Impact Factor
ISSN: 1449-1907
Int J Med Sci 2025; 22(11):2545-2559. doi:10.7150/ijms.108763 This issue Cite
Review
Unraveling the Role of N6-Methylation Modification: From Bone Biology to Osteoporosis
Laboratory of Endocrinology and Metabolism/Department of Endocrinology and Metabolism, Rare Disease Center, West China Hospital, Sichuan University, No. 37, Guoxue Xiang, Chengdu 610041, China
Received 2024-12-13; Accepted 2025-4-25; Published 2025-5-8
Abstract
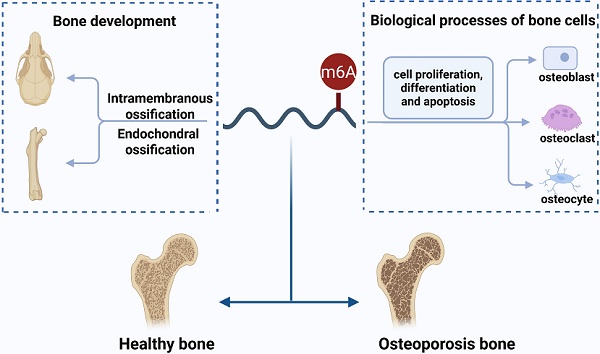
N6-methyladenosine (m6A) is the most abundant and reversible epitranscriptomic modification in eukaryotes, playing a pivotal role in regulating various RNA metabolic processes, including splicing, nuclear export, translation and degradation. Emerging evidence indicates that m6A modification is indispensable in biological processes of bone cells such as proliferation, differentiation and apoptosis. Given its pivotal influence on osteoblastogenesis and osteoclastogenesis, m6A modification, particularly via METTL3, has attracted considerable attention in osteoporosis (OP). In this review, we probe the function of m6A modification in intramembranous and endochondral ossification. Furthermore, we summarize the regulatory role of m6A modification in various biological processes in osteoblasts, osteoclasts and osteocytes, focusing on its potential signaling pathways in osteoblast and osteoclast differentiation. Specifically, m6A modulates osteoblast differentiation predominantly through signaling pathways such as Wnt/β-catenin, PI3K/AKT, and BMP/Smad. Concurrently, it regulates osteoclast differentiation and maturation via the RANKL/RANK pathway and its downstream signaling mechanisms. We also discuss recent discoveries that m6A modification regulates OP and further explore its potential clinical value in diagnosing and treating OP. Collectively, m6A modification serves as a crucial regulatory factor in bone metabolism, and a comprehensive understanding of the molecular mechanisms of m6A modification in bone biology is expected to provide new targets for treating OP.
Keywords: N6-methyladenosine, m6A modification, bone development, bone cells, osteoporosis
1. Introduction
Epigenetics refers to heritable changes in gene expression and function without alterations in the DNA sequence, mainly involving histone modifications, DNA methylation, and non-coding RNAs[1]. Recently, RNA methylation modifications, particularly N6-methyladenosine (m6A) methylation, have attracted widespread attention due to the popularity of high-throughput sequencing technologies[2]. m6A is the most abundant methylation modification in eukaryotic messenger RNAs (mRNAs) and also exists in non-coding RNAs such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and long non-coding RNAs (lncRNAs)[3]. Using sequencing technologies such as methylated RNA immunoprecipitation sequencing (MeRIP-Seq), researchers have discovered that m6A modification is significantly enriched in coding sequences (CDS), 3' untranslated regions (3' UTRs), near stop codons, and long internal exons[4-6]. m6A modification affects a variety of biological functions by regulating processes such as RNA transcription, splicing, translation and degradation[7]. Additionally, increasing evidence suggests that m6A modification plays essential roles in the pathogenesis of various diseases, including OP, osteoarthritis, and bone cancers, providing new therapeutic targets for these bone-related diseases[8, 9].
The skeleton supports and protects the body, with the ability of dynamic remodeling. This complex process depends on the coordination among bone cells, including osteoblasts, osteoclasts, and osteocytes[10]. Osteoblasts form and mineralize bone, while osteoclasts break down and resorb bone. Osteocytes, embedded within the bone matrix, regulate the activity of osteoblasts and osteoclasts by sensing mechanical stimuli and producing endocrine factors[11]. However, aging or pathological conditions can disrupt these regulatory mechanisms, leading to abnormal bone remodeling and eventually causing osteoporosis (OP)[12]. Studies have revealed that m6A modification plays a significant role in the pathophysiology of bone[13, 14]. Therefore, further research clarifying how m6A modification affects bone remodeling could help identify new targets for preventing and treating OP.
In this review, we explore the effect of m6A modification on the bone development and biological processes of bone cells (osteoblasts, osteoclasts, osteocytes). Particularly, we focus on the potential pathways involved in the regulation of bone cells by m6A modification. Moreover, we discuss the implications of m6A modification for the clinical diagnosis and treatment of OP.
2. m6A modification: writers, erasers, readers
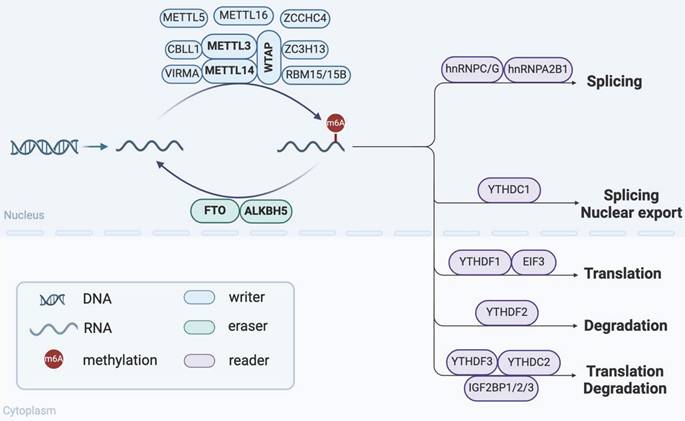
As a dynamic and reversible epigenetic mark, m6A is the most abundant post-transcriptional RNA modification in eukaryotes. m6A modification is regulated primarily by three types of regulators: m6A methyltransferases ("Writers"), m6A demethylases ("Erasers") and m6A-binding proteins ("Readers") (Figure 1)[15]. These regulators are responsible for adding, removing, and recognizing m6A methylation sites, respectively, thus playing crucial roles in various biological processes.
Components and regulatory mechanisms of m6A modification. The regulation of m6A modification depends on the coordination of m6A methyltransferases ("Writers"), m6A demethylases ("Erasers") and m6A-binding proteins ("Readers"). They collectively regulate various RNA biological processes, including splicing, nuclear export, translation, and degradation. (Created with BioRender.com.)
2.1 m6A methyltransferases (“Writers”)
The methyltransferase complex (MTC), which is essential for m6A modification, consists of core components and other regulators[15]. Methyltransferase like 3 (METTL3) is the only catalytic subunit of MTC that transfers the methyl group of S-adenosylmethionine (SAM) to adenosine[16]. The combination of methyltransferase like 14 (METTL14) and METTL3 forms a stable heterodimer complex to enhance its catalytic activity. Wilms tumor 1 associated protein (WTAP) recruits this heterodimer to mRNA specific sites to regulate m6A levels[17]. In addition, regulatory factors such as RNA-binding motif protein 15/15B (RBM15/15B), zinc finger CCCH-type containing 13 (ZC3H13), vir-like m6A methyltransferase associated (VIRMA), and Cbl proto-oncogene like 1 (CBLL1) also regulate this process[18]. Of note, Methyltransferase like 5 (METTL5), Methyltransferase like 16 (METTL16), and zinc finger CCHC-type containing 4 (ZCCHC4) function independently of the MTC. METTL5 and ZCCHC4 mediate m6A modification of 18S and 28S rRNA, respectively[19, 20], while METTL16 is involved in m6A modification of U6 small nuclear RNAs (snRNAs), lncRNAs and precursor messenger RNAs (pre-mRNAs)[21, 22]. The cooperative action of these methyltransferases ensures the efficient operation of m6A modification.
2.2 m6A demethylases (“Erasers”)
The m6A demethylases mainly include fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), which can reverse m6A modification and participate in the dynamic regulation of m6A levels. As the first reported m6A demethylase, FTO plays an important role in regulating obesity and fat metabolism[23]. FTO-deficient osteoblasts are more susceptible to genotoxic injury under high-fat diet-induced metabolic stress[24]. ALKBH5 is recognized as the second m6A demethylase, which significantly affects mRNA export and RNA metabolism by regulating m6A levels[25]. RNA binding protein RBM33 has been confirmed to form a complex with ALKBH5, thereby regulating the demethylase activity of ALKBH5[26].
