 0.620), (C) 43.6% and 47.3% for HPV negative population (P = 0.185). (D) For all patients, 5-year disease-specific survivals were 56.3% and 65.1% (P = 0.030), (E) 77.8% and 68.9% for HPV positive population (P = 0.778), (F) 55.0%% and 62.9% for HPV negative population (P = 0.123).
Univariate and multivariate logistic regression for moderate to poor histologic differentiation in all oral cancer patients
| All patients | ||
|---|---|---|
| Univariate | Multivariate | |
| Variable | OR (95% CI), P value | OR (95% CI), P value |
| Age (yrs) | ||
| ≥ 55 vs. <55 | 0.872(0.643-1.184), 0.382 | |
| Personal history | ||
| Cigarette smoking (yes vs. no) | 0.514(0.341-0.774), 0.001 | 0.665(0.411-1.078) |
| Alcohol drinking (yes vs. no) | 0.862(0.636-1.169), 0.340 | |
| Betel quid chewing (yes vs. no) | 0.567(0.401-0.801), 0.001 | 0.682(0.451-1.032) |
| Pathologic T staging | ||
| pT3/4 vs. pT1/2 | 1.146(0.850-1.545), 0.371 | |
| Pathologic N staging | ||
| pN+ vs. pN0 | 2.737(1.848-4.052), <0.001 | 2.594(1.744-3.857), <0.001 |
| Metastasis | ||
| M1 vs. M0 | 0.364(0.066-2.000), 0.245 | |
| rs4702373 | ||
| CC | Reference | Reference |
| CT | 1.441(1.015-2.045), 0.041 | 1.451(1.017-2.069), 0.040 |
| TT | 0.941(0.488-1.975), 0.872 | |
| CT+TT | 1.362(0.979-1.894), 0.066 | |
| rs166049 | ||
| TT | Reference | |
| TG | 0.831(0.606-1.139), 0.831 | |
| GG | 1.717(0.671-4.394), 0.259 | |
| TG+GG | 0.887(0.652-1.207), 0.447 | |
| rs13181449 | ||
| CC | Reference | |
| CT | 1.134(0.810-1.587), 0.463 | |
| TT | 0.840(0.547-1.288), 0.423 | |
| CT+TT | 1.046(0.764-1.433), 0.779 | |
| rs8192120 | ||
| CC | Reference | |
| CA | 1.183(0.863-1.621), 0.297 | |
| AA | 0.811(0.477-1.381), 0.441 | |
| CA+AA | 1.110(0.823-1.496), 0.494 |
Discussion
A total of 2514 participants were retrospectively enrolled in our study, with 52.4% belonging to the case group diagnosed with OCSCC and others as healthy control. NSUN2 rs13181449 allele TT was significantly associated with a lower OCSCC development. For cigarette smokers, the effect of NSUN2 rs13181449 decreasing the risk of OCSCC development was more obvious. In the prognostic role, the NSUN2 rs4702373 CT allele was an independent factor associated with the formation of advanced histological grade for the patients diagnosed with OCSCC. The published database validated that except for rs13181449 mutations associated with lower expression levels, the participants with NSUN2 polymorphism mutations significantly had higher expression than those without. For both subgroups with or without HPV associated, the patients with higher expression levels had worse clinical outcomes than those with lower expression. Future studies were needed.
There were several advantages in our study as follows. First, this was a large real-world population, with a total of 2514 participants, and 52.4% of them diagnosed with OCSCC. It was representative to discuss the role of NSUN2 in OCSCC. In addition, although genomic studies were important in the modern era, genetic roles in decreasing the risk of malignancies were limited [13]. In our study, NSUN2 rs13181449 independently reduced the risk of OCSCC development, especially for the subgroup with cigarette smoking. Third, NSUN2 expressions were various in several cancer types. In OCSCC, the patients with higher NSUN2 expression were associated with worse prognosis than those without. However, different allele mutations might be associated with opposite gene expressions. Future studies to validate this issue should be careful.
