Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(12):2992-3006. doi:10.7150/ijms.113226 This issue Cite
Review
An Overview of the Role of Genetic factors in Idiopathic Pulmonary Fibrosis: Insights from Epidemiology to Prognosis
1. Key Laboratory of Respiratory Disease of Zhejiang Province, Department of Respiratory and Critical Care Medicine, Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou 310009, Zhejiang, China.
2. Department of Endoscopy Center, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310009, Zhejiang, China.
Received 2025-3-4; Accepted 2025-5-22; Published 2025-6-12
Abstract
Idiopathic pulmonary fibrosis (IPF), a chronic progressive fibrosing interstitial lung disease with an unclear etiology, is characterized by progressive respiratory impairment and a median survival of 3-5 years. The pathophysiology associated with genetic factors in IPF remains largely unknown, despite the fact that both familial and sporadic IPF exhibit genetic susceptibility. In this review, we comprehensively examine genetic variations associated with the functional roles of mucin 5B (MUC5B), telomerase complex, surfactant proteins, cytokines, signaling pathways, and epigenetic mechanisms. A multifaceted perspective derived from genetic, epidemiological, and clinical studies demonstrates that genetic variations exert differential impacts on the development, progression, and prognosis of IPF. We advocate for the application of genetic knowledge to facilitate the refinement of diagnostic approaches, enhance the assessment of therapeutic strategies and prognostic outcomes, and underscore the significance of personalized therapy for IPF.
Keywords: genetics, idiopathic pulmonary fibrosis (IPF), MUC5B, surfactant, telomerase
1. Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic interstitial lung disease (ILD) that primarily affects adults and is characterized by fibrosis and clinical symptoms such as dyspnea and progressive deterioration of pulmonary function. IPF typically has an unfavorable prognosis with a median survival of approximately three to five years [10]. The disease manifests in both sporadic and familial forms, each exhibiting genetic susceptibility. Familial pulmonary fibrosis (FPF) is diagnosed when at least two first- or second-degree blood relatives are affected by ILD [11]. FPF accounts for 5-20% of all IPF cases [12, 13]. Approximately one-third of individuals with sporadic IPF have a family history of pulmonary fibrosis [14], with nearly a quarter of the genetic risk attributable to rare variants of known FPF-associated genes [13].
Genetic factors play a role in the development and progression of IPF (Figure 1). While the pathogenesis of IPF remains unclear, it is influenced by a complex interplay of environmental and host factors. Current genetic research has identified key contributors such as mucin 5B (MUC5B) [15], telomerase [5], surfactant proteins [17], cytokines, and related signaling pathways. Moreover, the role of epigenetic signaling pathways in regulating the development and progression of IPF has garnered increased attention. Advances in genetics have further deepened understanding of the pathophysiology of IPF and supported the development of personalized medical strategies. Treatment options for IPF are limited and currently consist of two anti-fibrotic drugs, pirfenidone and nintedanib, which slow disease progression but fail to reverse established fibrosis [10, 18, 19]. Lung transplantation may be considered for patients with end-stage disease. Notably, genetic variations, including MUC5B promoter polymorphisms and the Desmoplakin (DSP) rs2076295 genotype, have been linked to differential therapeutic responses to pirfenidone and nintedanib [20, 21]. The genetic perspective improves both the effectiveness of treatments and the quality of life of patients through targeted therapeutic interventions.
In summary, advancements in genetics have provided new perspectives for a deeper understanding of IPF. In this review, we examine the complex role that genetic factors have in the pathogenesis, progression, and prognosis of the disease. While this review emphasizes genetic and epigenetic drivers of IPF, we contextualize these findings with select transcriptomic and proteomic studies that elucidate functional consequences of genetic perturbations. In addition, we present the latest concepts that are useful for researchers to discover new diagnostic and therapeutic pathways.
2. Epidemiology
The global incidence of IPF demonstrates marked geographic heterogeneity, with multinational registry studies highlighting distinct epidemiological patterns [25]. Notably, adjusted incidence estimates range from 3.5 to 13 per 100,000 individuals in the Asia-Pacific region compared to 0.9-4.9 in Europe and 7.5-9.3 in North America [26]. While occupational exposures and environmental factors have been associated with disease risk [27], the strong familial clustering observed in 5-20% of cases provides compelling evidence for genetic predisposition [11].
Graphical summary. This graphical summary provides an overview of the main topics covered in our review. The “Pathogenesis” section examines several key genetic variations associated with primary profibrotic mechanisms. As shown in Figure 2, genetic factors are closely associated with the progression of IPF. The “Clinical Manifestations” section illustrates the impact of genetic variations on the clinical presentation of IPF patients, particularly regarding lung function. The “Diagnostic Assessment” section examines the importance of genetic factors in IPF diagnosis and predicts their role in future diagnostic strategies. In the “Treatment” section, we analyze the impact of genetic variations on pharmacological and transplantation therapies and evaluate the potential of gene therapy. Finally, the “Prognosis” section proposes innovative methods for developing prognostic models and biomarkers based on genetic factors.
Emerging genetic epidemiology reveals population-specific risk architectures. For instance, the frequency of the MUC5B rs35705950 minor allele differs significantly across populations: 0.007 in East Asian, 0.02 in African/African American, and 0.11 in European populations. While this variant confers stronger association in European populations [28], its attenuated association in Asian cohorts like the South Korean IPF registry [29] suggests modifier loci or environmental interactions may shape ethnic-specific risk profiles. These genetic differences parallel clinical disparities. Multicenter studies have shown that African American patients with pulmonary fibrosis are diagnosed, hospitalized, and die at earlier ages than patients of European or Latino ancestry [30]. These differences in clinical outcomes among different ethnic groups require a thorough examination of underlying causes, particularly focusing on the impact of genetic variations on the development and progression of IPF.
3. Pathogenesis
3.1. Mucin and cell adhesion
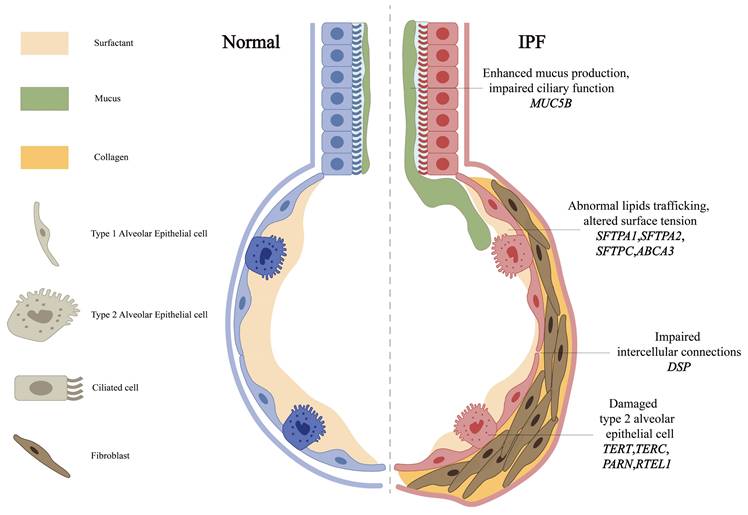
The MUC5B gene, encoding a mucin critical for airway defense, is expressed in the mucous glands of the terminal and conducting airways, alveolar type II epithelial cells (AECII), and the epithelial cells of alveolar cysts [31]. The T/G variant rs35705950 in its promoter is the strongest genetic risk factor for IPF (Table 1) [32], with epigenetic studies revealing this risk allele is associated with MUC5B promoter hypomethylation and transcriptional activation [2]. Epigenome-wide studies reinforce DNA methylation's role, such as MUC5B promoter hypomethylation, and highlight Solute Carrier Family 6 Member 6 (SLC6A6) rs112271207, a taurine transporter with putative epigenetic effects, as a candidate gene [2]. While its minor allele frequency varies, functional studies reveal conserved pathogenic mechanisms: excessive production of MUC5B may impair mucosal defense mechanisms and reduce the effectiveness of mucociliary clearance in removing inhaled particles and microorganisms (Figure 2). Elevated levels of MUC5B mucin can disrupt normal surfactant function in the alveoli and distal airways, potentially resulting in alveolar collapse and inflammatory reactions [33]. Although these mechanisms are important in the pathogenesis of IPF, they are not exclusive determinants, and the precise role of MUC5B in IPF remains unclear. Recent multi-ancestry meta-analyses have also implicated Mucin 1 (MUC1), encoding another transmembrane mucin, as a novel susceptibility locus, highlighting mucin dysregulation as a broader mechanism in IPF pathogenesis [34]. While Nitrogen Permease Regulator 3-like Protein (NPRL3) rs74614704, a regulator of mTORC1 signaling, further links mucin pathways to fibrotic remodeling [3, 35].
DSP, an integral component of desmosomal structures, is crucial for maintaining intercellular connections and tissue integrity. Mechanistically, DSP variants, including rs2076295, disrupt alveolar epithelial integrity (Figure 2) [4]. Such dysfunction can trigger abnormal extracellular matrix deposition and accelerate the progression of pulmonary fibrosis [4]. Notably, the genetic variations in MUC5B and DSP genes exhibit significant correlations with DNA methylation [36], suggesting that multiple genetic factors may interact synergistically to influence the development and progression of IPF. These findings position mucin and adhesion pathways as interconnected drivers of IPF pathogenesis.
3.2. Telomere shortening and dysfunction
Telomeres, protective nucleoprotein complexes at chromosome termini, progressively shorten with cell division. Critical telomere attrition triggers DNA damage response (DDR) pathways. In IPF, sustained DDR activation may compromise the function of AECs, inducing alveolar epithelial cell senescence [37]. Mutations in telomerase reverse transcriptase (TERT), telomerase RNA component (TERC) [6], poly(A)-specific ribonuclease (PARN), and regulator of telomere length 1 (RTEL1) affect telomere length and stability and are associated with the development of pulmonary fibrosis (Table 1) [38]. Notably, TERT expression in lung fibroblasts correlates with acetylated histone H3K9 binding at its promoter [39], suggesting epigenetic regulation of telomerase activity.