2.3 m6A-binding proteins (“Readers”)
The m6A-binding protein, also known as m6A reader, affects a variety of metabolic processes of target RNA by specifically recognizing and binding to the m6A site. YT521-B homology (YTH) family proteins are essential members of the m6A readers. In the cytoplasm, YTHDF1 binds to initiation factors in the cytoplasm to promote mRNA translation, while YTHDF2 plays a dominant role in accelerating mRNA degradation[27, 28]. YTHDF3 is considered a collaborator of YTHDF1 and YTHDF2, playing a pivotal role in regulating mRNA dynamic balance[29]. Eukaryotic initiation factor 3 (EIF3), as a translation initiation factor, regulates translation efficiency by recognizing m6A-modified mRNA[30]. Moreover, nucleus-located YTHDC1 promotes selective splicing and nuclear export of m6A-modified mRNA by interacting with splicing factor SRSF3[31, 32]. YTHDC2 is closely associated with mammalian spermatogenesis, promoting translation and reducing the stability of target mRNA[33]. Besides the YTH family proteins, the insulin-like growth factor 2 mRNA binding proteins (IGF2BP)1/2/3 also maintains target mRNA stability and translational efficiency in an m6A-dependent manner[34]. It has been reported that IGF2BP3 binds to and stabilizes Beclin mRNA, which promotes the osteogenic potential of bone marrow mesenchymal stem cells (BMSCs)[35]. Heterogeneous nuclear ribonucleoproteins (hnRNPs) family also plays important roles as m6A readers. hnRNPA2B1 facilitates the processing of pri-miRNAs by interacting with DGCR8[36]. HNRNPC and HNRNPG participate in splicing pre-mRNA via m6A-mediated RNA structural switches[37, 38].
Taken together, the functions of m6A regulatory factors are complex and diverse, mainly involving various biological processes such as cell proliferation, immune responses, and tumorigenesis. However, the known m6A regulatory factors are limited, and their specific mechanisms and interactions in various cell types and biological processes still require further research.
3. m6A modification and bone development
m6A modification regulates RNA post-transcriptional processes, influencing cell fate, tissue development and the maintenance of organ function[39]. Accumulating studies documented that m6A modification is crucial in embryonic development, with its dysregulation potentially leading to severe developmental defects or even embryonic lethality. Several pieces of evidence have identified that homozygous deletions of m6A regulatory factors, such as METTL3, METTL14 or WTAP, result in embryonic lethality in mice[40, 41]. The deletion of METTL3, METTL5 or METTL14 impairs the differentiation ability of mouse embryonic stem cells (mESCs), highlighting the pivotal role of m6A modification in maintaining mESCs proliferation and differentiation[42-44]. Furthermore, abnormal m6A modification also affects the development of multiple organs, including the liver, nerve and bone. For example, the conditional knockout of METTL3 in the embryonic liver impairs liver development and maturation[45]. The m6A reader YTHDF2 is involved in neural development and differentiation[46]. These findings offer new insights into the mechanisms underlying organ development.
Bone development is mainly achieved through intramembranous and endochondral ossification[47]. Intramembranous ossification primarily occurs in the cranium, facial bones, and clavicles. Several studies have confirmed that m6A modification is crucial in craniofacial development. METTL3-mediated m6A modification of PSEN1 regulates craniofacial development in vertebrates via the Wnt/β-catenin signaling pathway[48]. Mutations in the FTO gene result in patients with multiple malformations, which are characterized by postnatal growth retardation, craniofacial anomalies and finger deformities[49]. FTO knockout mice also exhibit a similar growth retardation phenotype, including decreased bone mineral density (BMD)[50]. In addition, normal closure of cranial sutures is crucial for intramembranous ossification of the skull[51]. METTL5 mutations have been identified as a trigger for autosomal recessive skeletal dysplasia in patients[52]. Lei et al. further showed that METTL5-deficient mice exhibit delayed suture closure and cleidocranial dysplasia[53]. Xu et al. demonstrated that cranial sutures coordinate the intramembranous ossification process of the cranial vault through METTL3. Specifically, the deletion of METTL3 in Ctsk+ calvarial stem cells resulted in delayed suture closure and skull development by inhibiting the Hedgehog signaling pathway[54]. Endochondral ossification is a process driven by the proliferation, differentiation and hypertrophy of chondrocytes, and dysfunction of chondrocytes leads to abnormal bone development[55]. In recent years, studies have found that m6A modification significantly influences the chondrocyte physiological processes and endochondral ossification. He et al. revealed that the METTL3/m6A/YTHDF1/Dmp1 axis is involved in endochondral ossification. METTL3 upregulates the expression of the target gene Dmp1 through YTHDF1-mediated m6A modification, thus promoting chondrocyte proliferation and hypertrophic differentiation[56]. METTL3 methylates circRNA3634 and upregulates the downstream target gene mitogen-activated protein kinase 1 (MAPK1), promoting the proliferation and differentiation of antler chondrocytes[57]. Moreover, METTL3 is essential for maintaining endochondral ossification of condylar cartilage. METTL3 deficiency in Acan+ chondrocytes leads to abnormal mandibular condylar chondrogenesis by upregulating the expression of Lats1, a key molecule of the Hippo/Yap1 pathway[58].
Bone development is regulated by key transcription factors such as Runx2, Osterix, SOX9, and ATF4[59]. Numerous studies have found that m6A modification regulates these transcription factors via m6A regulatory factors, affecting osteogenesis and chondrogenesis commitment. Specifically, METTL3, METTL14, WTAP, ALKBH5, YTHDF1 and YTHDF3 promote osteoblast differentiation by directly or indirectly upregulating Runx2 expression[60-65]. Additionally, m6A modification mediated by METTL3 and YTHDF3 also enhances the expression of Osterix, which cooperates with Runx2 to regulate the osteogenic process[63, 66]. However, FTO and YTHDC2 negatively regulate Runx2 expression, inhibiting osteogenic differentiation[67, 68]. SOX9 is a key transcription factor mediating chondrogenic differentiation and bone development. METTL3 has been proven to target the 3'UTR of SOX9 mRNA and regulate SOX9 translation during cartilage differentiation[69]. Furthermore, another study reported that tension-stimulated METTL3 inhibited extracellular matrix synthesis in endplate chondrocytes by mediating SOX9 m6A modification and inducing its degradation[70].
Collectively, m6A modification regulates intramembranous and endochondral ossification and maintains normal bone development by modulating relevant transcription factors and signaling pathways.
4. The regulatory effect of m6A modification in bone cells
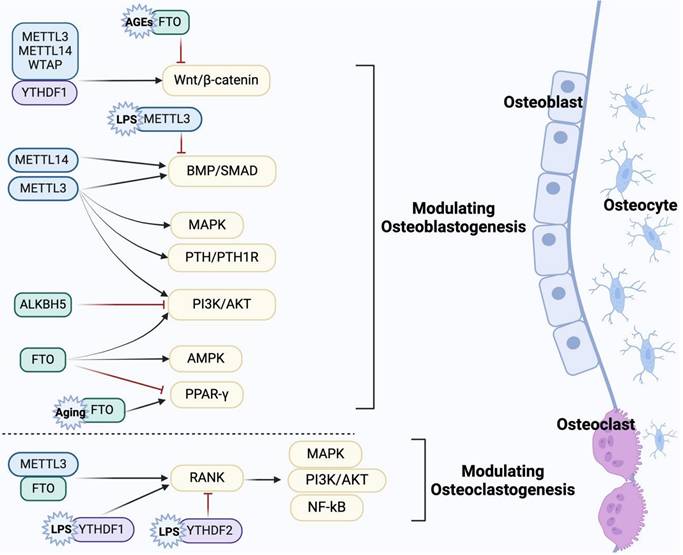
The coordination of bone cells (osteoblasts, osteoclasts, osteocytes) is responsible for the dynamic remodeling of bone. Existing evidence suggests that m6A modification not only regulates the proliferation and apoptosis of bone cells (Table 1), but also participates in the differentiation of osteoblasts and osteoclasts via multiple signaling pathways (Figure 2). Hence, clarifying the precise regulatory mechanism of m6A modification on the biological processes of bone cells contributes to a better understanding of bone metabolism and bone-related diseases.
The role of m6A regulatory factors in the biological processes of bone cells
| m6A regulators | Expression | Target genes | Function | References |
|---|---|---|---|---|
| Osteoblast | ||||
| METTL3 | ↓ | Grp78 | Promoting osteoblast apoptosis and inhibiting proliferation and differentiation | [71] |
| FTO | ↑ | Hspa1a/NF-κB | Inhibiting osteoblast genotoxic-induced apoptosis | [24] |
| Osteoclast | ||||
| FTO | ↑ | CDK2, Cyclin A2 and DNA damage-related proteins | Promoting osteoclast proliferation and inhibiting apoptosis | [116] |
| METTL3 | ↓ | Nos2 | Inhibiting osteoclast differentiation and promoting apoptosis | [117] |
| METTL3/YTHDF2 | ↓ | Atp6v0d2 | Inhibiting osteoclast differentiation and activity | [118] |
| METTL3 | ↑ | CTSK | Promoting osteoclast migration | [119] |
| Osteocyte | ||||
| METTL3 | ↓ | - | Inhibiting the number and development of osteocyte | [54] |
The pathways involved in the regulation of osteoblast and osteoclast differentiation by m6A modification. m6A regulators are involved in osteoblast differentiation via Wnt/β-catenin, BMP/Smad, PI3K/AKT, and other potential signaling pathways, while regulating osteoclast differentiation via RANKL/RANK and its downstream pathways. In addition, under pathological conditions such as aging, AGE and LPS, m6A regulators also affect bone-resorbing osteoclasts and bone-forming osteoblasts through relevant pathways. (Created with BioRender.com.)