DNA methylation has been reported as an important regulator of gene transcription, and its carcinogenetic role is also an interesting issue in the last few years. Alterations in DNA methylation are common in a variety of tumors as well as in development [37]. Recently, the roles of RNA methylation have been discussed widely [38, 39]. The Biological significance of NSUN2 upregulating cell proliferation and metastasis was reported, and the TCGA database also showed that in tumor specimens, NSUN2 expressions are higher than those in normal tissue across several cancer types [25, 40, 41]. For HNSCC, Lu et al. study presented that NSUN2 expression was 1.99-fold upregulation versus normal tissue. The patients associated with higher NSUN2 were correlated with worse survival than those with lower [42]. NSUN2 expression was also reported as a potential biomarker that immune checkpoint therapy might benefit patients with higher expression and less effective for those with lower expression [43]. Our study reported that for OCSCC, patients with higher NSUN2 expression were associated with worsened survival. In addition, NSUN2 rs4702373 allele mutation, related to higher expression, was independent of advanced histological grade. This finding was correlative to previous studies. However, the expression levels might vary between different alleles. Compared with normal tissue, the expression level of the rs13181449 mutation was lower, and conversely, the expression level of the rs4702373 and rs166049 mutations was higher. This point needed to be advanced validated.
Although the mechanism that rs13181449 associated with lower expression needed to be studied, patients with rs13181449 mutation seemed to have a lower risk of cancer development. In Lin et al. study, NSUN2 rs13181449 was significantly associated with lower neuroblastoma risk (CT/TT vs. CC: AOR [95% CI]: 0.69 [0.50-0.97]) [22]. In our cohort, NSUN2 rs13181449 also reduced the development of OCSCC, especially for cigarette smokers. Although detailed mechanisms between NSUN2 mutation, cigarette smokers, and cancer development are unknown, it might be because cigarette smoking might increase HIF-1-alpha levels, which mediated RNA modification through complex regulation, such as cell cycle, PI3K-AKT pathway, and others [44-46]. Therefore, NSUN2 rs13181449-associated lower expression might decrease the activation of the cell cycle and PI3K-AKT pathway, protecting cancer development more obviously for patients with cigarette smoking. Advanced experiment studies are needed.
Several limitations were presented in this study. Although this was a large cohort study, with more than two thousand five hundred participants enrolled, functional experiments in vitro and in vivo were needed to validate. In addition, the anonymous and delinked policy due to ethical issues caused detailed clinicopathological information not available. This information was also important for validation. Third, expression levels of different NSUN2 polymorphisms were various in the published database. Detailed mechanisms were suggested to support our findings, especially for the relationship between NSUN2 rs13181449, cigarette smokers, and cancer development.
In conclusion, the roles of NSUN2 polymorphisms in OCSCC development and prognosis were rarely discussed. In our study, NSUN2 rs13181449 was independent in decreasing the risk of OCSCC, especially for cigarette smokers. However, based on the published database, expression levels of different polymorphisms might be various. In the prognostic role, the patients with higher expression were associated with advanced histological grade and worsened survival. In vitro and in vivo advanced functional studies were needed to validate the results.
Funding
This study was supported by Chung Shan Medical University Hospital (CSH-2023-C-006).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Su SC, Chang LC, Huang HD, Peng CY, Chuang CY, Chen YT, Lu MY, Chiu YW, Chen PY, Yang SF. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. 2021;42:127-35
2. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH, Lee YC, Yang SF. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. J Pineal Res. 2021;71:e12760