In patients with IPF, a novel mutation in the NHP2 ribonucleoprotein (NHP2) gene (p.Y24N) disrupts the nuclear import of the NHP2 protein, thereby reducing the levels of proteins critical for telomere maintenance and telomerase activity [40]. Mechanistically, telomere dysfunction impairs AECII progenitor function: dysfunctional telomeres in AECII (Figure 2) disrupt differentiation and regenerative capacity, promoting alveolar collapse and fibrosis [41]. Genome-wide association studies (GWAS) now extend telomere-related genetics beyond core telomerase components. For example, the Spindle Apparatus Coiled-coil Protein 1 (SPDL1) rs116483731, encoding a mitotic spindle assembly protein, has been identified as a novel risk factor that accelerates telomere attrition and cellular senescence, likely via mitotic errors that exacerbate replicative stress [8]. Similarly, Kinetochore Scaffold 1 (KNL1) rs12912339 and Stathmin 3 (STMN3) rs76537958 variants, implicated in spindle assembly, suggest that mitotic errors contribute to alveolar stem cell dysfunction [3, 32].
Genetic variants associated with idiopathic pulmonary fibrosis
| Gene | Variant | Chr | Impact on IPF | Reference |
|---|---|---|---|---|
| MUC5B | rs35705950 | 11 | Strongest genetic risk factor; promoter hypomethylation increases mucin production, impairing mucosal defense. Associated with higher CT fibrosis scores | [1] |
| SLC6A6 | rs112271207 | 3 | Its role as a taurine transporter suggests epigenetic regulation of IPF pathogenesis | [2] |
| NPRL3 | rs74614704 | 16 | Regulates mTORC1 signaling, linking mucin pathways (e.g., MUC5B) to fibrotic remodeling. May modulate cellular energy metabolism and fibrotic signaling cascades | [3] |
| DSP | rs2076295 | 6 | Disrupts alveolar epithelial integrity, accelerating fibrosis. Correlates with differential responses to nintedanib therapy | [4] |
| TERT | rs4449583 | 5 | Telomere shortening induces alveolar epithelial senescence. Linked to early-onset IPF, hematologic complications post-transplant, and reduced survival | [5] |
| TERC | rs2293607 | 3 | [6] | |
| PARN | / | / | [7] | |
| RTEL1 | rs41308092 | 20 | [5] | |
| SPDL1 | rs116483731 | 5 | Risk factor accelerating telomere attrition through mitotic errors | [8] |
| KNL1 | rs12912339 | 15 | Mitotic spindle assembly protein variant implicated in mitotic errors and replicative stress. Contributes to alveolar stem cell dysfunction and cellular senescence | [3] |
| STMN3 | rs112087793 | 20 | Variant associated with cytoskeletal reorganization via spindle assembly defects. Promotes mitotic stress, impairing alveolar epithelial repair and regeneration | [3] |
| SFTPA1 | rs1215316727 | / | Mutations cause surfactant dysfunction, leading to alveolar collapse and fibrosis. Associated with atypical radiological patterns and rapid lung function decline | [9] |
| SFTPA2 | rs371035540 | / | [9] | |
| SFTPC | / | / | [16] | |
| ABCA3 | / | / | [17] | |
| AKAP13 | rs62025270 | 15 | RhoA regulator exacerbates TGF-β dysregulation, increasing fibrotic remodeling | [7] |
| TOLLIP | rs3750920 | 11 | Modifies therapeutic response to N-acetylcysteine; TT genotype benefits while CC genotype may worsen outcomes | [22] |
| PCSK6 | rs35647788 | 15 | Associated with reduced transplant-free survival via dysregulated proteolytic processing of profibrotic mediators | [23] |
| PKN2 | rs115982800 | 1 | Correlates with rapid FVC decline; regulates cytoskeletal remodeling and fibroblast activation | [24] |
Chr = chromosome.
Some genes related to the main pro-fibrotic mechanisms. The primary profibrotic mechanisms are associated with both mutations and polymorphisms in genes such as MUC5B, DSP, telomerase-associated genes (including TERT, TERC, PARN and RTEL1) and surfactant proteins (including SFTPA1, SFTPA2, SFTPC and ABCA3). Overexpression of MUC5B may impair mucosal defense by reducing the efficiency of ciliary clearance of inhaled particles and microorganisms. Mutations within the DSP gene reduce its expression, while DSP is essential for maintaining cell-cell connections and tissue structural integrity. Reduced telomerase activity, due to mutations in TERT, TERC, PARN and RTEL1, leads to telomere shortening. Surfactant proteins play a critical role in modulating host defense functions, including the production of pro-inflammatory cytokines, cellular chemotaxis, and tissue repair, while also maintaining alveolar stability.
While these mutations are associated with IPF, they are not detected in the majority of patients [42]. Strikingly, in IPF patients homozygous for the non-risk MUC5B rs35705950 allele, rare functional variants in TERT, PARN, TERC, or RTEL1 are enriched [7], suggesting a potential genetic interaction between MUC5B and telomerase-related genes. It highlights the need for stratified genetic testing: MUC5B-centric screening may overlook telomerase-driven subtypes requiring distinct management. These findings underscore the interplay between genetic, epigenetic, and mitotic stress mechanisms in IPF pathogenesis.
3.3. Surfactants
Pulmonary surfactants, lipid-protein complexes synthesized by AECII, maintain alveolar integrity by reducing surface tension. Surfactant dysfunction directly contributes to IPF pathogenesis through AECII injury and aberrant repair [43] (Figure 2), leading to decreased alveolar stability, alveolar collapse, and the development of fibrosis. Beyond structural roles, surfactant proteins modulate host defense by regulating pro-inflammatory cytokines, chemotaxis factors, and tissue repair processes [43].
Four surfactant-associated genes—surfactant protein A1 (SFTPA1), surfactant protein A2 (SFTPA2), surfactant protein C (SFTPC) and the ATP-binding cassette-type family A member 3 transporter (ABCA3)—are implicated in familial and sporadic pulmonary fibrosis (Table 1) [17, 44]. Distinct functional roles of SFTPA1 and SFTPA2 may arise from altered expression ratios that modulate surfactant activity [9]. Autosomal dominant SFTPC mutations, like SFTPC-I73T, disrupt lamellar body maturation in AECII, causing surfactant accumulation and oxidative stress [16]. Recessive ABCA3 variants, localized to lamellar body membranes, are frequently linked to pediatric interstitial lung disease, underscoring surfactant dysregulation as a pan-age mechanism [17].
3.4. Cytokine and signaling pathway dysregulation
The transforming growth factor-β (TGF-β) signaling pathway constitutes a central axis in IPF pathogenesis, with both genetic predisposition and downstream effector mechanisms contributing to fibrotic progression. TGF-β1 induces fibroblast-to-myofibroblast differentiation through canonical Smad2/3 activation [45, 46], with epigenetic mechanisms critically modulating this process. Specifically, TGF-β1 promotes methylation of the Thy-1 promoter and recruits methyl-CpG binding domain protein 2 (MBD2) to activate the TGF-β-Smad pathway, creating a feed-forward loop that sustains fibroblast activation [47]. Furthermore, TGF-β1 modulates H3K9me2/3 and H3K4me1/2/3 histone marks at Collagen Type I Alpha 1 (Col1A1), Connective Tissue Growth Factor (CTGF), and Plasminogen Activator Inhibitor 1 (PAI-1) promoters, enhancing the transcriptional activity of fibrosis-associated genes [48]. Nuclear accumulation of Smad proteins enables their function as transcriptional regulators of fibrosis-associated genes. Smad complexes also interact with histone deacetylases to remodel chromatin, facilitating extracellular matrix (ECM) protein expression [49]. Experimental evidence indicates that Bone Morphogenetic Protein 4 (BMP4) antagonizes TGF-β1-driven effects by activating Smad1/5/9, thereby suppressing Smad2/3 phosphorylation and inhibiting myofibroblast differentiation and ECM synthesis [46]. In addition, miR-17-92 and miR-29 inhibit TGF-β-driven fibrosis: miR-17-92 maintains alveolar homeostasis by blocking fibroblast activation [50], while miR-29 suppresses ECM synthesis via Yes-associated protein (YAP) signaling [51].
Canonical Wnt/β-catenin signaling exhibits spatial specificity in IPF, with alveolar epithelial-specific activation driving IL-1β-mediated TGF-β amplification [52]. In addition, the non-canonical Wnt signaling pathway, exemplified by WNT5A, initiates cytoskeletal reorganization via the JNK and ROCK signaling pathways. F-actin-generated biomechanical tension facilitates proteolytic activation of latent TGF-β via integrin αv [53]. Furthermore, genetic variants in A-kinase anchoring protein 13 (AKAP13), a kind of RhoA regulator, exacerbate TGF-β dysregulation and increased IPF susceptibility, underscoring the role of genetic predisposition in fibrotic remodeling [54]. Notably, miR-26a downregulation in fibrotic environments exacerbates TGF-β1-induced ECM deposition, whereas its overexpression attenuates fibrosis [55], suggesting miRNA-based modulation of Wnt-TGF-β crosstalk. Emerging evidence positions mTOR signaling as both a TGF-β effector and independent genetic risk modulator. TGF-β-induced mTOR signaling depends on the canonical Smad signaling pathway and is independent of Phosphoinositide 3-kinase (PI3K)/AKT activity [56]. Notably, genetic polymorphisms in DEP domain-containing mTOR-interacting protein (DEPTOR), an endogenous mTOR inhibitor, and other mTOR pathway components, like Regulatory Associated Protein of mTOR Complex 1 (RPTOR), have been linked to altered IPF risk, emphasizing the genetic modulation of mTOR-driven fibrosis [57]. This multilayered interplay underscores the need for integrated therapeutic approaches to address signaling dysregulation in IPF.