4.1 The regulatory effect of m6A modification on osteoblasts
Osteoblasts undergo multiple stages during bone formation, including proliferation, differentiation, mineralization and apoptosis. m6A modification is involved in maintaining the dynamic balance of bone by regulating these processes. It was reported that METTL3 knockdown mediates osteoblast proliferation, differentiation and apoptosis by activating the endoplasmic reticulum stress signaling pathway[71]. FTO targets the HSPA1a/NF-κB signaling axis, inhibiting genotoxicity-induced osteoblast apoptosis[24]. Osteoblast progenitor cells differentiate into mature osteoblasts through four stages: proliferation, differentiation, matrix synthesis, and mineralization. During this process, osteoblasts synthesize and secrete the bone matrix, facilitating mineral deposition to form new bone tissue[72]. Osteoblast differentiation is critical for proper bone formation. Key transcription factors, such as Runx2 and Osterix, play central roles in this process, with their functions driven by signaling pathways such as Wnt/β-catenin and bone morphogenic protein (BMP)[73]. Proper osteoblast differentiation ensures bone matrix synthesis and mineralization, whereas its dysfunction may lead to decreased bone mass or skeletal developmental defects[73, 74]. Of note, m6A modification plays a crucial role in the differentiation of osteoblasts. Herein, we systematically summarize the osteogenic differentiation pathways related to m6A modification.
4.1.1 m6A regulates osteogenic differentiation by Wnt/β-catenin pathway
It is well known that the Wnt signaling pathway plays a pivotal role in osteoblast-mediated bone formation, and β-catenin is an essential regulatory factor in the canonical Wnt pathway[75]. Accumulating evidence suggests that activation of the Wnt/β-catenin signaling pathway promotes the proliferation, differentiation and maintenance of osteoblasts[76]. Recent studies have emphasized the potential role of m6A modification in osteogenic differentiation by regulating the Wnt/β-catenin pathway. Wu et al. demonstrated that METTL3 overexpression partially rescues the osteogenic potential of BMSCs in OP rats by activating the Wnt signaling pathway[77]. METTL3 can also activate the Wnt/β-catenin/c-Myc signaling pathway in a YTHDF2-dependent manner in the inflammation induced by lipopolysaccharides (LPS), contributing to the biological behaviors of osteoblasts[78]. METTL14 upregulates TCF1 expression by m6A modification. As a key transcription factor in the Wnt/β-catenin pathway, TCF1 enhances Runx2 expression to alleviate OP, suggesting that METTL14 may regulate the Wnt/β-catenin pathway[65]. WTAP-mediated m6A modification of miR-181a and miR-181c contributes to the osteogenesis of BMSCs by suppressing the expression of the target secretory frizzled-related protein 1 (SFRP1)[79]. SFRP1 has been confirmed as a negative regulator of the Wnt pathway involved in the lineage differentiation of BMSCs[80, 81]. This suggests that WTAP may indirectly modulate the Wnt signaling pathway, thereby playing a significant role in bone homeostasis. Furthermore, m6A erasers and readers also engage in the osteogenic differentiation related to the Wnt pathway. Gao et al. showed that YTHDF1 activates Wnt/β-catenin signaling through autophagy, thereby promoting osteogenic differentiation and proliferation[82]. FTO methylates sclerostin (SOST) transcripts, thereby mediating osteogenic impairment induced by advanced glycation end products (AGEs) via the Wnt signaling pathway[83].
4.1.2 m6A regulates osteogenic differentiation by BMP/Smad pathway
The BMP signaling pathway plays a critical biological role in bone development and fracture healing[84]. As one of the important subtypes of the BMP family, BMP2 binds to its receptor BMPR to activate downstream Smad1/5/8 phosphorylation, which mediates osteoblast differentiation by up-regulating the expression of osteogenic-specific genes such as Runx2, Osterix[85, 86]. Liu et al. found that BMP2 is a downstream target of METTL3, and m6A activity is inhibited by piRNA-36741. piRNA-36741 can enhance the expression of BMP2 by binding to METTL3, thereby promoting the osteogenic differentiation of BMSCs[87]. Meanwhile, METTL3 positively correlates with receptor BMPR1B expression. METTL3 promotes competitive binding of LINC00657 to miR-144-3p and targets the LINC00657/miR-144-3p/BMPR1B axis to initiate osteoblast differentiation and bone formation[88]. In addition, the low METTL14 can downregulate the expression of Smad1, Smad5 and Smad8 in BMSCs, impairing osteogenic differentiation by inhibiting the m6A modification of Smad1[89]. Smad7 and Smurf1 are inhibitors of the BMP signaling pathway. Smad7 recruits E3 ubiquitin ligases such as Smurf1, leading to ubiquitinated degradation of the phosphorylated receptor[90, 91]. METTL3 knockdown promotes the stability of Smad7 and Smurf1 in MC3T3-E1 cells, thereby negatively regulating Smad-dependent signaling and osteogenic differentiation under inflammatory conditions[92]. These studies revealed that m6A modification in osteogenic differentiation is related to BMP/Smad pathways.
4.1.3 m6A regulates osteogenic differentiation by PI3K/AKT pathway
The phosphoinositide 3-kinase/AKT (PI3K/AKT) pathway is a key signaling pathway that regulates cell proliferation, differentiation and survival[93]. PI3K phosphorylates PIP2 to produce PIP3, which acts as a second messenger to recruit and activate the downstream serine/threonine-protein kinase AKT, mediating various cellular physiological and pathological processes[94, 95]. In the skeletal system, activation of the PI3K/AKT pathway is involved in osteoblast proliferation, differentiation and apoptosis, thereby facilitating the progression of OP[96, 97]. For example, an in vitro study suggested that lower levels of METTL3 downregulate the PI3K/AKT pathway, triggering impaired osteogenic differentiation in BMSCs[98]. Li et al. found that mice with specific knockout of ALKBH5 in BMSCs have significantly higher bone mass than littermate controls. They revealed that ALKBH5 negatively regulates osteogenic differentiation in BMSCs by targeting protein arginine methyltransferase 6 (PRMT6), which inactivates the downstream PI3K/AKT pathway[99]. Furthermore, overexpressed FTO in ovariectomy (OVX) mice is inhibited by miR-22-3p derived from BMSCs, which promotes osteogenic differentiation through the MYC/PI3K/AKT pathway[100]. Besides, the intact insulin-like growth factor (IGF)-induced PI3K/AKT signaling cascade is crucial for osteoblast differentiation and bone development[101]. As a key component of the IGF signaling pathway, IGF-2 mediates signal transduction through the IGF receptor, subsequently activating the PI3K/AKT signaling pathway[101]. It has been reported recently that the glutamine-αKG axis suppresses the translation efficiency of IGF2 in an m6A modification-dependent manner, thereby modulating the osteo/odontogenic differentiation of mesenchymal adult stem cells (m-ASCs)[102]. These results indicate that m6A plays an important role in osteogenic differentiation by the PI3K/AKT signaling pathway.
4.1.4 m6A regulates osteogenic differentiation by MAPK pathway
The MAPK pathway is crucial for receiving extracellular stimuli and conveying these signals into the intracellular environment[103]. ERKs, JNKs and p38 constitute the classical MAPK signaling pathway, collaboratively engaging in cellular responses to diverse external stimuli via distinct functions and regulatory mechanisms[104, 105]. In MC3T3-E1 cells, METTL3 depletion significantly increases the phosphorylation levels of ERK, JNK and p38 within the MAPK signaling pathway, thereby activating the inflammatory response of osteoblasts[92]. Intriguingly, Song et al. reported that METTL3-mediated m6A modification can increase the phosphorylation levels of p38, JNK and ERK and subsequently activate the MAPK signaling pathway, participating in the osteogenic differentiation of human adipose derived stem cells (ASCs)[106]. Lin et al. revealed that overexpression of METTL3 involves in osteoblast ferroptosis by activating the ASK1/p38 signaling pathway under high glucose and high fat conditions[107]. In contrast, they found inhibition of p38 phosphorylation significantly alleviated osteoblast dysfunction[107]. These findings suggest that m6A modification may be involved in the pathological processes of osteoblasts and the development of bone diseases through abnormal dysregulation of ERKs, JNKs and p38. This provides evidence to support further exploration of the precise mechanisms by which the m6A and MAPK pathways are involved in osteoblast differentiation, as well as the identification of novel therapeutic targets.
4.1.5 m6A regulates osteogenic differentiation by other potential pathways
Notably, m6A modification is also involved in osteogenic differentiation by regulating other pathways such as adenosine monophosphate-activated protein kinase (AMPK) and parathyroid hormone/parathyroid hormone 1 receptor (PTH/PTH1R). An in vitro study confirmed that the m6A demethylase FTO promoted the osteogenic differentiation of C3H10T1/2 cells by activating the AMPK signaling pathway and inducing mild endoplasmic reticulum stress through a BMP2-induced FTO/p-AMPK positive feedback loop[108]. Chen et al. reported that FTO overexpression during aging inhibits osteoblast differentiation and OP progress by increasing the expression of peroxisome proliferator-activated receptor γ (PPARγ)[109]. Nevertheless, another study showed that FTO could target the 3' UTR of PPARγ mRNA and negatively regulate the PPARγ signaling pathway, thereby increasing the expression of osteogenic marker genes such as alkaline phosphatase (ALP) and osteopontin (OPN)[110]. The inconsistency requires further studies to explore the interaction between FTO and PPARγ signaling pathway in OP. In osteoblasts and osteocytes, the PTH signaling pathway is essential for regulating bone metabolism and alleviating age-related bone loss[111, 112]. METTL3 deficiency in BMSCs has been shown to inhibit osteogenesis in mice. Mechanistically, the deficiency of METTL3 can block PTH/PTH1R signaling by reducing the translation efficiency of PTH1R, ultimately leading to the differentiation of BMSCs into adipocytes instead of osteoblasts[113]. These findings provide new mechanistic insights into the role of m6A modification in osteogenic differentiation.