3. Lee YA, Li S, Chen Y, Li Q, Chen CJ, Hsu WL, Lou PJ, Zhu C, Pan J, Shen H, Ma H, Cai L, He B, Wang Y, Zhou X, Ji Q, Zhou B, Wu W, Ma J, Boffetta P, Zhang ZF, Dai M, Hashibe M. Tobacco smoking, alcohol drinking, betel quid chewing, and the risk of head and neck cancer in an East Asian population. Head Neck. 2019;41:92-102
4. da Silva AM, Falcão MML, Freitas VS, Vieira AR. Genetic and environmental contributions for the relationship between tooth loss and oral potentially malignant disorders and oral squamous cell carcinoma. Head Neck. 2024;46:1417-27
5. da Silva AM, Freitas VS, Vieira AR. Polymorphisms associated with oral clefts as potential markers for oral pre and malignant disorders. Oral Dis. 2024;30:2985-90
6. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY, Hsieh MJ, Yang SF. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-52
7. Yeh JC, Chen YT, Chou YE, Su SC, Chang LC, Chen YL, Lin CW, Yang SF. Interactive effects of CDKN2B-AS1 gene polymorphism and habitual risk factors on oral cancer. J Cell Mol Med. 2023;27:3395-403
8. Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP, Yang WE, Su CW, Chuang CY, Li WH, Chung WH, Yang SF. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7:1088-99
9. Burtness B, Harrington KJ, Greil R, Soulieres D, Tahara M, de Castro G Jr, Psyrri A, Baste N, Neupane P, Bratland A, Fuereder T, Hughes BGM, Mesia R, Ngamphaiboon N, Rordorf T, Wan Ishak WZ, Hong RL, Gonzalez Mendoza R, Roy A, Zhang Y, Gumuscu B, Cheng JD, Jin F, Rischin D, Investigators K-. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915-28
10. Su CW, Chang YC, Chien MH, Hsieh YH, Chen MK, Lin CW, Yang SF. Loss of TIMP3 by promoter methylation of Sp1 binding site promotes oral cancer metastasis. Cell Death Dis. 2019;10:793
11. Colomer R, Mondejar R, Romero-Laorden N, Alfranca A, Sanchez-Madrid F, Quintela-Fandino M. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine. 2020;25:100487
12. Lu HJ, Su CW, Su SC, Chang LC, Wu MF, Lin CW, Yang SF. Prognostic impact of caspase-8 mutation in oral cavity squamous cell carcinoma. Oral Dis. 2025;31:769-81
13. Momozawa Y, Sasai R, Usui Y, Shiraishi K, Iwasaki Y, Taniyama Y, Parsons MT, Mizukami K, Sekine Y, Hirata M, Kamatani Y, Endo M, Inai C, Takata S, Ito H, Kohno T, Matsuda K, Nakamura S, Sugano K, Yoshida T, Nakagawa H, Matsuo K, Murakami Y, Spurdle AB, Kubo M. Expansion of Cancer Risk Profile for BRCA1 and BRCA2 Pathogenic Variants. JAMA Oncol. 2022;8:871-8
14. van Veen EM, Brentnall AR, Byers H, Harkness EF, Astley SM, Sampson S, Howell A, Newman WG, Cuzick J, Evans DGR. Use of Single-Nucleotide Polymorphisms and Mammographic Density Plus Classic Risk Factors for Breast Cancer Risk Prediction. JAMA Oncol. 2018;4:476-82
15. Chen PJ, Lu YT, Yang WE, Su CW, Chang LC, Yang SF, Lin CW, Chou YE. The Impact of MET Variants in Oral Cancer Progression and Clinicopathological Characteristics. J Cancer. 2025;16:1747-53
16. Yang L, Tang L, Min Q, Tian H, Li L, Zhao Y, Wu X, Li M, Du F, Chen Y, Li W, Li X, Chen M, Gu L, Sun Y, Xiao Z, Shen J. Emerging role of RNA modification and long noncoding RNA interaction in cancer. Cancer Gene Ther. 2024;31:816-30
17. Lin CW, Yang WE, Su CW, Lu HJ, Su SC, Yang SF. IGF2BP2 promotes cell invasion and epithelial-mesenchymal transition through Src-mediated upregulation of EREG in oral cancer. Int J Biol Sci. 2024;20:818-30
18. Su SC, Lin CW, Ju PC, Chang LC, Chuang CY, Liu YF, Hsieh MJ, Yang SF. Association of LINC00673 Genetic Variants with Progression of Oral Cancer. J Pers Med. 2021;11:468
19. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM, Yang SF. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J Dent Res. 2018;97:717-24
20. Tung MC, Lin CY, Wen YC, Chang LC, Yang SF, Chien MH. Associations of the Expression Levels and Risk Variants of CDKN2B-AS1 Long Noncoding RNA With the Susceptibility and Progression of Prostate Cancer. J Cell Mol Med. 2024;28:e70264
21. Lin CW, Lu JW, Chuang CY, Hsieh WY, Tsai YJ, Yang SF, Lin SH. Clinical significance of long non-coding RNA MIR155HG genetic variants and susceptibility to oral cancer. Sci Rep. 2025;15:9956
22. Lin L, Deng C, Zhou C, Zhang X, Zhu J, Liu J, Wu H, He J. NSUN2 gene rs13181449 C>T polymorphism reduces neuroblastoma risk. Gene. 2023;854:147120
23. Wang H, Feng J, Zeng C, Liu J, Fu Z, Wang D, Wang Y, Zhang L, Li J, Jiang A, He M, Cao Y, Yan K, Tang H, Guo D, Xu K, Zhou X, Zhou L, Lan K, Zhou Y, Chen Y. NSUN2-mediated M(5)c methylation of IRF3 mRNA negatively regulates type I interferon responses during various viral infections. Emerg Microbes Infect. 2023;12:2178238