The pathogenesis of IPF involves genetic susceptibility, alveolar epithelial dysfunction, and dysregulated signaling pathways. Mucin/adhesion abnormalities, telomere attrition, and surfactant dysregulation drive injury and senescence. Concurrently, cytokine/signaling imbalances and epigenetic modifications establish fibrotic cascades through genetic-epigenetic crosstalk. These mechanisms underscore the need for stratified therapies targeting molecular subtypes.
4. Clinical manifestations and diagnostic assessment
4.1. Clinical manifestations
IPF is a chronic and progressive lung disease characterized by a range of clinical symptoms. Initially, patients often experience exertional dyspnea, which may progress to persistent dry cough, weight loss, and other systemic symptoms as the disease advances. On physical examination, findings may include clubbing of the fingers and basal inspiratory crackles or rales, indicating impaired gas exchange and progressive pulmonary fibrosis. High-resolution computed tomography (HRCT) images typically reveal characteristic IPF findings, such as reticular opacities and honeycomb changes. Pulmonary function tests often demonstrate restrictive ventilatory defects and reduced carbon monoxide diffusion capacity. These tests are crucial for diagnosing IPF and monitoring disease progression.
With the advancement of precision medicine, genetic testing of patients with IPF is increasing. Genetic backgrounds can significantly influence the range and severity of clinical manifestations. For example, the MUC5B gene encodes key mucins present in the honeycomb cysts of patients with IPF [31]. Carriers of the MUC5B promoter variant rs35705950 often exhibit more severe coughing symptoms and higher quantitative CT fibrosis scores, which may help quantify disease risk for relatives [58]. These variants also correlate with distinct prognoses [59]. In addition, further advances in imaging technology may lead to more accurate prognoses for patients. Observational studies show that patients with SFTPC or other surfactant-related gene mutations present with atypical radiological patterns, including cystic changes characteristic of interstitial pneumonia [60]. These patients are often younger (average age 45 years) and exhibit lower forced vital capacity (FVC) and lung diffusion capacity for carbon monoxide (DLCO) compared to those with familial or sporadic IPF [61]. Carriers of TERT mutations, even asymptomatic individuals, display significantly reduced DLCO and impaired DLCO response during exercise. HRCT scans in these patients reveal signs of pulmonary fibrosis and increased lung tissue volume fractions. Compared to non-carriers, TERT mutation carriers with IPF show a more pronounced reduction in lung diffusion capacity [62].
4.2. Diagnostic assessment
The 2018 guidelines [63] jointly published by the American Thoracic Society (ATS), the European Respiratory Society (ERS), the Japanese Respiratory Society (JRS), and the Asociación Latinoamericana de Torax (ALAT), recommend an initial assessment for suspected cases of IPF, with a focus on identifying possible known causes of ILD. After identifying a potential cause of ILD, a comprehensive evaluation is necessary to identify or rule out conditions such as hypersensitivity pneumonitis, connective tissue disease, pneumoconiosis, and iatrogenic problems. If diagnostic uncertainty persists after standard assessments, a multidisciplinary discussion (MDD) should be convened. This MDD should integrate clinical and HRCT findings to confirm or rule out the diagnosis. A definitive IPF diagnosis can be established by correlating appropriate HRCT and histopathological patterns.
Given the overlapping clinical features of chronic respiratory diseases, early differential diagnosis and the identification of relatives at risk for IPF are of significant value in genetic research. A machine learning model that uses gene expression data from peripheral blood mononuclear cells to predict IPF has been developed [64]. This 44-gene model can accurately predict IPF in healthy controls and patients with tuberculosis, HIV, and asthma. Moreover, the model also allows subtyping of IPF subtypes. Thus, the model shows promise as a non-invasive diagnostic tool [64]. Clinical research has shown that pathogenic variations in the telomerase complex genes are found in approximately 10% of patients with IPF, regardless of family history, suggesting the need for genetic counseling for all patients with IPF [65].
When assessing lung diseases, HRCT provides finer images than traditional CT scans. However, interpreting the morphology and extent of lesions using HRCT remains a relatively subjective process that requires a solid foundation in imaging knowledge, and robust quantitative diagnostic methods are lacking. Additionally, cost and radiation exposure must also be considered. Future genetic research could complement HRCT by establishing quantitative standards for the diagnosis of IPF [58, 66]. The MUC5B gene is particularly interesting in this context. Polygenic risk scores for IPF and interstitial lung anomalies have been developed using data from a GWAS in IPF, confirming the predictive value of MUC5B in identifying individuals at risk for pulmonary fibrosis [67].
5. Treatment
5.1. Drug therapy
Pirfenidone and nintedanib are anti-fibrotic drugs approved by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for the treatment of IPF. Clinical studies have shown that pirfenidone can slow the decline in FVC [18]. However, its clinical effectiveness may vary among patients with IPF and different genetic backgrounds. In particular, a subset of patients with IPF who have high expression of genes related to ciliogenic epithelial cells responds more positively to pirfenidone [68]. Nintedanib is a small-molecule tyrosine kinase inhibitor that reduces lung fibroblast proliferation, migration, and differentiation by inhibiting multiple growth factor receptors. Nintedanib may slow the rate of FVC decline in patients with IPF and prolong their survival [19]. In a cohort analysis, patients carrying the DSP rs2076295 G allele experienced greater benefits in overall survival and lung function when treated with nintedanib compared with TT homozygous patients [69]. To the best of our knowledge, no studies have specifically examined the association between genetic differences and adverse reactions to these two anti-fibrotic drugs in patients with IPF.
Although clinical trials have confirmed the effectiveness of these drugs in slowing lung function decline [18, 19], they cannot reverse or resolve existing fibrosis. Advances in genetic research have provided new insights into the antifibrotic mechanisms of pirfenidone and nintedanib, and evidence suggests that different gene expression patterns can influence drug efficacy [70]. For instance, the Toll-interacting protein (TOLLIP) rs3750920 polymorphism has been shown to modify responses to N-acetylcysteine (NAC), with TT genotype carriers deriving therapeutic benefit while CC genotype carriers may experience harm [22]. Similarly, the MUC5B rs35705950 variant has been linked to differential outcomes in IPF therapies [71]. These findings underscore the potential of pharmacogenomics to guide personalized treatment strategies, as exemplified by the ongoing PRECISIONS trial (NCT04300920), which stratifies IPF patients by TOLLIP genotypes to optimize NAC therapy [22]. Identifying genotypes associated with drug responses could enable the prediction of individual patient responses to these drugs. Furthermore, this may facilitate the development of personalized treatment strategies, including tailored drug selection, dosage adjustment, and combination therapies.
5.2. Lung transplantation
The pharmacological treatment of patients with IPF is primarily aimed at slowing the progression of fibrosis and providing palliative care for those in advanced stages. However, despite optimal therapeutic interventions, lung function in patients with IPF may still progressively deteriorate. IPF can reach a terminal stage characterized by severely impaired lung function that is unresponsive to medical treatment. For these patients, lung transplantation (LT) represents the sole therapeutic option capable of significantly prolonging their survival and enhancing quality of life. The 5-year survival rate following lung transplantation approaches 50% [72]. Nonetheless, LT faces several obstacles, including the shortage of donor lungs, stringent technical requirements, and the substantial costs associated with the procedure. Additionally, genetic factors have been shown to influence both the success and quality of LT outcomes.
The risk of complications following LT is associated with post-transplant survival and represents a crucial consideration when assessing LT candidacy [73]. Clinical observations indicate that lung transplant recipients with IPF (IPF-LTRs) have circulating T cells suggestive of immunodeficiency [74], thereby increasing their risk of hematological complications [75]. Moreover, IPF-LTRs are more susceptible to rare telomere-related genetic variants and shorter telomere lengths compared to non-transplant individuals [76]. Some researchers have recommended incorporating genetic factors into LT evaluations, such as measuring telomere length prior to transplantation and conducting genetic testing for telomere gene variants. Such tests may help identify IPF transplant recipients who are at higher risk of hematologic complications [75].
The results of a retrospective study indicate that LT is appropriate even in patients with telomerase-related gene mutations, provided there are no myelodysplastic symptoms and a systematic hematological evaluation is performed [77]. In addition, genetic factors influence decisions regarding the administration of immunosuppressants after LT. The evidence suggests that standard immunosuppressive therapy should be maintained in young transplant recipients with shorter telomeres, even if immune deficiency is present [74].
6. Prognosis
Along with FVC and DLCO, age and gender are important prognostic indicators during the stable phase of IPF. Men generally exhibit worse survival rates than women [78]. The GAP staging system integrates age, gender, and lung function parameters (FVC, DLCO) to predict mortality risk, though its sensitivity for short-term (1-year) outcomes remains limited [79]. Biomarkers and genetic variations provide complementary prognostic insights and may enhance existing prognostic assessment tools by addressing their limitations.
A retrospective study involving a European cohort of 1751 patients with IPF revealed that the MUC5B T allele is a significant independent predictor of patient survival. No significant correlation was observed in patients under 56 years of age, whereas in the older cohort, individuals with the T allele exhibited better survival rates [80]. Conversely, in another retrospective case-control study conducted within the Portuguese population, no associations were found between MUC5B variations and disease survival rates [81]. These inconsistencies likely stem from confounding factors that were not accounted for in the study design or analysis. However, the preponderance of evidence suggests that the MUC5B minor T allele is associated with improved patient survival rates [81-83], independent of age, gender, FVC, and DLCO. This paradoxical association (increased disease risk but better survival) may be influenced by index bias, as studies predominantly including prevalent rather than incident IPF cases could disproportionately select for resilient individuals with the MUC5B risk allele, thereby inflating survival estimates [84]. This underscores the significance of the MUC5B genotype in a survival prognosis model for patients with IPF [83]. Telomere length has been independently associated with transplant-free survival in patients with IPF, as confirmed by observational cohort studies, further highlighting its role in the prognosis of IPF [85]. Ethnic-specific outcomes are evident in the prognosis of IPF patients. Rare TERT variants enriched in Latin American IPF patients correlate with aggressive disease trajectories, though mechanistic links require further study [86]. Japanese patients show distinct causes of death, like acute exacerbations, and prognoses compared to other ethnic groups [87].