4.2 The regulatory effect of m6A modification on osteoclasts
Osteoclasts are critical cells in bone resorption and are responsible for bone development, growth, and remodeling[114]. Notably, mononuclear osteoclast precursors have limited resorptive capacity, which significantly increases only after they undergo a series of complex processes, including migration, recognition, intercellular adhesion, and membrane fusion, to differentiate into multinucleated mature osteoclasts[114]. This differentiation is primarily regulated by the RANK/RANKL/OPG signaling pathway. Mature osteoclasts establish a sealing zone and secrete acidic substances and proteases to degrade the bone matrix, thus completing the resorption process[115]. A recent study has demonstrated that FTO is mainly involved in the proliferation and apoptosis of osteoclast precursors by upregulating the expression of CDK2 and cyclin A2 while inhibiting the expression of DNA damage-related proteins[116]. However, FTO inhibitor treatment significantly reduces the number of multinucleated osteoclasts and bone resorption ability [116]. Li et al. found that METTL3 inhibits osteoclast apoptosis via iNOS/NO-mediated mitochondrial dysfunction under inflammatory conditions[117]. Intriguingly, an in vitro study showed that knockdown of METTL3 and YTHDF2 upregulated the stability of Atp6v0d2 mRNA, leading to the formation of multinucleated osteoclasts, without affecting the proliferation of osteoclast precursor[118]. These findings suggest that m6A modification is involved in the regulation of osteoclast proliferation, apoptosis and activity. Migration of osteoclasts during bone resorption is critical for the progress of OP. Recently, an in vitro study reported that follicle-stimulating hormone (FSH) enhances the m6A activity of METTL3 through cyclic-AMP response element-binding protein (CREB), which increases cathepsin K (CTSK) mRNA stability and subsequently promotes osteoclast migration[119]. This result provides a new target for utilizing m6A in the treatment of postmenopausal OP. Moreover, m6A modification is closely associated with osteoclast differentiation under different pathological conditions. Early growth response 1 (EGR1) was shown to promote METTL3 transcription and enhance the chitinase 3-like protein 1 (CHI3L1) m6A methylation, thus stimulating osteoclast differentiation in OVX mice[120]. Shen et al. reported that toll-like receptor 4 (TLR4) promotes high glucose and high fat-induced osteoclast differentiation by inhibiting FTO-mediated m6A modification, suggesting that m6A modification participated in developing diabetic bone loss[121]. Specifically, METTL14 inhibited bone-resorbing osteoclasts by binding to the methylation functional site of nuclear factor of activated T-cells cytoplasmic 1 (NFATc1)[122]. In addition, METTL14 enhances glutathione peroxidase 4 (GPX4) mRNA stability under the influence of human antigen R (HuR), thereby inhibiting osteoclast formation and bone resorption[123]. Similarly, YTHDC1 enhanced protein tyrosine phosphatase non-receptor type 6 (PTPN6) expression in an m6A-HuR-dependent manner to inhibit osteoclast differentiation and OP progression[124]. Liu et al. reported that knockdown of WTAP significantly increases the expression of osteoclast-related genes[125], indicating that WTAP-mediated m6A modification serves an inhibitory role in osteoclast differentiation.
Receptor activator of NF-κB ligand (RANKL) is a critical factor in osteoclastogenesis. Upon binding to its receptor activator of NF-κB (RANK) on osteoclast precursors, RANKL recruits various adaptor molecules, particularly tumor necrosis factor receptor-associated factor 6 (TRAF6). This recruitment activates downstream signaling pathways, including NF-κB, MAPK and PI3K/AKT, thus promoting the maturation of osteoclasts and facilitating bone resorption[126, 127]. He et al. found that METTL3 knockdown prevents the nuclear export of TRAF6 mRNA, leading to the inactivation of downstream signaling pathways of RANKL/RANK and inhibiting the differentiation and formation of osteoclasts[118]. They also demonstrated that YTHDF1 is involved in inflammatory osteoclastogenesis. YTHDF1 depletion can inactivate the NF-κB, MAPK and PI3K/AKT pathways by affecting the stability of the RANK mRNA and ultimately inhibits LPS-induced osteoclastogenesis[128]. In contrast, an in vitro study reported that the YTHDF2 knockdown in osteoclast precursor promotes osteoclastogenesis under inflammatory stimuli, primarily by activating the NF-κB and MAPK signaling pathways[129]. Of note, the NF-κB pathway is one of the most important pathways mediating osteoclast differentiation following the binding of RANK and RANKL[130]. FTO activates the NF-κB signaling pathway, promoting osteoclast differentiation and bone resorption. However, treatment with NF-κB inhibitor attenuates the positive regulatory effect of FTO[131]. These studies provide evidence that m6A modification is involved in regulating osteoclast differentiation via the RANKL/RANK and its downstream pathways. Additionally, Osteoprotegerin (OPG) acts as a decoy receptor for RANKL, inhibiting osteoclastogenesis by preventing the binding of RANKL to RANK[115]. Thus, the RANKL/OPG ratio is essential for regulating bone resorption. Studies have shown that m6A modification influences this ratio. Specifically, overexpression of METTL14 decreases RANKL levels while increasing OPG expression in osteoblasts, thereby enhancing osteoclast differentiation[65]. IGF2BP2 has also been shown to affect the RANKL/OPG ratio under inflammatory conditions[132].
4.3 The regulatory effect of m6A modification on osteocytes
Osteocytes, the most abundant cell type in bone tissue, possess unique dendritic structures that interconnect through a network of canaliculi[133]. These structures enable osteocytes to sense and respond to mechanical loads to maintain the dynamic balance of bone mass. Studies have indicated that epigenetic modification, particularly histone modifications, plays a significant role in the development and function of osteocytes. For example, Datta and his team established the initial link between mechanical loading and epigenetic changes in bone. They found that mechanical loading epigenetically regulates the expression of the zinc finger of the cerebellum 1 (ZIC1), which influences the transcriptional activity of osteocyte markers[134]. Stegen et al. revealed that the absence of the oxygen sensor prolyl hydroxylase 2 (PHD2) in osteocytes downregulates SOST expression through SIRT1-mediated epigenetic modification, thereby activating the Wnt/β-catenin signaling pathway to increase bone mass[135]. Moreover, the histone demethylase Utx in osteocytes positively regulates their differentiation and maturation by removing H3K27me3-mediated histone modification[136]. Recently, m6A modification has been shown to regulate osteocyte structure and function. For example, Xu et al. demonstrated that the specific knockout of METTL3 in non-osteoclastic Ctsk+ lineage cells resulted in a reduced number of osteocytes with impaired development, characterized by fewer canaliculi[54]. Nevertheless, the specific regulatory mechanism of m6A modification in osteocytes remains largely unknown. As such, further research is required to clarify the complex relationship between m6A modification and osteocytes, which could pave the way for novel therapeutic strategies targeting OP and other skeletal diseases.
4.4 The regulatory effect of m6A modification on other cells
Besides osteoblasts and osteoclasts, other cell types, such as vascular endothelial cells and immune cells, also play crucial roles in bone remodeling. Macrophages, as a special cell type within bone tissue, significantly influence immune regulation and inflammatory responses through their polarization[137]. Under inflammatory conditions, downregulation of WTAP can promote macrophage polarization towards the M2 phenotype, thereby enhancing osteogenic differentiation of BMSCs[138]. METTL3 regulates macrophage polarization by mediating m6A modification of HDAC5 and further participates in bone repair processes[139]. These evidences suggest that m6A modification plays a critical role in coordinating macrophage-mediated bone immune responses. Furthermore, pyroptosis contributes to the development of OP by modulating the inflammatory immune microenvironment[140]. Tang et al. reported that overexpression of METTL14 inhibits macrophage-osteoclast differentiation and macrophage pyroptosis. Mechanistically, METTL14 targeting the METTL14/HOXA5/WNK1 axis to suppress NLRP3-dependent pyroptosis, thereby alleviating OP[141]. Yang et al. applied a diabetes-associated periodontitis model to demonstrate that METTL3 induces macrophage pyroptosis by methylating NLRP3 mRNA, thereby accelerating alveolar bone loss[142].
Angiogenesis, which is closely linked to osteogenesis, is essential for maintaining bone homeostasis[143]. The vascular endothelial growth factor A (VEGFA) signaling pathway mediated by endothelial cells plays a key role in bone vascularization[144]. An in vitro study indicated that METTL3 modulates the expression of VEGFA and its splice variants during osteogenic differentiation of BMSCs, highlighting a potential role for m6A in regulating angiogenesis through endothelial cell-associated genes[98]. Jiang et al. further revealed that METTL3 is a critical regulator of angiogenesis in endothelial progenitor cells (EPCs). METTL3 participates in the proliferation, migration, and tube formation of EPCs through the PI3K/AKT signaling pathway, ultimately promoting distraction osteogenesis-related ossification[145]. Studies should further investigate the specific role of m6A modification in bone remodeling across these cell types, particularly its interactions with bone cells.