24. Shinoda S, Kitagawa S, Nakagawa S, Wei FY, Tomizawa K, Araki K, Araki M, Suzuki T, Suzuki T. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8734-45
25. Chellamuthu A, Gray SG. The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer. Cells. 2020;9:1758
26. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303-22
27. Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, Shi LH, Zhao GH, Wang MH, Wu LS, Wei JF. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020;11:270
28. Zhang X, An K, Ge X, Sun Y, Wei J, Ren W, Wang H, Wang Y, Du Y, He L, Li O, Zhou S, Shi Y, Ren T, Yang YG, Kan Q, Tian X. NSUN2/YBX1 promotes the progression of breast cancer by enhancing HGH1 mRNA stability through m(5)C methylation. Breast Cancer Res. 2024;26:94
29. Zheng L, Li M, Wei J, Chen S, Xue C, Duan Y, Tang F, Li G, Xiong W, She K, Deng H, Zhou M. NOP2/Sun RNA methyltransferase 2 is a potential pan-cancer prognostic biomarker and is related to immunity. PLoS One. 2023;18:e0292212
30. Lin NC, Hsu JT, Tsai KY. Difference between Female and Male Patients with Oral Squamous Cell Carcinoma: A Single-Center Retrospective Study in Taiwan. Int J Environ Res Public Health. 2020;17:3978
31. Lu HJ, Chuang CY, Su CW, Chen MK, Yang WE, Yeh CM, Tang CH, Lin CW, Yang SF. Role of TNFSF15 variants in oral cancer development and clinicopathologic characteristics. J Cell Mol Med. 2022;26:5452-62
32. Lu HJ, Chuang CY, Chen MK, Su CW, Yang WE, Yeh CM, Lai KM, Tang CH, Lin CW, Yang SF. The impact of ALDH7A1 variants in oral cancer development and prognosis. Aging (Albany NY). 2022;14:4556-71
33. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-4
34. Chou CH, Chang CY, Lu HJ, Hsin MC, Chen MK, Huang HC, Yeh CM, Lin CW, Yang SF. IGF2BP2 Polymorphisms Are Associated with Clinical Characteristics and Development of Oral Cancer. Int J Mol Sci. 2020;21:5662
35. Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank. 2015;13:307-8
36. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
37. Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632-42
38. Jo H, Shim K, Jeoung D. Roles of RNA Methylations in Cancer Progression, Autophagy, and Anticancer Drug Resistance. Int J Mol Sci. 2023;24:4225
39. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613
40. Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, Sun X, Chen Z, Shi X, Hu Y, Shen X, Xue X, Lu M. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842
41. Tong X, Xiang Y, Hu Y, Hu Y, Li H, Wang H, Zhao KN, Xue X, Zhu S. NSUN2 Promotes Tumor Progression and Regulates Immune Infiltration in Nasopharyngeal Carcinoma. Front Oncol. 2022;12:788801
42. Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA Transferase NSUN2 Gene Expression is Associated with Poor Prognosis in Head and Neck Squamous Carcinoma. Cancer Invest. 2018;36:246-53
43. Lu L, Gaffney SG, Cannataro VL, Townsend J. Transfer RNA methyltransferase gene NSUN2 mRNA expression modifies the effect of T cell activation score on patient survival in head and neck squamous carcinoma. Oral Oncol. 2020;101:104554
44. Yang Y, Cheng C, He B, Du X, Liu J, Xia H, Wang P, Wu M, Wu H, Liu Q. Cigarette smoking, by accelerating the cell cycle, promotes the progression of non-small cell lung cancer through an HIF-1alpha-METTL3-m(6)A/CDK2AP2 axis. J Hazard Mater. 2023;455:131556
45. Du X, Cheng C, Yang Y, Fan B, Wang P, Xia H, Ni X, Liu Q, Lu L, Wei L. NSUN2 promotes lung adenocarcinoma progression through stabilizing PIK3R2 mRNA in an m(5)C-dependent manner. Mol Carcinog. 2024;63:962-76
46. Verghese M, Wilkinson E, He YY. Role of RNA modifications in carcinogenesis and carcinogen damage response. Mol Carcinog. 2023;62:24-37
Author contact
![]() Corresponding authors: Shun-Fa Yang, Ph.D. or Hsueh-Ju Lu, MD., Ph.D., Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel.: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw (Shun-Fa Yang); E-mail: hsuehju0311com (Hsueh-Ju Lu).
Corresponding authors: Shun-Fa Yang, Ph.D. or Hsueh-Ju Lu, MD., Ph.D., Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Tel.: +886-4-24739595 ext. 34253; Fax: +886-4-24723229; E-mail: ysfedu.tw (Shun-Fa Yang); E-mail: hsuehju0311com (Hsueh-Ju Lu).

 Global reach, higher impact
Global reach, higher impact