A comprehensive proteomic analysis of multiple IPF patient cohorts has yielded a model based on the different expression levels of osteopontin (OPN), serum protein D (SPD), intercellular adhesion molecule 1 (ICAM1), and matrix metalloproteinase 7 (MMP7), allowing robust differentiation between progressive and stable IPF [88]. These circulating serum proteins are significantly associated with clinical outcomes, increased mortality rates, and greater disease severity, highlighting the feasibility of developing serology-based methods to assess IPF progression [88]. For instance, baseline serum levels of cathepsin B (CTSB) strongly correlate with the extent of lung function decline at one year. Patients with elevated serum CTSB levels are more likely to exhibit a progressive IPF phenotype, irrespective of GAP stage [89]. Given the growing number of potential biomarkers, developing robust methods to evaluate their clinical utility is critical. The innovative progression index offers a quantitative measure of biomarkers' influence on clinical progress [88].
Recent studies have identified polymorphisms in TGF-β1, proprotein convertase subtilisin/kexin type 6 (PCSK6), and protein kinase N2 (PKN2) as critical determinants of disease progression and survival outcomes in IPF. The TGF-β1 T869C variant has been implicated in disease severity, with the TT genotype linked to reduced PaO₂ and increased D(A-a)O₂ at diagnosis, suggesting a role in accelerating functional decline [90, 91]. Similarly, the PCSK6 rs35647788 variant has been associated with reduced transplantation-free survival, potentially through dysregulated proteolytic processing of profibrotic mediators [23]. The PKN2 rs115982800 variant, located in the antisense RNA PKN2-AS1, correlates with rapid FVC decline, highlighting its role in cytoskeletal remodeling and fibroblast activation [24]. These findings underscore the importance of genetic variants in modulating IPF progression.
Genes associated with endoplasmic reticulum stress [92], macrophage function [93], and mitochondrial dynamics [93] are implicated in the development and progression of IPF. These genes exhibit robust associations with canonical signaling pathways, including the apoptosis signaling pathway and the PI3K/AKT pathway, which collectively modulate the pulmonary immune microenvironment. Prognostic signature genes derived from the synergistic expression of m5C-regulated genes and immune-associated genes are likely to exert significant influence over immune and inflammatory responses, enabling precise prediction of survival outcomes in IPF patients [94]. Continued advancements in genetic research are anticipated to unveil novel genetic determinants, thereby facilitating the development of refined prognostic models and elucidating the underlying mechanisms of IPF pathogenesis.
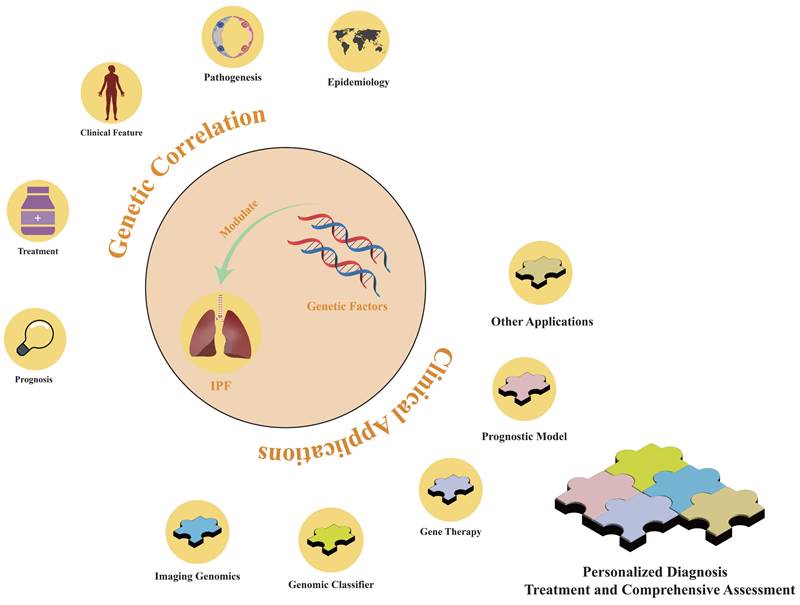
The role of genetic factors in idiopathic pulmonary fibrosis. Genetic factors are implicated in the epidemiology, pathogenesis, clinical manifestations, diagnostic approaches, therapeutic modalities, and prognostic outcomes of IPF. By leveraging genetic factors, personalized diagnostic methods and comprehensive assessment models for treatment and prognostic benefits can be developed.
7. Genetic-Based Diagnosis and Treatment Models
Genetic testing serves principally as an auxiliary tool within the classical diagnostic and therapeutic strategies for IPF. Although genetic testing exhibits limited independent utility for IPF diagnosis, treatment, or prognosis, its integration as a central analytical framework in clinical practice remains feasible. This strategy is particularly advantageous for patient subgroups at elevated risk of disease progression and adverse outcomes, as it enables the customization of therapeutic strategies according to individual genetic profiles.
Genetic studies have delineated distinct patient subgroups for IPF diagnosis that can be used to identify high-risk populations. The 2022 ATS/ERS/JRS/ALAT guidelines emphasize the significance of genetic factors in stratifying the clinical severity of IPF and discuss their clinical applications. Despite guideline recommendations against using genomic classifiers as a standard diagnostic tool for usual interstitial pneumonia due to a lack of consensus [10], genetic factors remain essential in the personalized treatment strategies of precision medicine. As advancements in genetic etiology and cost-effective genetic testing continue, identifying patients who would benefit from genetic testing will grow increasingly critical. This will aid in developing intervention strategies to slow disease progression and selecting the most appropriate screening and management protocols (Figure 3). The Envision Genomic Classifier for IPF diagnosis, a clinically validated tool derived from whole transcriptome mRNA sequencing in transbronchial biopsy samples, integrates clinical factors and HRCT imaging, demonstrating high diagnostic sensitivity [95]. Moreover, detecting the differential expression of the pirfenidone response gene across IPF subgroups is important, and sophisticated machine learning techniques can facilitate the development of classifiers that reflect cell-type characteristics and gene expression patterns [68].
Genetic variations in patients with IPF influence their responsiveness to pharmacotherapies, necessitating the integration of genetic insights into drug development (Figure 3). Modern genomics offers precise molecular targets for disease diagnosis and therapeutic intervention. Integrating gene expression profiles and pathological characteristics of patients with IPF into computational methods can expedite drug development [96]. Artificial intelligence platforms that identify drug targets optimize the drug development process by streamlining target discovery [97]. For instance, computational simulations have demonstrated that a plant-derived microRNA, osa-miR172d-5p, downregulates the expression of TAK1-binding protein 1 and fibrosis-related genes in TGF-β-stimulated pulmonary fibroblasts [98].
In murine studies, gene therapy has demonstrated the potential to arrest the progression of pulmonary fibrosis. Utilizing an adeno-associated virus serotype 9 (AAV9)-Tert vector for gene therapy, reactivation of telomerase in the lung can delay disease progression in murine models of pulmonary fibrosis [99]. Prophylactic intratracheal administration of AAV9-Tspyl2 delays the onset of bleomycin-induced pulmonary fibrosis in mice by inhibiting the TGF-β/Smad3 signaling pathway [100]. BIX01294, an exceptionally selective G9a histone methyltransferase inhibitor, reduces TGF-β-induced H3K9 methylation and matrix stiffness via upregulation of the PPARGC1A gene [101], thereby diminishing collagen deposition in the lungs of mice following bleomycin injury. These results suggest that therapeutic interventions targeting epigenetic repression mechanisms hold promise. Studies on the relaxin/RXFP1 axis [102] and microRNA-144-3p [103] have revealed their promising anti-fibrotic properties in patients with IPF, offering new avenues for IPF therapeutics.
The identification of genetic factors enables the stratification of IPF cases, the correlation of genetic and phenotypic profiles, and prediction of treatment effectiveness and patient outcomes (Figure 3). The molecular signatures of IPF-associated fibroblastic subtypes and their prognostic implications have been characterized through machine learning and single-cell analyses [104]. New risk assessment models can be developed using bioinformatics and machine learning algorithms to facilitate the stratification of patient subgroups and refine personalized therapeutic strategies.
8. Discussion
Genetic factors significantly contribute to the epidemiological variance observed in IPF and are significant to its pathogenesis. The incidence and clinical outcomes of IPF exhibit variability across ethnicities and regions, primarily due to genetic predispositions. For instance, in Asian and North American populations, the minor allele frequency of the T allele in the MUC5B gene rs35705950 is significantly correlated with the IPF incidence. However, the lower incidence of IPF in the European population, despite a higher frequency of the minor T allele, suggests that additional factors affect disease development. In addition to genetic factors, environmental determinants, including occupational exposure and gender differences, have been implicated in the occurrence and progression of IPF (Figure 3). A broad consensus exists that environmental and host factors exert cumulative effects on IPF risk [105], and family history research offers an opportunity to elucidate the genetic contributions to IPF.
Family studies have revealed the so-called anticipation phenomenon, characterized by an earlier onset of pulmonary fibrosis symptoms in successive generations of families with TERT mutations. This phenomenon correlates with shorter telomeres in the offspring of families with telomerase mutations, which have been validated as a risk factor for IPF and are associated with adverse clinical outcomes [106]. Although anticipation has not been observed for other genetic factors implicated in the etiology and progression of IPF (such as MUC5B, DSP, and surfactant proteins), evidence suggests potential interactions among these factors [7, 33]. Epigenetic mechanisms may act both as independent contributors to IPF [36] and as mediators in its pathogenesis [36, 39]. A bioinformatics-driven network of genetic interactions may systematically elucidate the role of genetic factors in IPF pathogenesis.