5. m6A modification and osteoporosis
5.1 Evidence from animal and cellular studies
OP is a metabolic bone disease characterized by reduced bone mass and deterioration of bone microarchitecture[146]. With the global aging population, the incidence of OP is rising, significantly affecting the quality of life in patients[146, 147]. The pathogenesis of OP is closely related to the dysregulation of bone metabolism caused by the osteoblast-osteoclast uncoupling and imbalance of BMSCs differentiation[148]. Researchers suggest that m6A modification plays a vital role in OP. Based on extensive studies on animal models and cell cultures, Liang et al. summarized the regulatory mechanisms of m6A in osteoblasts, osteoclasts and BMSCs, emphasizing its crucial role in OP[149]. Specifically, m6A modification regulates bone homeostasis through various mechanisms. It influences osteoclast differentiation to modulate bone resorption, controls osteoblast activity for new bone formation, and directs BMSCs differentiation toward osteogenesis while inhibiting adipogenesis. Key m6A regulators, such as METTL3 and METTL14, orchestrate these processes, positioning them as promising targets for OP treatment[149]. Recent studies further reveal new insights into m6A and senile OP. Wang et al. discovered that the expression of METTL3 was significantly decreased in the senile OP mouse model. METTL3 partially reversed the aging of osteoblasts and subsequent age-related bone loss by increasing the stability of Hspa1a mRNA[150]. Cell culture study also confirmed that downregulation of METTL3 promotes the degradation of LINC01013, thereby inhibiting the differentiation of pre-osteoblasts during aging[151]. However, another study showed that METTL3 facilitates osteoblast senescence via the METTL3/IGF2BP2/Slc1a5 axis, thereby exacerbating age-related bone loss[152]. Additionally, an in vitro study demonstrated that ALKBH5 regulates cellular senescence and osteogenic differentiation in age-related OP by modulating the m6A modification of voltage-dependent anion channel 3 (VDAC3)[153]. The discrepancies observed in these researches may result from differing experimental conditions, emphasizing the necessity for further exploration of how m6A affects OP in different pathological conditions.
5.2 Evidence from human studies
In recent years, m6A epigenetic modification has emerged as a burgeoning field, providing new perspectives for understanding the pathophysiology of bone diseases[13]. Several clinical studies have provided evidence supporting a close association between m6A regulatory factors and metrics indicative of bone health, particularly BMD. Deng et al. observed low levels of METTL14 in the serum of postmenopausal women with OP, which were positively correlated with BMD[123]. Similarly, Serum YTHDC1 levels were positively correlate with lumbar spine BMD in OP patients[124]. Notably, single nucleotide polymorphisms (SNPs) have been associated with a variety of disease susceptibilities, and m6A-related SNPs can also influence the progression of OP. Han and his team identified a large number of m6A-SNPs were associated with BMD through genome-wide association studies (GWAS)[154, 155]. Meanwhile, several follow-up studies confirmed these findings, revealing significant associations between FTO SNPs and hip BMD as well as osteoporotic fracture risk[156-158]. Additionally, numerous studies have shown that osteoporotic bone tissue exhibits significantly lower levels of m6A compared to normal healthy individuals, primarily because of decreased expression of m6A methyltransferases METTL3[66, 88, 150, 159], METTL14[65, 89] and WTAP[79, 125], as well as increased expression of the m6A demethylase FTO[67, 110]. However, evidence provided by Dong et al. indicates that the levels of m6A and METTL14 are upregulated in the bone tissue of patients with OP[160]. Therefore, future clinical studies with larger sample sizes are necessary to accurately reveal the specific relationship between m6A regulators and OP.
Potential anti-osteoporosis drugs targeting m6A modification.
| Potential drugs | Target | Mechanism | Pharmacological function | References |
|---|---|---|---|---|
| Icariin | METTL14 | METTL14/P4HB | Promoting osteogenesis | [167] |
| Chinese Ecliptae herba | METTL3 | HIF-1α, PI3K/AKT and Hippo pathway | Promoting osteogenesis | [168] |
| Qianggu Decoction | METTL3 | METTL3/Runx2 | Promoting osteogenesis | [169] |
| Xianling Gubao Capsule | METTL3 | - | Promoting osteogenesis | [170] |
| Specnuezhenide | METTL3 | METTL3/Runx2 METTL3/SLIT3 | Promoting LepR+ BMSCs-dependent osteogenesis and angiogenesis | [171] |
5.3 Clinical value of m6A in osteoporosis
Due to the lack of reliable biomarkers for the prediction and diagnosis of OP, the discovery of novel biomarkers using histological techniques, especially the epitranscriptome, could be useful for early screening and clinical decision-making in OP[161]. m6A modification as an important RNA epigenetic modification shows potential value in the prediction and diagnosis of OP. For example, a diagnostic model for OP constructed based on the Gene Expression Omnibus (GEO) dataset identified four m6A regulator factors: METTL16, CBLL1, YTHDF2, and FTO. Notably, CBLL1 and YTHDF2 were recognized as protective factors, whereas METTL16 and FTO were classified as risk factors[162, 163]. Zhang et al. identified seven m6A regulators, including FTO, FMR1, YTHDC2, HNRNPC, RBM15, RBM15B and WTAP in human peripheral blood mononuclear cells (PBMCs) by bioinformatics analysis, which may serve as potential biomarkers for diagnosis of postmenopausal OP[164]. These findings provide new insights into the exploration of biomarkers associated with OP. However, further validation of their clinical reliability and practicality is necessary.
In terms of treatment, although anti-osteoporosis medications offer significant therapeutic benefits for patients, their potential risks and side effects may limit long-term use[165, 166]. Therefore, developing novel medications with fewer side effects for treating OP remains a crucial strategy. As mentioned previously, m6A regulates the proliferation, differentiation and maturation of osteoblasts and osteoclasts, which provides strong evidence for targeting m6A as a therapeutic approach in OP. Recently, several studies have identified potential drugs for treating OP by modulating m6A (Table 2). Jin et al. revealed that icariin enhances the m6A levels of P4HB through METTL14, thereby promoting the osteogenic differentiation of BMSCs[167]. Chinese Ecliptae herba extract and its component wedelolactone can target METTL3, activating various pathways such as HIF-1α, PI3K, and Hippo to alleviate OP[168]. Xianling Gubao Capsule and Qianggu Decoction also rescue the decreased osteogenic potential of BMSCs in OP via METTL3-mediated m6A modification[169, 170]. Intriguingly, Wei et al. identified Specnuezhenide as a promising candidate for OP treatment through single-cell sequencing, showing its ability to enhance angiogenesis and osteogenesis in LepR+ BMSCs by activating METTL3[171]. In addition, several clinical studies have confirmed the positive effects of these drugs in the treatment of OP. For example, meta-analysis based on randomized clinical trials revealed that Xianling Gubao capsules have the potential to improve quality of life and alleviate bone pain in the treatment of primary OP[172]. A 24-month randomized controlled clinical trial provided evidence that Epimedium-derived compounds, primarily consisting of icariin, significantly attenuated bone loss in postmenopausal women[173]. Yong et al. further demonstrated that the purified Epimedium extract exhibited a favorable safety profile following continuous administration for 6 weeks in postmenopausal women[174]. These results suggest that targeting m6A shows excellent potential in the treatment of OP. Nevertheless, therapeutic agents targeting m6A remain in the early phases of clinical application. Although several drugs have shown promising efficacy in preclinical studies, large-scale clinical trials are still scarce. In the future, as more clinical studies targeting m6A progress, further development of novel m6A-based targeted drugs is expected to provide more effective and personalized clinical benefits to patients with OP.
Furthermore, numerous studies have confirmed that m6A plays an essential role in the prognosis of cancers (i.e., liver cancer, breast cancer and osteosarcoma)[175-177], which indicates that m6A regulatory factors may serve as potential biomarkers in evaluating the prognosis of OP. Taken together, m6A methylation modification has a broad application in the prediction, diagnosis, treatment, and prognostic management of OP.
6. Conclusion and future perspectives
In summary, m6A modification is a key contributor to the biological processes of bone cells, particularly by regulating various signaling pathways that affect their differentiation and activity. Specifically, m6A regulatory factors modulate osteoblast differentiation and mineralization through signaling pathways such as Wnt/β-catenin, PI3K/AKT, and BMP/Smad. Meanwhile, they regulate osteoclast differentiation and bone resorption via the RANKL/RANK pathway and its downstream signaling mechanisms. Moreover, the number and development of osteocytes are also influenced by m6A modification. Notably, abnormal m6A modification may impair the bone remodeling, contributing to the progression of metabolic bone diseases such as OP. However, the specific mechanisms of m6A modification in bone pathophysiology remain largely unknown.
Therefore, future research should prioritize the following aspects: First, the onset of OP may involve the collaborative effects of multiple m6A regulatory proteins, requiring further exploration of novel m6A regulators and their interactions in bone pathophysiology. Second, it is essential to clarify the specific roles of m6A modification in different bone cells, especially osteoclasts and osteocytes. Creating specific knockout mouse models lacking m6A regulatory factors will be essential for advancing this research. Finally, given the complex relationship between m6A modification and bone remodeling, future efforts should develop novel anti-osteoporotic drugs targeting m6A and translate these findings into effective clinical diagnostic and therapeutic tools for OP.
Abbreviations
m6A: N6-methyladenosine; OP: Osteoporosis; mRNAs: messenger RNAs; tRNAs: transfer RNAs; rRNAs: ribosomal RNAs; lncRNAs: long non-coding RNAs; MeRIP-Seq: Methylated RNA immunoprecipitation sequencing; CDS: Coding sequences; 3' UTRs: 3' untranslated regions; MTC: Methyltransferase complex; METTL3: Methyltransferase like 3; SAM: S-adenosylmethionine; METTL14: Methyltransferase like 14; WTAP: Wilms tumor 1 associated protein; RBM15/15B: RNA-binding motif protein 15/15B; ZC3H13: Zinc finger CCCH-type containing 13; VIRMA: Vir-like m6A methyltransferase associated; CBLL1: Cbl proto-oncogene like 1; METTL5: Methyltransferase like 5; METTL16: Methyltransferase like 16; ZCCHC4: Zinc finger CCHC-type containing 4; snRNAs: Small nuclear RNAs; pre-mRNAs: precursor messenger RNAs; FTO: Fat mass and obesity-associated protein; ALKBH5: AlkB homolog 5; YTH: YT521-B homology; EIF3: Eukaryotic initiation factor 3; IGF2BP: Insulin-like growth factor 2 mRNA binding proteins; BMSCs: Bone marrow mesenchymal stem cells; hnRNPs: Heterogeneous nuclear ribonucleoproteins; mESCs: Mouse embryonic stem cells; BMD: Bone mineral density; MAPK: Mitogen-activated protein kinase; BMP: Bone morphogenic protein; LPS: Lipopolysaccharides; SFRP1: Secretory frizzled-related protein 1; SOST: Sclerostin; AGEs: Advanced glycation end products; PI3K: Phosphoinositide 3-kinase; PRMT6: Protein arginine methyltransferase 6; OVX: Ovariectomy; IGF: Insulin-like growth factor; m-ASCs: Mesenchymal adult stem cells; ASCs: Adipose derived stem cells; AMPK: Adenosine monophosphate-activated protein kinase; PTH: Parathyroid hormone; PTH1R: Parathyroid hormone 1 receptor; PPARγ: Peroxisome proliferator-activated receptor γ; ALP: Alkaline phosphatase; OPN: Osteopontin; FSH: Follicle-stimulating hormone; CREB: Cyclic-AMP response element-binding protein; CTSK: Cathepsin K; EGR1: Early growth response 1; CHI3L1: Chitinase 3-like protein 1; TLR4: Toll-like receptor 4; NFATc1: Nuclear factor of activated T-cells cytoplasmic 1; GPX4: Glutathione peroxidase 4; HuR: Human antigen R; PTPN6: Protein tyrosine phosphatase non-receptor type 6; RANKL: Receptor activator of NF-κB ligand; RANK: Receptor activator of NF-κB; TRAF6: Tumor necrosis factor receptor-associated factor 6; OPG: Osteoprotegerin; ZIC1: Zinc finger of the cerebellum 1; PHD2: Prolyl hydroxylase 2; VEGFA: vascular endothelial growth factor A; EPCs: Endothelial progenitor cells; VDAC3: Voltage-dependent anion channel 3; SNPs: Single nucleotide polymorphisms; GWAS: Genome-wide association studies; GEO: Gene Expression Omnibus; PBMCs: Peripheral blood mononuclear cells.
Acknowledgements
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82273294) and the 1.3.5 project for discipline of excellence, West China Hospital, Sichuan University (No. 2020HXFH008, No. ZYJC18003).
Author contributions
All authors made a significant contribution to the work reported, whether in the conception, study design, acquisition of data, analysis and interpretation. JL drafted the manuscript. XC and XY critically revised the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol. 2018;19:774-90
2. Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176-89
3. Zhou Y, Kong Y, Fan W, Tao T, Xiao Q, Li N. et al. Principles of RNA methylation and their implications for biology and medicine. Biomed Pharmacother. 2020;131:110731
4. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
5. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N. et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48:6251-64
6. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
7. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608-24
8. Han J, Kong H, Wang X, Zhang XA. Novel insights into the interaction between N6-methyladenosine methylation and noncoding RNAs in musculoskeletal disorders. Cell Prolif. 2022;55:e13294
9. Ghafouri-Fard S, Shoorei H, Hussen BM, Dong P, Zhai T, Taheri M. et al. The significance of N6-methyladenosine-modified non-coding RNAs in different disorders. Eur J Pharmacol. 2023;946:175644
10. Zaidi M, Kim SM, Mathew M, Korkmaz F, Sultana F, Miyashita S. et al. Bone circuitry and interorgan skeletal crosstalk. Elife. 2023 12
11. Robling AG, Bonewald LF. The Osteocyte: New Insights. Annu Rev Physiol. 2020;82:485-506
12. Johnston CB, Dagar M. Osteoporosis in Older Adults. Med Clin North Am. 2020;104:873-84
13. Yang C, Dong Z, Ling Z, Chen Y. The crucial mechanism and therapeutic implication of RNA methylation in bone pathophysiology. Ageing Res Rev. 2022;79:101641
14. Gu Y, Song Y, Pan Y, Liu J. The essential roles of m(6)A modification in osteogenesis and common bone diseases. Genes Dis. 2024;11:335-45
15. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet. 2014;15:293-306
16. Corbeski I, Vargas-Rosales PA, Bedi RK, Deng J, Coelho D, Braud E. et al. The catalytic mechanism of the RNA methyltransferase METTL3. Elife. 2024 12
17. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
18. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74
19. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719-33
20. Pinto R, Vågbø CB, Jakobsson ME, Kim Y, Baltissen MP, O'Donohue MF. et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020;48:830-46
21. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C. et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004-14
22. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP. et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824-35.e14
23. Yang Z, Yu GL, Zhu X, Peng TH, Lv YC. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2022;9:51-61
24. Zhang Q, Riddle RC, Yang Q, Rosen CR, Guttridge DC, Dirckx N. et al. The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc Natl Acad Sci U S A. 2019;116:17980-9
25. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
26. Yu F, Zhu AC, Liu S, Gao B, Wang Y, Khudaverdyan N. et al. RBM33 is a unique m(6)A RNA-binding protein that regulates ALKBH5 demethylase activity and substrate selectivity. Mol Cell. 2023;83:2003-19.e6
27. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
28. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
29. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-28
30. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O. et al. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999-1010
31. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF. et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507-19
32. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y. et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017 6
33. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y. et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115-27
34. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
35. He M, Lei H, He X, Liu Y, Wang A, Ren Z. et al. METTL14 Regulates Osteogenesis of Bone Marrow Mesenchymal Stem Cells via Inducing Autophagy Through m6A/IGF2BPs/Beclin-1 Signal Axis. Stem Cells Transl Med. 2022;11:987-1001
36. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299-308
37. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
38. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051-63
39. Zhang M, Zhai Y, Zhang S, Dai X, Li Z. Roles of N6-Methyladenosine (m(6)A) in Stem Cell Fate Decisions and Early Embryonic Development in Mammals. Front Cell Dev Biol. 2020;8:782
40. Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q. et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27:1216-30
41. Horiuchi K, Umetani M, Minami T, Okayama H, Takada S, Yamamoto M. et al. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci U S A. 2006;103:17278-83
42. Xing M, Liu Q, Mao C, Zeng H, Zhang X, Zhao S. et al. The 18S rRNA m(6) A methyltransferase METTL5 promotes mouse embryonic stem cell differentiation. EMBO Rep. 2020;21:e49863
43. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707-19
44. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191-8
45. Xu Y, Zhou Z, Kang X, Pan L, Liu C, Liang X. et al. Mettl3-mediated mRNA m(6)A modification controls postnatal liver development by modulating the transcription factor Hnf4a. Nat Commun. 2022;13:4555
46. Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z. et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69