Genetic variations are important factors in the lung function and radiological features of patients with IPF, influencing the spectrum and severity of clinical phenotypes. Most patients with IPF experience a gradual decline in clinical, functional, and radiographic status, but some undergo acute respiratory exacerbations (AE-IPF). Research indicates that shortened telomere length is associated with an increased risk of AE-IPF or mortality in these patients [107], and that expression of the S100A8/A12 genes is linked to the etiology of AE-IPF [108]. Genetic factors provide important insights into the stratification of clinical severity in patients with IPF, as acknowledged in the ATS/ERS/JRS/ALAT clinical guidelines, which recognize the clinical application of genetic classifiers [10]. Physicians are encouraged to thoroughly investigate the optimal application of genetic factors to enhance disease diagnosis and treatment, in accordance with current clinical guidelines.
Genetic imaging, a multidisciplinary field integrating genetics and neuroimaging, assesses changes in brain morphology and function to determine the impact of genetic variations on individual behaviors and diseases. Imaging techniques have identified links between genetic variations and brain structures, for example, a correlation has been found between the SNP rs42352 in the Semaphorin 5A (SEMA5A) gene and bilateral hippocampal volume [109]. Incorporating imaging data to examine associations between genetic variations and disease enhances diagnostic confidence and demonstrates the viability of a new diagnostic paradigm that leverages genetic factors to interpret imaging data and clinical phenotypes. This approach is particularly relevant in the IPF field, which relies heavily on diagnostic imaging tools (Figure 3). Studying imaging variants caused by genetic factors can highlight the role of imaging data in tracking disease progression. In addition, genetic factors can serve as quantifiable benchmarks and provide complementary assessment support in challenging imaging diagnoses.
In the realm of pharmacotherapy for IPF, FDA-approved treatments such as pirfenidone and nintedanib lack evidence of reversing pulmonary fibrosis, underscoring significant market opportunities for the development of novel IPF therapies. Understanding the genetic factors involved in IPF pathogenesis provides a theoretical basis for leveraging gene therapy to treat the disease. For example, Luxturna, the groundbreaking in vivo gene therapy for hereditary retinal dystrophy, transfers the RPE65 gene to retinal cells using an AAV vector, and its successful approval confirmed the feasibility of gene therapy [110]. While experimental data from clinical trials are limited, AAV-based in vivo gene therapy has demonstrated the potential to halt the progression of pulmonary fibrosis in murine models, representing a promising avenue for IPF treatment.
Prognostic assessments significantly influence physicians' clinical decision-making. In the context of IPF, a disease characterized by complex genetic mechanisms and diverse clinical manifestations, machine learning and bioinformatics may be leveraged to develop personalized prognostic models to overcome the limitations of the GAP tier system in short-term risk prediction. The identification of additional biomarkers that can predict disease progression would be clinically useful, enabling timely adjustments to treatment protocols in response to individual patient progression. Current biomarkers exhibit variable reliability for predicting IPF progression, and identifying robust markers remains a challenge. Computer-assisted techniques that assign weights to each marker in disease progression could streamline the development of future clinical guidelines and facilitate the integration of new biomarkers.
8.1. Limitations
This review has several limitations. First, it is not a systematic review and has not undergone a formal quality assessment. Second, the articles included are limited to English-language publications. Third, we focused on describing the role of certain genetic factors in IPF, although some pathogenic mechanisms have not been studied in detail. Fourth, relevant articles may have been overlooked.
9. Conclusions
Genetic factors are of utmost importance in the etiology and progression of IPF. They significantly influence the epidemiological profile of IPF and its pathogenesis, clinical presentation, and prognostic outcomes. Current genetic research is poised to refine existing diagnostic frameworks and provide essential quantitative benchmarks for the development of personalized therapeutic strategies. By recognizing the importance of genetic factors and refining diagnostic and therapeutic methods tailored to these influences, it will be possible to better categorize therapeutic interventions and improve the accuracy of prognostic assessments. This approach aligns with the emerging paradigm of precision medicine.
Abbreviations
AAV: Adeno-associated virus
AAV9: Adeno-associated virus serotype 9
ABCA3: ATP-binding cassette-type family A member 3 transporter
AEC: Alveolar epithelial cell
AECII: Alveolar type II epithelial cells
AE-IPF: Acute exacerbation of IPF
AKAP13: A-kinase anchoring protein 13
ALAT: Asociación Latinoamericana de Torax
ATS: American Thoracic Society
BMP4: Bone morphogenetic protein 4
Col1A1: Collagen type I alpha 1
CTGF: Connective tissue growth factor
CTSB: Cathepsin B
DDR: DNA damage response
DEPTOR: DEP domain-containing mTOR-interacting protein
DLCO: Diffusion capacity for carbon monoxide
DSP: Desmoplakin
ECM: Extracellular matrix
EMA: European Medicines Agency
ERS: European Respiratory Society
FDA: Food and Drug Administration
FPF: Familial pulmonary fibrosis
FVC: Forced vital capacity
GAP: Gender-age-physiology (staging system)
GWAS: Genome-wide association study
HRCT: High-Resolution Computed Tomography
ICAM1: Intercellular adhesion molecule 1
ILD: Interstitial lung disease
IPF: Idiopathic pulmonary fibrosis
IPF-LTRs: Lung transplant recipients with IPF
JRS: Japanese Respiratory Society
KNL1: Kinetochore scaffold 1
LT: Lung transplantation
MBD2: Methyl-CpG binding domain protein 2
MDD: Multidisciplinary discussion
MMP7: Matrix metalloproteinase 7
mTOR: Mechanistic target of rapamycin
MUC1: Mucin 1
MUC5B: Mucin 5B
NAC: N-acetylcysteine
NHP2: NHP2 ribonucleoprotein
NPRL3: Nitrogen permease regulator 3-like protein
OPN: Osteopontin
PAI-1: Plasminogen activator inhibitor 1
PARN: Poly(A)-specific ribonuclease
PBMC: Peripheral blood mononuclear cells
PCSK6: Proprotein convertase subtilisin/kexin type 6
PI3K: Phosphatidylinositol 3-kinase
PI3K: Phosphoinositide 3-Kinase
PKN2: Protein kinase N2
RPTOR: Regulatory associated protein of mTOR complex 1
RTEL1: Regulator of telomere length 1
SEMA5A: Semaphorin 5A
SFTPA1: Surfactant protein A1
SFTPA2: Surfactant protein A2
SFTPC: Surfactant protein C
SLC6A6: Solute carrier family 6 member 6
SPD: Serum protein D
SPDL1: Spindle apparatus coiled-coil protein 1
STMN3: Stathmin 3
TERC: Telomerase RNA component
TERT: Telomerase reverse transcriptase
TGF-β: Transforming growth factor-β
TOLLIP: Toll-interacting protein
YAP: Yes-associated protein
Acknowledgements
Funding
This work was supported in part by grants from Technology Project of Zhejiang Provincial Health Commission (grant no. 2022482758) and Interdisciplinary Frontier Research Fund in Medicine of Zhejiang University.
Author contributions
Jiahao Liu: Writing - original draft, Visualization, Conceptualization, Writing - review & editing. Zihan Yi: Writing - review & editing. Ting Chen: Writing - review & editing. Yinghua Ying: Writing - review & editing, Supervision. Yue Hu: Writing - review & editing, Supervision, Funding acquisition.