47. Berendsen AD, Olsen BR. Bone development. Bone. 2015;80:14-8
48. Ma L, Zhou X, Yao S, Zhang X, Mao J, Vona B. et al. METTL3-dependent m(6)A modification of PSEN1 mRNA regulates craniofacial development through the Wnt/β-catenin signaling pathway. Cell Death Dis. 2024;15:229
49. Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS. et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85:106-11
50. Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005
51. Opperman LA. Cranial sutures as intramembranous bone growth sites. Dev Dyn. 2000;219:472-85
52. Richard EM, Polla DL, Assir MZ, Contreras M, Shahzad M, Khan AA. et al. Bi-allelic Variants in METTL5 Cause Autosomal-Recessive Intellectual Disability and Microcephaly. Am J Hum Genet. 2019;105:869-78
53. Lei K, Xu R, Wang Q, Xiong Q, Zhou X, Li Q. et al. METTL5 regulates cranial suture fusion via Wnt signaling. Fundam Res. 2023;3:369-76
54. Xu R, Sheng R, Lin W, Jiang S, Zhang D, Liu L. et al. METTL3 Modulates Ctsk(+) Lineage Supporting Cranial Osteogenesis via Hedgehog. J Dent Res. 2024;103:734-44
55. Liu CF, Samsa WE, Zhou G, Lefebvre V. Transcriptional control of chondrocyte specification and differentiation. Semin Cell Dev Biol. 2017;62:34-49
56. He Y, Wang W, Luo P, Wang Y, He Z, Dong W. et al. Mettl3 regulates hypertrophic differentiation of chondrocytes through modulating Dmp1 mRNA via Ythdf1-mediated m(6)A modification. Bone. 2022;164:116522
57. Song M, Yao H, Sun Z, Chen D, Xu X, Long G. et al. METTL3/YTHDC1-medicated m6A modification of circRNA3634 regulates the proliferation and differentiation of antler chondrocytes by miR-124486-5-MAPK1 axis. Cell Mol Biol Lett. 2023;28:101
58. Sheng R, Meng W, Zhang Z, Yin Q, Jiang S, Li Q. et al. METTL3 regulates cartilage development and homeostasis by affecting Lats1 mRNA stability in an m(6)A-YTHDF2-dependent manner. Cell Rep. 2024;43:114535
59. Salhotra A, Shah HN, Levi B, Longaker MT. Mechanisms of bone development and repair. Nat Rev Mol Cell Biol. 2020;21:696-711
60. Zhou S, Zhang G, Wang K, Yang Z, Tan Y. METTL3 potentiates osteogenic differentiation of bone marrow mesenchymal stem cells via IGF2BP1/m6A/RUNX2. Oral Dis. 2024;30:1313-21
61. Feng L, Fan Y, Zhou J, Li S, Zhang X. The RNA demethylase ALKBH5 promotes osteoblast differentiation by modulating Runx2 mRNA stability. FEBS Lett. 2021;595:2007-14
62. Liu T, Zheng X, Wang C, Wang C, Jiang S, Li B. et al. The m(6)A "reader" YTHDF1 promotes osteogenesis of bone marrow mesenchymal stem cells through translational control of ZNF839. Cell Death Dis. 2021;12:1078
63. Feng ZW, Xiao HF, Wang XW, Niu YK, Zhao DC, Tian C. et al. Unraveling Key m(6)A Modification Regulators Signatures in Postmenopausal Osteoporosis through Bioinformatics and Experimental Verification. Orthop Surg. 2024;16:1418-33
64. Gao W, Miao X, Xu T. Wilms tumor 1-associated protein mediated m6A modification promotes osteogenic differentiation of stem cells from human exfoliated deciduous teeth. J Dent Sci. 2024;19:2305-14
65. Wang X, Zou C, Li M, Hou C, Jiang W, Bian Z. et al. METTL14 upregulates TCF1 through m6A mRNA methylation to stimulate osteogenic activity in osteoporosis. Hum Cell. 2023;36:178-94
66. Feng ZW, Peng B, Wang SH, Zhao DC, Wang YB, Yang A. et al. METTL3-mediated m(6)A modification of SOX4 regulates osteoblast proliferation and differentiation via YTHDF3 recognition. Cell Signal. 2024;115:111038
67. Wang J, Fu Q, Yang J, Liu JL, Hou SM, Huang X. et al. RNA N6-methyladenosine demethylase FTO promotes osteoporosis through demethylating Runx2 mRNA and inhibiting osteogenic differentiation. Aging (Albany NY). 2021;13:21134-41
68. Ma B, Cao P, Zhang L, Zhu H, Ye X, Wang L. et al. YTHDC2 inhibits rat bone mesenchymal stem cells osteogenic differentiation by accelerating RUNX2 mRNA degradation via m6A methylation. Heliyon. 2023;9:e18876
69. Yang L, Ren Z, Yan S, Zhao L, Liu J, Zhao L. et al. Nsun4 and Mettl3 mediated translational reprogramming of Sox9 promotes BMSC chondrogenic differentiation. Commun Biol. 2022;5:495
70. Xiao L, Hu B, Ding B, Zhao Q, Liu C, Öner FC. et al. N(6)-methyladenosine RNA methyltransferase like 3 inhibits extracellular matrix synthesis of endplate chondrocytes by downregulating sex-determining region Y-Box transcription factor 9 expression under tension. Osteoarthritis Cartilage. 2022;30:613-25
71. Kong Y, Zhang Y, Cai Y, Li D, Yi B, Xu Q. METTL3 mediates osteoblast apoptosis by regulating endoplasmic reticulum stress during LPS-induced inflammation. Cell Signal. 2022;95:110335
72. Capulli M, Paone R, Rucci N. Osteoblast and osteocyte: games without frontiers. Arch Biochem Biophys. 2014;561:3-12
73. Ponzetti M, Rucci N. Osteoblast Differentiation and Signaling: Established Concepts and Emerging Topics. Int J Mol Sci. 2021 22
74. Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387:1657-71
75. Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19:179-92
76. Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231-44
77. Wu T, Tang H, Yang J, Yao Z, Bai L, Xie Y. et al. METTL3-m(6) A methylase regulates the osteogenic potential of bone marrow mesenchymal stem cells in osteoporotic rats via the Wnt signalling pathway. Cell Prolif. 2022;55:e13234
78. Zhang Y, Kong Y, Zhang W, He J, Zhang Z, Cai Y. et al. METTL3 promotes osteoblast ribosome biogenesis and alleviates periodontitis. Clin Epigenetics. 2024;16:18
79. You Y, Liu J, Zhang L, Li X, Sun Z, Dai Z. et al. WTAP-mediated m(6)A modification modulates bone marrow mesenchymal stem cells differentiation potential and osteoporosis. Cell Death Dis. 2023;14:33
80. Boudin E, Fijalkowski I, Piters E, Van Hul W. The role of extracellular modulators of canonical Wnt signaling in bone metabolism and diseases. Semin Arthritis Rheum. 2013;43:220-40
81. Taipaleenmäki H, Abdallah BM, AlDahmash A, Säämänen AM, Kassem M. Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Exp Cell Res. 2011;317:745-56
82. Gao X, Wang J, Wang Y, Li W, Pan Z. The m(6)A Reader YTHDF1 Accelerates the Osteogenesis of Bone Marrow Mesenchymal Stem Cells Partly via Activation of the Autophagy Signaling Pathway. Stem Cells Int. 2023;2023:5563568
83. Zhou J, Zhu Y, Ai D, Zhou M, Li H, Li G. et al. Advanced glycation end products impair bone marrow mesenchymal stem cells osteogenesis in periodontitis with diabetes via FTO-mediated N(6)-methyladenosine modification of sclerostin. J Transl Med. 2023;21:781
84. Salazar VS, Gamer LW, Rosen V. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12:203-21
85. Rosen V. BMP2 signaling in bone development and repair. Cytokine Growth Factor Rev. 2009;20:475-80
86. Nickel J, Mueller TD. Specification of BMP Signaling. Cells. 2019 8
87. Liu J, Chen M, Ma L, Dang X, Du G. piRNA-36741 regulates BMP2-mediated osteoblast differentiation via METTL3 controlled m6A modification. Aging (Albany NY). 2021;13:23361-75
88. Peng J, Zhan Y, Zong Y. METTL3-mediated LINC00657 promotes osteogenic differentiation of mesenchymal stem cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. 2022;388:301-12
89. Huang C, Wang Y. Downregulation of METTL14 improves postmenopausal osteoporosis via IGF2BP1 dependent posttranscriptional silencing of SMAD1. Cell Death Dis. 2022;13:919
90. Yan X, Liao H, Cheng M, Shi X, Lin X, Feng XH. et al. Smad7 Protein Interacts with Receptor-regulated Smads (R-Smads) to Inhibit Transforming Growth Factor-β (TGF-β)/Smad Signaling. J Biol Chem. 2016;291:382-92
91. Deng Z, Fan T, Xiao C, Tian H, Zheng Y, Li C. et al. TGF-β signaling in health, disease, and therapeutics. Signal Transduct Target Ther. 2024;9:61
92. Zhang Y, Gu X, Li D, Cai L, Xu Q. METTL3 Regulates Osteoblast Differentiation and Inflammatory Response via Smad Signaling and MAPK Signaling. Int J Mol Sci. 2019 21
93. Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050-60
94. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell. 2017;170:605-35
95. Carnero A, Paramio JM. The PTEN/PI3K/AKT Pathway in vivo, Cancer Mouse Models. Front Oncol. 2014;4:252
96. Deng S, Dai G, Chen S, Nie Z, Zhou J, Fang H. et al. Dexamethasone induces osteoblast apoptosis through ROS-PI3K/AKT/GSK3β signaling pathway. Biomed Pharmacother. 2019;110:602-8