Consent for publication
All authors have reviewed and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE. et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503-12 doi:10.1056/NEJMoa1013660
2. Chin D, Hernandez-Beeftink T, Donoghue L, Guillen-Uio B, Leavy OC, Adegunsoye A. et al. Genome-wide association study of Idiopathic Pulmonary Fibrosis susceptibility using clinically-curated European-ancestry datasets. medRxiv. 2025 doi:10.1101/2025.01.30.25321017
3. Allen RJ, Stockwell A, Oldham JM, Guillen-Guio B, Schwartz DA, Maher TM. et al. Genome-wide association study across five cohorts identifies five novel loci associated with idiopathic pulmonary fibrosis. Thorax. 2022;77:829-33 doi:10.1136/thoraxjnl-2021-218577
4. Hao Y, Bates S, Mou H, Yun JH, Pham B, Liu J. et al. Genome-Wide Association Study: Functional Variant rs2076295 Regulates Desmoplakin Expression in Airway Epithelial Cells. American Journal of Respiratory and Critical Care Medicine. 2020;202:1225-36 doi:10.1164/rccm.201910-1958OC
5. Peljto AL, Blumhagen RZ, Walts AD, Cardwell J, Powers J, Corte TJ. et al. Idiopathic Pulmonary Fibrosis Is Associated with Common Genetic Variants and Limited Rare Variants. American journal of respiratory and critical care medicine. 2023;207:1194-202 doi:10.1164/rccm.202207-1331OC
6. Moore C, Blumhagen RZ, Yang IV, Walts A, Powers J, Walker T. et al. Resequencing Study Confirms That Host Defense and Cell Senescence Gene Variants Contribute to the Risk of Idiopathic Pulmonary Fibrosis. American Journal of Respiratory and Critical Care Medicine. 2019;200:199-208 doi:10.1164/rccm.201810-1891OC
7. Dressen A, Abbas AR, Cabanski C, Reeder J, Ramalingam TR, Neighbors M. et al. Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study. The Lancet Respiratory Medicine. 2018;6:603-14 doi:10.1016/S2213-2600(18)30135-8
8. Dhindsa RS, Mattsson J, Nag A, Wang Q, Wain LV, Allen R. et al. Identification of a missense variant in SPDL1 associated with idiopathic pulmonary fibrosis. Commun Biol. 2021;4:392 doi:10.1038/s42003-021-01910-y
9. Floros J, Thorenoor N, Tsotakos N, Phelps DS. Human Surfactant Protein SP-A1 and SP-A2 Variants Differentially Affect the Alveolar Microenvironment, Surfactant Structure, Regulation and Function of the Alveolar Macrophage, and Animal and Human Survival Under Various Conditions. Frontiers in Immunology. 2021;12:681639 doi:10.3389/fimmu.2021.681639
10. Raghu G, Remy-Jardin M, Richeldi L, Thomson CC, Inoue Y, Johkoh T. et al. Idiopathic Pulmonary Fibrosis (an Update) and Progressive Pulmonary Fibrosis in Adults: An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med. 2022;205:e18-e47 doi:10.1164/rccm.202202-0399ST
11. Borie R, Kannengiesser C, Antoniou K, Bonella F, Crestani B, Fabre A. et al. European Respiratory Society statement on familial pulmonary fibrosis. Eur Respir J. 2023;61:2201383 doi:10.1183/13993003.01383-2022
12. Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK. et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. American Journal of Respiratory and Critical Care Medicine. 2011;183:788-824 doi:10.1164/rccm.2009-040GL
13. Liu Q, Zhou Y, Cogan JD, Mitchell DB, Sheng Q, Zhao S. et al. The Genetic Landscape of Familial Pulmonary Fibrosis. American Journal of Respiratory and Critical Care Medicine. 2023;207:1345-57 doi:10.1164/rccm.202204-0781OC
14. Moss BJ, Ryter SW, Rosas IO. Pathogenic Mechanisms Underlying Idiopathic Pulmonary Fibrosis. Annual Review of Pathology: Mechanisms of Disease. 2022;17:515-46 doi:10.1146/annurev-pathol-042320-030240
15. Evans CM, Fingerlin TE, Schwarz MI, Lynch D, Kurche J, Warg L. et al. Idiopathic Pulmonary Fibrosis: A Genetic Disease That Involves Mucociliary Dysfunction of the Peripheral Airways. Physiol Rev. 2016;96:1567-91 doi:10.1152/physrev.00004.2016
16. Dickens JA, Rutherford EN, Abreu S, Chambers JE, Ellis MO, van Schadewijk A. et al. Novel insights into surfactant protein C trafficking revealed through the study of a pathogenic mutant. The European respiratory journal. 2022;59:2100267 doi:10.1183/13993003.00267-2021
17. Sutton RM, Bittar HT, Sullivan DI, Silva AG, Bahudhanapati H, Parikh AH. et al. Rare surfactant-related variants in familial and sporadic pulmonary fibrosis. Human Mutation. 2022;43:2091-101 doi:10.1002/humu.24476
18. Noble PW, Albera C, Bradford WZ, Costabel U, Glassberg MK, Kardatzke D. et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet (London, England). 2011;377:1760-9 doi:10.1016/S0140-6736(11)60405-4
19. Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U. et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. The New England Journal of Medicine. 2014;370:2071-82 doi:10.1056/NEJMoa1402584
20. Biondini D, Cocconcelli E, Bernardinello N, Lorenzoni G, Rigobello C, Lococo S. et al. Prognostic role of MUC5B rs35705950 genotype in patients with idiopathic pulmonary fibrosis (IPF) on antifibrotic treatment. Respir Res. 2021;22:98 doi:10.1186/s12931-021-01694-z
21. Provenzani A, Leonardi Vinci D, Alaimo M, Di Maria S, Tuzzolino F, Floridia G. et al. Real-world insights into safety, tolerability, and predictive factors of adverse drug reactions in treating idiopathic pulmonary fibrosis with pirfenidone and nintedanib. Ther Adv Drug Saf. 2025;16:20420986251341645 doi:10.1177/20420986251341645
22. Oldham JM, Ma SF, Martinez FJ, Anstrom KJ, Raghu G, Schwartz DA. et al. TOLLIP, MUC5B, and the Response to N-Acetylcysteine among Individuals with Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2015;192:1475-82 doi:10.1164/rccm.201505-1010OC
23. Oldham JM, Allen RJ, Lorenzo-Salazar JM, Molyneaux PL, Ma SF, Joseph C. et al. PCSK6 and Survival in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2023;207:1515-24 doi:10.1164/rccm.202205-0845OC
24. Allen RJ, Oldham JM, Jenkins DA, Leavy OC, Guillen-Guio B, Melbourne CA. et al. Longitudinal lung function and gas transfer in individuals with idiopathic pulmonary fibrosis: a genome-wide association study. Lancet Respir Med. 2023;11:65-73 doi:10.1016/s2213-2600(22)00251-x
25. Hutchinson J, Fogarty A, Hubbard R, McKeever T. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. The European Respiratory Journal. 2015;46:795-806 doi:10.1183/09031936.00185114
26. Maher TM, Bendstrup E, Dron L, Langley J, Smith G, Khalid JM. et al. Global incidence and prevalence of idiopathic pulmonary fibrosis. Respiratory Research. 2021;22:197 doi:10.1186/s12931-021-01791-z
27. Pauchet A. et al. Idiopathic Pulmonary Fibrosis: What do we Know about the Role of Occupational and Environmental Determinants? A Systematic Literature Review and Meta-Analysis. Journal of toxicology and environmental health Part B, Critical reviews. 2022;25:372-392 doi:10.1080/10937404.2022.2131663
28. Wu X, Li W, Luo Z, Chen Y. The minor T allele of the MUC5B promoter rs35705950 associated with susceptibility to idiopathic pulmonary fibrosis: a meta-analysis. Scientific reports. 2021;11:24007 doi:10.1038/s41598-021-03533-z
29. Peljto AL, Selman M, Kim DS, Murphy E, Tucker L, Pardo A. et al. The MUC5B promoter polymorphism is associated with idiopathic pulmonary fibrosis in a Mexican cohort but is rare among Asian ancestries. Chest. 2015;147:460-4 doi:10.1378/chest.14-0867
30. Adegunsoye A, Freiheit E, White EN, Kaul B, Newton CA, Oldham JM. et al. Evaluation of Pulmonary Fibrosis Outcomes by Race and Ethnicity in US Adults. JAMA network open. 2023;6:e232427 doi:10.1001/jamanetworkopen.2023.2427
31. Kurche JS, Cool CD, Blumhagen RZ, Dobrinskikh E, Heinz D, Herrera JA. et al. MUC5B Idiopathic Pulmonary Fibrosis Risk Variant Promotes a Mucosecretory Phenotype and Loss of Small Airway Secretory Cells. Am J Respir Crit Care Med. 2024;210:517-21 doi:10.1164/rccm.202311-2111LE
32. Partanen JJ, Häppölä P, Zhou W, Lehisto AA, Ainola M, Sutinen E. et al. Leveraging global multi-ancestry meta-analysis in the study of idiopathic pulmonary fibrosis genetics. Cell Genom. 2022;2:100181 doi:10.1016/j.xgen.2022.100181
33. Asakura T, Okuda K, Chen G, Dang H, Kato T, Mikami Y. et al. Proximal and Distal Bronchioles Contribute to the Pathogenesis of Non-Cystic Fibrosis Bronchiectasis. American Journal of Respiratory and Critical Care Medicine. 2024;209:374-89 doi:10.1164/rccm.202306-1093OC
34. Milara J, Ballester B, Montero P, Escriva J, Artigues E, Alós M. et al. MUC1 intracellular bioactivation mediates lung fibrosis. Thorax. 2020;75:132-42 doi:10.1136/thoraxjnl-2018-212735
35. Donoghue LJ, Stockwell AD, Neighbors M, Sheng RX, Prabhakaran R, Wolters PJ. et al. Identification of a Genetic Susceptibility Locus for Idiopathic Pulmonary Fibrosis in the 16p Subtelomere Using Whole-Genome Sequencing. Am J Respir Crit Care Med. 2023;207:941-4 doi:10.1164/rccm.202206-1139LE
36. Borie R, Cardwell J, Konigsberg IR, Moore CM, Zhang W, Sasse SK. et al. Colocalization of Gene Expression and DNA Methylation with Genetic Risk Variants Supports Functional Roles of MUC5B and DSP in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2022;206:1259-70 doi:10.1164/rccm.202110-2308OC
37. McDonough JE, Martens DS, Tanabe N, Ahangari F, Verleden SE, Maes K. et al. A role for telomere length and chromosomal damage in idiopathic pulmonary fibrosis. Respiratory research. 2018;19:132 doi:10.1186/s12931-018-0838-4
38. Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP. et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613-20 doi:10.1038/ng.2609
39. Liu T, Ullenbruch M, Young Choi Y, Yu H, Ding L, Xaubet A. et al. Telomerase and telomere length in pulmonary fibrosis. American journal of respiratory cell and molecular biology. 2013;49:260-8 doi:10.1165/rcmb.2012-0514OC
40. Liu L, Sheng Y, Wang C-Y, Liu X, Guo T, Peng H. et al. A novel mutation (p.Y24N) in NHP2 leads to idiopathic pulmonary fibrosis and lung carcinoma chronic obstructive lung disease by disrupting the expression and nucleocytoplasmic localization of NHP2. Biochimica et biophysica acta Molecular basis of disease. 2023;1869:166692 doi:10.1016/j.bbadis.2023.166692
41. Jackson S-R, Lee J, Reddy R, Williams GN, Kikuchi A, Freiberg Y. et al. Partial pneumonectomy of telomerase null mice carrying shortened telomeres initiates cell growth arrest resulting in a limited compensatory growth response. American Journal of Physiology Lung Cellular and Molecular Physiology. 2011;300:L898-909 doi:10.1152/ajplung.00409.2010
42. Wang H, Xu H, Lyu W, Xu Q, Fan S, Chen H. et al. KLF4 regulates TERT expression in alveolar epithelial cells in pulmonary fibrosis. Cell death & disease. 2022;13:435 doi:10.1038/s41419-022-04886-7
43. Frerking I, Günther A, Seeger W, Pison U. Pulmonary surfactant: functions, abnormalities and therapeutic options. Intensive Care Medicine. 2001;27:1699-717 doi:10.1007/s00134-001-1121-5
44. Legendre M, Butt A, Borie R, Debray MP, Bouvry D, Filhol-Blin E. et al. Functional assessment and phenotypic heterogeneity of SFTPA1 and SFTPA2 mutations in interstitial lung diseases and lung cancer. Eur Respir J. 2020;56:2002806 doi:10.1183/13993003.02806-2020
45. Hata A, Chen Y-G. TGF-β Signaling from Receptors to Smads. Cold Spring Harbor Perspectives in Biology. 2016;8:a022061 doi:10.1101/cshperspect.a022061
46. Guan R, Yuan L, Li J, Wang J, Li Z, Cai Z. et al. Bone morphogenetic protein 4 inhibits pulmonary fibrosis by modulating cellular senescence and mitophagy in lung fibroblasts. The European respiratory journal. 2022;60:2102307 doi:10.1183/13993003.02307-2021
47. Wang Y, Zhang L, Huang T, Wu G-R, Zhou Q, Wang F-X. et al. The methyl-CpG-binding domain 2 facilitates pulmonary fibrosis by orchestrating fibroblast to myofibroblast differentiation. The European Respiratory Journal. 2022;60:2003697 doi:10.1183/13993003.03697-2020
48. Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. Journal of the American Society of Nephrology: JASN. 2010;21:2069-80 doi:10.1681/ASN.2010060633
49. Ohkouchi S, Kanehira M, Saigusa D, Ono M, Tazawa R, Terunuma H. et al. Metabolic and Epigenetic Regulation of SMAD7 by STC1 Ameliorates Lung Fibrosis. American journal of respiratory cell and molecular biology. 2022;67:320-33 doi:10.1165/rcmb.2021-0445OC
50. Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G. et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. American journal of respiratory and critical care medicine. 2013;187:397-405 doi:10.1164/rccm.201205-0888OC
51. Herrera J, Beisang DJ, Peterson M, Forster C, Gilbertsen A, Benyumov A. et al. Dicer1 Deficiency in the Idiopathic Pulmonary Fibrosis Fibroblastic Focus Promotes Fibrosis by Suppressing MicroRNA Biogenesis. American journal of respiratory and critical care medicine. 2018;198:486-96 doi:10.1164/rccm.201709-1823OC
52. Königshoff M, Balsara N, Pfaff E-M, Kramer M, Chrobak I, Seeger W. et al. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3:e2142 doi:10.1371/journal.pone.0002142
53. Trinh-Minh T, Chen C-W, Tran Manh C, Li Y-N, Zhu H, Zhou X. et al. Noncanonical WNT5A controls the activation of latent TGF-β to drive fibroblast activation and tissue fibrosis. The Journal of clinical investigation. 2024;134:e159884 doi:10.1172/JCI159884
54. Allen RJ, Porte J, Braybrooke R, Flores C, Fingerlin TE, Oldham JM. et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med. 2017;5:869-80 doi:10.1016/s2213-2600(17)30387-9
55. Liang H, Xu C, Pan Z, Zhang Y, Xu Z, Chen Y. et al. The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22:1122-33 doi:10.1038/mt.2014.42
56. Woodcock HV, Eley JD, Nanthakumar C, Maher TM, Mercer PF, Chambers RC. S51 MTOR regulates TGF-β induced pro-fibrotic gene expression in primary human lung fibroblasts. Thorax. 2016;71:A31-A doi:10.1136/thoraxjnl-2016-209333.57
57. Allen RJ, Guillen-Guio B, Oldham JM, Ma SF, Dressen A, Paynton ML. et al. Genome-Wide Association Study of Susceptibility to Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2020;201:564-74 doi:10.1164/rccm.201905-1017OC
58. Mathai SK, Humphries S, Kropski JA, Blackwell TS, Powers J, Walts AD. et al. MUC5B variant is associated with visually and quantitatively detected preclinical pulmonary fibrosis. Thorax. 2019;74:1131-9 doi:10.1136/thoraxjnl-2018-212430
59. Cocconcelli E, Bernardinello N, Giraudo C, Castelli G, Greco C, Polverosi R. et al. Radiological Assessment in Idiopathic Pulmonary Fibrosis (IPF) Patients According to MUC5B Polymorphism. International journal of molecular sciences. 2022;23:15890 doi:10.3390/ijms232415890
60. van Moorsel CHM, van Oosterhout MFM, Barlo NP, de Jong PA, van der Vis JJ, Ruven HJT. et al. Surfactant protein C mutations are the basis of a significant portion of adult familial pulmonary fibrosis in a dutch cohort. American journal of respiratory and critical care medicine. 2010;182:1419-25 doi:10.1164/rccm.200906-0953OC
61. Klay D, Grutters JC, van der Vis JJ, Platenburg MGJP, Kelder JC, Tromp E. et al. Progressive Disease With Low Survival in Adult Patients With Pulmonary Fibrosis Carrying Surfactant-Related Gene Mutations: An Observational Study. Chest. 2023;163:870-80 doi:10.1016/j.chest.2022.11.002
62. Diaz de Leon A, Cronkhite JT, Yilmaz C, Brewington C, Wang R, Xing C. et al. Subclinical lung disease, macrocytosis, and premature graying in kindreds with telomerase (TERT) mutations. Chest. 2011;140:753-63 doi:10.1378/chest.10-2865
63. Raghu G, Remy-Jardin M, Myers JL, Richeldi L, Ryerson CJ, Lederer DJ. et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. American Journal of Respiratory and Critical Care Medicine. 2018;198:e44-e68 doi:10.1164/rccm.201807-1255ST
64. Shapanis A, Jones MG, Schofield J, Skipp P. Topological data analysis identifies molecular phenotypes of idiopathic pulmonary fibrosis. Thorax. 2023;78:682-9 doi:10.1136/thorax-2022-219731
65. Šelb J, Osolnik K, Kern I, Korošec P, Rijavec M. Utility of Telomerase Gene Mutation Testing in Patients with Idiopathic Pulmonary Fibrosis in Routine Practice. Cells. 2022;11:372 doi:10.3390/cells11030372
66. Wang H, Zhuang Y, Peng H, Cao M, Li Y, Xu Q. et al. The relationship between MUC5B promoter, TERT polymorphisms and telomere lengths with radiographic extent and survival in a Chinese IPF cohort. Scientific reports. 2019;9:15307 doi:10.1038/s41598-019-51902-6
67. Moll M, Peljto AL, Kim JS, Xu H, Debban CL, Chen X. et al. A Polygenic Risk Score for Idiopathic Pulmonary Fibrosis and Interstitial Lung Abnormalities. American journal of respiratory and critical care medicine. 2023;208:791-801 doi:10.1164/rccm.202212-2257OC
68. Karman J, Wang J, Bodea C, Cao S, Levesque MC. Lung gene expression and single cell analyses reveal two subsets of idiopathic pulmonary fibrosis (IPF) patients associated with different pathogenic mechanisms. PLoS One. 2021;16:e0248889 doi:10.1371/journal.pone.0248889
69. Doubkova M, Kriegova E, Littnerova S, Schneiderova P, Sterclova M, Bartos V. et al. DSP rs2076295 variants influence nintedanib and pirfenidone outcomes in idiopathic pulmonary fibrosis: a pilot study. Therapeutic advances in respiratory disease. 2021;15:17534666211042529 doi:10.1177/17534666211042529
70. Wilson AC, Chiles J, Ashish S, Chanda D, Kumar PL, Mobley JA. et al. Integrated bioinformatics analysis identifies established and novel TGFβ1-regulated genes modulated by anti-fibrotic drugs. Scientific reports. 2022;12:3080 doi:10.1038/s41598-022-07151-1
71. Blumhagen RZ, Humphries SM, Peljto AL, Lynch DA, Cardwell J, Bang TJ. et al. MUC5B Genotype and Other Common Variants Are Associated with Computational Imaging Features of Usual Interstitial Pneumonia. Ann Am Thorac Soc. 2025;22:533-40 doi:10.1513/AnnalsATS.202401-022OC
72. Chambers DC, Yusen RD, Cherikh WS, Goldfarb SB, Kucheryavaya AY, Khusch K. et al. The Registry of the International Society for Heart and Lung Transplantation: Thirty-fourth Adult Lung And Heart-Lung Transplantation Report-2017; Focus Theme: Allograft ischemic time. The Journal of Heart and Lung Transplantation: The Official Publication of the International Society for Heart Transplantation. 2017;36:1047-59 doi:10.1016/j.healun.2017.07.016
73. Silhan LL, Shah PD, Chambers DC, Snyder LD, Riise GC, Wagner CL. et al. Lung transplantation in telomerase mutation carriers with pulmonary fibrosis. The European respiratory journal. 2014;44:178-87 doi:10.1183/09031936.00060014
74. Snyder ME, Anderson MR, Benvenuto LJ, Sutton RM, Bondonese A, Koshy R. et al. Impact of age and telomere length on circulating T cells and rejection risk after lung transplantation for idiopathic pulmonary fibrosis. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 2023;42:1666-77 doi:10.1016/j.healun.2023.08.001
75. Hannan SJ, Iasella CJ, Sutton RM, Popescu ID, Koshy R, Burke R. et al. Lung transplant recipients with telomere-mediated pulmonary fibrosis have increased risk for hematologic complications. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2023;23:1590-602 doi:10.1016/j.ajt.2023.06.014
76. Alder JK, Sutton RM, Iasella CJ, Nouraie M, Koshy R, Hannan SJ. et al. Lung transplantation for idiopathic pulmonary fibrosis enriches for individuals with telomere-mediated disease. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 2022;41:654-63 doi:10.1016/j.healun.2021.11.008
77. Phillips-Houlbracq M, Mal H, Cottin V, Gauvain C, Beier F, Sicre de Fontbrune F. et al. Determinants of survival after lung transplantation in telomerase-related gene mutation carriers: A retrospective cohort. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2022;22:1236-44 doi:10.1111/ajt.16893
78. Zaman T, Moua T, Vittinghoff E, Ryu JH, Collard HR, Lee JS. Differences in Clinical Characteristics and Outcomes Between Men and Women With Idiopathic Pulmonary Fibrosis: A Multicenter Retrospective Cohort Study. Chest. 2020;158:245-51 doi:10.1016/j.chest.2020.02.009
79. Ley B, Ryerson CJ, Vittinghoff E, Ryu JH, Tomassetti S, Lee JS. et al. A multidimensional index and staging system for idiopathic pulmonary fibrosis. Annals of Internal Medicine. 2012;156:684-91 doi:10.7326/0003-4819-156-10-201205150-00004
80. van der Vis JJ, Prasse A, Renzoni EA, Stock CJW, Caliskan C, Maher TM. et al. MUC5B rs35705950 minor allele associates with older age and better survival in idiopathic pulmonary fibrosis. Respirology (Carlton, Vic). 2023;28:455-64 doi:10.1111/resp.14440
81. Mota PC, Soares ML, Vasconcelos CD, Ferreira AC, Lima BA, Manduchi E. et al. Predictive value of common genetic variants in idiopathic pulmonary fibrosis survival. Journal of molecular medicine (Berlin, Germany). 2022;100:1341-53 doi:10.1007/s00109-022-02242-y
82. van der Vis JJ, Snetselaar R, Kazemier KM, ten Klooster L, Grutters JC, van Moorsel CHM. Effect of Muc5b promoter polymorphism on disease predisposition and survival in idiopathic interstitial pneumonias. Respirology (Carlton, Vic). 2016;21:712-7 doi:10.1111/resp.12728
83. Al P, Y Z, Te F, Sf M, Jg G, Tj R. et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA. 2013;309:2232-9 doi:10.1001/jama.2013.5827
84. Cai S, Allen RJ, Wain LV, Dudbridge F. Reassessing the association of MUC5B with survival in idiopathic pulmonary fibrosis. Ann Hum Genet. 2023;87:248-53 doi:10.1111/ahg.12522
85. Stuart BD, Lee JS, Kozlitina J, Noth I, Devine MS, Glazer CS. et al. Effect of telomere length on survival in patients with idiopathic pulmonary fibrosis: an observational cohort study with independent validation. The Lancet Respiratory medicine. 2014;2:557-65 doi:10.1016/S2213-2600(14)70124-9
86. Zhang D, Povysil G, Newton CA, Maher TM, Molyneaux PL, Noth I. et al. Genome-wide Enrichment of TERT Rare Variants in Idiopathic Pulmonary Fibrosis Patients of Latino Ancestry. American journal of respiratory and critical care medicine. 2022;206:903-5 doi:10.1164/rccm.202203-0622LE
87. Saito S, Lasky JA, Hagiwara K, Kondoh Y. Ethnic differences in idiopathic pulmonary fibrosis: The Japanese perspective. Respiratory investigation. 2018;56:375-83 doi:10.1016/j.resinv.2018.06.002
88. Clynick B, Corte TJ, Jo HE, Stewart I, Glaspole IN, Grainge C. et al. Biomarker signatures for progressive idiopathic pulmonary fibrosis. The European respiratory journal. 2022;59:2101181 doi:10.1183/13993003.01181-2021
89. Yeo HJ, Ha M, Shin DH, Lee HR, Kim YH, Cho WH. Development of a Novel Biomarker for the Progression of Idiopathic Pulmonary Fibrosis. International journal of molecular sciences. 2024;25:599 doi:10.3390/ijms25010599
90. Son JY, Kim SY, Cho SH, Shim HS, Jung JY, Kim EY. et al. TGF-β1 T869C polymorphism may affect susceptibility to idiopathic pulmonary fibrosis and disease severity. Lung. 2013;191:199-205 doi:10.1007/s00408-012-9447-z
91. Xaubet A, Marin-Arguedas A, Lario S, Ancochea J, Morell F, Ruiz-Manzano J. et al. Transforming growth factor-beta1 gene polymorphisms are associated with disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2003;168:431-5 doi:10.1164/rccm.200210-1165OC
92. Liu B, Zhang X, Liu Z, Pan H, Yang H, Wu Q. et al. A novel model for predicting prognosis in patients with idiopathic pulmonary fibrosis based on endoplasmic reticulum stress-related genes. Cell biology international. 2024;48:483-95 doi:10.1002/cbin.12121
93. Bao Y, Yang S, Zhao H, Wang Y, Li K, Liu X. et al. A prognostic model of idiopathic pulmonary fibrosis constructed based on macrophage and mitochondria-related genes. BMC pulmonary medicine. 2024;24:176 doi:10.1186/s12890-024-02976-0
94. Huang T, Zhou H-F. A Novel 5-Methylcytosine- and Immune-Related Prognostic Signature Is a Potential Marker of Idiopathic Pulmonary Fibrosis. Computational and mathematical methods in medicine. 2022;2022:1685384 doi:10.1155/2022/1685384
95. Lasky JA, Case A, Unterman A, Kreuter M, Scholand MB, Chaudhary S. et al. The Impact of the Envisia Genomic Classifier in the Diagnosis and Management of Patients with Idiopathic Pulmonary Fibrosis. Annals of the American Thoracic Society. 2022;19:916-24 doi:10.1513/AnnalsATS.202107-897OC
96. Wang Y, Yella JK, Ghandikota S, Cherukuri TC, Ediga HH, Madala SK. et al. Pan-transcriptome-based candidate therapeutic discovery for idiopathic pulmonary fibrosis. Therapeutic advances in respiratory disease. 2020;14:1753466620971143 doi:10.1177/1753466620971143
97. Han S, Lee JE, Kang S, So M, Jin H, Lee JH. et al. Standigm ASK™: knowledge graph and artificial intelligence platform applied to target discovery in idiopathic pulmonary fibrosis. Briefings in bioinformatics. 2024;25:bbae035 doi:10.1093/bib/bbae035
98. Kumazoe M, Ogawa F, Hikida A, Shimada Y, Yoshitomi R, Watanabe R. et al. Plant miRNA osa-miR172d-5p suppressed lung fibrosis by targeting Tab1. Scientific reports. 2023;13:2128 doi:10.1038/s41598-023-29188-6
99. Piñeiro-Hermida S, Autilio C, Martínez P, Bosch F, Pérez-Gil J, Blasco MA. Telomerase treatment prevents lung profibrotic pathologies associated with physiological aging. The Journal of cell biology. 2020;219:e202002120 doi:10.1083/jcb.202002120
100. Zhang S, Tong X, Liu S, Huang J, Zhang L, Zhang T. et al. AAV9-Tspyl2 gene therapy retards bleomycin-induced pulmonary fibrosis by modulating downstream TGF-β signaling in mice. Cell death & disease. 2023;14:389 doi:10.1038/s41419-023-05889-8
101. Ligresti G, Caporarello N, Meridew JA, Jones DL, Tan Q, Choi KM. et al. CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI insight. 2019;5:127111 doi:10.1172/jci.insight.127111
102. Tan J, Tedrow JR, Dutta JA, Juan-Guardela B, Nouraie M, Chu Y. et al. Expression of RXFP1 Is Decreased in Idiopathic Pulmonary Fibrosis. Implications for Relaxin-based Therapies. American journal of respiratory and critical care medicine. 2016;194:1392-402 doi:10.1164/rccm.201509-1865OC
103. Bahudhanapati H, Tan J, Dutta JA, Strock SB, Sembrat J, Àlvarez D. et al. MicroRNA-144-3p targets relaxin/insulin-like family peptide receptor 1 (RXFP1) expression in lung fibroblasts from patients with idiopathic pulmonary fibrosis. The Journal of biological chemistry. 2019;294:5008-22 doi:10.1074/jbc.RA118.004910
104. Hou J, Yang Y, Han X. Machine Learning and Single-Cell Analysis Identify Molecular Features of IPF-Associated Fibroblast Subtypes and Their Implications on IPF Prognosis. International journal of molecular sciences. 2023;25:2200957 doi:10.3390/ijms25010094
105. Cui F, Sun Y, Xie J, Li D, Wu M, Song L. et al. Air pollutants, genetic susceptibility and risk of incident idiopathic pulmonary fibrosis. The European respiratory journal. 2023;61:2200777 doi:10.1183/13993003.00777-2022
106. van der Vis JJ, van der Smagt JJ, Hennekam FAM, Grutters JC, van Moorsel CHM. Pulmonary Fibrosis and a TERT Founder Mutation With a Latency Period of 300 Years. Chest. 2020;158:612-9 doi:10.1016/j.chest.2020.03.069
107. Tomos I, Karakatsani A, Manali ED, Kottaridi C, Spathis A, Argentos S. et al. Telomere length across different UIP fibrotic-Interstitial Lung Diseases: a prospective Greek case-control study. Pulmonology. 2022;28:254-61 doi:10.1016/j.pulmoe.2020.11.005
108. Arai N, Nakajima M, Matsuyama M, Matsumura S, Yazaki K, Sakai C. et al. Variations in S100A8/A12 Gene Expression Are Associated with the Efficacy of Nintedanib and Acute Exacerbation Development in Idiopathic Pulmonary Fibrosis Patients. American journal of respiratory cell and molecular biology. 2023;69:242-6 doi:10.1165/rcmb.2023-0054LE
109. Zhu B, Chen C, Xue G, Moyzis RK, Dong Q, Chen C. et al. The SEMA5A gene is associated with hippocampal volume, and their interaction is associated with performance on Raven's Progressive Matrices. Neuroimage. 2014;88:181-7 doi:10.1016/j.neuroimage.2013.11.035
110. Maguire AM, Russell S, Wellman JA, Chung DC, Yu Z-F, Tillman A. et al. Efficacy, Safety, and Durability of Voretigene Neparvovec-rzyl in RPE65 Mutation-Associated Inherited Retinal Dystrophy: Results of Phase 1 and 3 Trials. Ophthalmology. 2019;126:1273-85 doi:10.1016/j.ophtha.2019.06.017
Author contact
![]() Corresponding authors: Yue Hu, Yinghua Ying; Emails: huyue88edu.cn; yingyinghuaedu.cn.
Corresponding authors: Yue Hu, Yinghua Ying; Emails: huyue88edu.cn; yingyinghuaedu.cn.

 Global reach, higher impact
Global reach, higher impact