97. Liu Y, Zhao L, He X, Shen Y, Wang N, Hu S. et al. Jintiange proteins promote osteogenesis and inhibit apoptosis of osteoblasts by enhancing autophagy via PI3K/AKT and ER stress pathways. J Ethnopharmacol. 2023;311:116399
98. Tian C, Huang Y, Li Q, Feng Z, Xu Q. Mettl3 Regulates Osteogenic Differentiation and Alternative Splicing of Vegfa in Bone Marrow Mesenchymal Stem Cells. Int J Mol Sci. 2019 20
99. Li Z, Wang P, Li J, Xie Z, Cen S, Li M. et al. The N(6)-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6. Cell Death Dis. 2021;12:578
100. Zhang X, Wang Y, Zhao H, Han X, Zhao T, Qu P. et al. Extracellular vesicle-encapsulated miR-22-3p from bone marrow mesenchymal stem cell promotes osteogenic differentiation via FTO inhibition. Stem Cell Res Ther. 2020;11:227
101. Chen L, Jiang W, Huang J, He BC, Zuo GW, Zhang W. et al. Insulin-like growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic differentiation and bone formation. J Bone Miner Res. 2010;25:2447-59
102. Tian Q, Gao S, Li S, Wan M, Zhou X, Du W. et al. Glutamine-αKG axis affects dentin regeneration and regulates osteo/odontogenic differentiation of mesenchymal adult stem cells via IGF2 m6A modification. Stem Cell Res Ther. 2024;15:479
103. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50-83
104. Franceschi RT, Ge C. Control of the Osteoblast Lineage by Mitogen-Activated Protein Kinase Signaling. Curr Mol Biol Rep. 2017;3:122-32
105. Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911-2
106. Song Y, Pan Y, Wu M, Sun W, Luo L, Zhao Z. et al. METTL3-Mediated lncRNA m(6)A Modification in the Osteogenic Differentiation of Human Adipose-Derived Stem Cells Induced by NEL-Like 1 Protein. Stem Cell Rev Rep. 2021;17:2276-90
107. Lin Y, Shen X, Ke Y, Lan C, Chen X, Liang B. et al. Activation of osteoblast ferroptosis via the METTL3/ASK1-p38 signaling pathway in high glucose and high fat (HGHF)-induced diabetic bone loss. Faseb j. 2022;36:e22147
108. Son HE, Min HY, Kim EJ, Jang WG. Fat Mass and Obesity-Associated (FTO) Stimulates Osteogenic Differentiation of C3H10T1/2 Cells by Inducing Mild Endoplasmic Reticulum Stress via a Positive Feedback Loop with p-AMPK. Mol Cells. 2020;43:58-65
109. Shen GS, Zhou HB, Zhang H, Chen B, Liu ZP, Yuan Y. et al. The GDF11-FTO-PPARγ axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim Biophys Acta Mol Basis Dis. 2018;1864:3644-54
110. Chen LS, Zhang M, Chen P, Xiong XF, Liu PQ, Wang HB. et al. The m(6)A demethylase FTO promotes the osteogenesis of mesenchymal stem cells by downregulating PPARG. Acta Pharmacol Sin. 2022;43:1311-23
111. Saini V, Marengi DA, Barry KJ, Fulzele KS, Heiden E, Liu X. et al. Parathyroid hormone (PTH)/PTH-related peptide type 1 receptor (PPR) signaling in osteocytes regulates anabolic and catabolic skeletal responses to PTH. J Biol Chem. 2013;288:20122-34
112. Uda Y, Saini V, Petty CA, Alshehri M, Shi C, Spatz JM. et al. Parathyroid hormone signaling in mature osteoblasts/osteocytes protects mice from age-related bone loss. Aging (Albany NY). 2021;13:25607-42
113. Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y. et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772
114. Yahara Y, Nguyen T, Ishikawa K, Kamei K, Alman BA. The origins and roles of osteoclasts in bone development, homeostasis and repair. Development. 2022 149
115. Veis DJ, O'Brien CA. Osteoclasts, Master Sculptors of Bone. Annu Rev Pathol. 2023;18:257-81
116. He J, Zhao Y, Zhang Y, Zhang Z, Li D, Xu Q. FTO regulates osteoclast development by modulating the proliferation and apoptosis of osteoclast precursors in inflammatory conditions. Cell Signal. 2024;117:111098
117. Li D, He J, Fang C, Zhang Y, He M, Zhang Z. et al. METTL3 Regulates Osteoclast Biological Behaviors via iNOS/NO-Mediated Mitochondrial Dysfunction in Inflammatory Conditions. Int J Mol Sci. 2023 24
118. Li D, Cai L, Meng R, Feng Z, Xu Q. METTL3 Modulates Osteoclast Differentiation and Function by Controlling RNA Stability and Nuclear Export. Int J Mol Sci. 2020 21
119. Li X, Fan C, Wang J, Li P, Xu X, Guo R. et al. Follicle-stimulating hormone accelerates osteoclast migration by enhancing methyltransferase-like 3-mediated m6A methylation of cathepsin K. J Mol Endocrinol. 2024 72
120. Wang C, Zhang X, Chen R, Zhu X, Lian N. EGR1 mediates METTL3/m(6)A/CHI3L1 to promote osteoclastogenesis in osteoporosis. Genomics. 2023;115:110696
121. Shen X, Lan C, Lin Y, Zhang F, Zhang Y, Chen M. et al. Suppression of TLR4 prevents diabetic bone loss by regulating FTO-mediated m(6)A modification. Int Immunopharmacol. 2023;122:110510
122. Yang JG, Sun B, Wang Z, Li X, Gao JH, Qian JJ. et al. Exosome-targeted delivery of METTL14 regulates NFATc1 m6A methylation levels to correct osteoclast-induced bone resorption. Cell Death Dis. 2023;14:738
123. Deng M, Luo J, Cao H, Li Y, Chen L, Liu G. METTL14 represses osteoclast formation to ameliorate osteoporosis via enhancing GPX4 mRNA stability. Environ Toxicol. 2023;38:2057-68
124. Zhang M, Guan J, Yu S, Zhang Y, Cheng L, Zhang Y. YTHDC1 inhibits osteoclast differentiation to alleviate osteoporosis by enhancing PTPN6 messenger RNA stability in an m6A-hUR-dependent manner. J Leukoc Biol. 2024;115:1154-64
125. Liu J, You Y, Sun Z, Zhang L, Li X, Dai Z. et al. WTAP-Mediated m6A RNA Methylation Regulates the Differentiation of Bone Marrow Mesenchymal Stem Cells via the miR-29b-3p/HDAC4 Axis. Stem Cells Transl Med. 2023;12:307-21
126. Shi JH, Sun SC. Tumor Necrosis Factor Receptor-Associated Factor Regulation of Nuclear Factor κB and Mitogen-Activated Protein Kinase Pathways. Front Immunol. 2018;9:1849
127. Wada T, Nakashima T, Hiroshi N, Penninger JM. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol Med. 2006;12:17-25
128. He M, Li D, Fang C, Xu Q. YTHDF1 regulates endoplasmic reticulum stress, NF-κB, MAPK and PI3K-AKT signaling pathways in inflammatory osteoclastogenesis. Arch Biochem Biophys. 2022;732:109464
129. Fang C, He M, Li D, Xu Q. YTHDF2 mediates LPS-induced osteoclastogenesis and inflammatory response via the NF-κB and MAPK signaling pathways. Cell Signal. 2021;85:110060
130. Abu-Amer Y. NF-κB signaling and bone resorption. Osteoporos Int. 2013;24:2377-86
131. Zhuang J, Ning H, Wang M, Zhao W, Jing Y, Liu X. et al. Downregulated fat mass and obesity-associated protein inhibits bone resorption and osteoclastogenesis by nuclear factor-kappa B inactivation. Cell Signal. 2021;87:110137
132. Ma XX, Zhou XY, Feng MG, Ji YT, Song FF, Tang QC. et al. Dual Role of IGF2BP2 in Osteoimmunomodulation during Periodontitis. J Dent Res. 2024;103:208-17
133. Delgado-Calle J, Bellido T. The osteocyte as a signaling cell. Physiol Rev. 2022;102:379-410
134. Datta HK, Kringen MK, Tuck SP, Salpingidou G, Olstad OK, Gautvik KM. et al. Mechanical-Stress-Related Epigenetic Regulation of ZIC1 Transcription Factor in the Etiology of Postmenopausal Osteoporosis. Int J Mol Sci. 2022 23
135. Stegen S, Stockmans I, Moermans K, Thienpont B, Maxwell PH, Carmeliet P. et al. Osteocytic oxygen sensing controls bone mass through epigenetic regulation of sclerostin. Nat Commun. 2018;9:2557
136. Xia Y, Ikedo A, Lee JW, Iimura T, Inoue K, Imai Y. Histone H3K27 demethylase, Utx, regulates osteoblast-to-osteocyte differentiation. Biochem Biophys Res Commun. 2022;590:132-8
137. Miron RJ, Bosshardt DD. OsteoMacs: Key players around bone biomaterials. Biomaterials. 2016;82:1-19
138. Li Y, Yang Y, Niu Y, Li Y, Hu Z, Sun S. et al. The role of WTAP in regulating macrophage-mediated osteoimmune responses and tissue regeneration in periodontitis. Front Immunol. 2024;15:1423378
139. Lei H, He M, He X, Li G, Wang Y, Gao Y. et al. METTL3 induces bone marrow mesenchymal stem cells osteogenic differentiation and migration through facilitating M1 macrophage differentiation. Am J Transl Res. 2021;13:4376-88
140. Chen T, Jin L, Li J, Liu Y. Pyroptosis mediates osteoporosis via the inflammation immune microenvironment. Front Immunol. 2024;15:1371463
141. Tang H, Du Y, Tan Z, Li D, Xie J. METTL14-mediated HOXA5 m(6)A modification alleviates osteoporosis via promoting WNK1 transcription to suppress NLRP3-dependent macrophage pyroptosis. J Orthop Translat. 2024;48:190-203
142. Yang Q, Xiao J, Liu Y, Yang Z, Wang C, Sun J. et al. METTL3-mediated m6A modifications of NLRP3 accelerate alveolar bone resorption through enhancing macrophage pyroptosis. Cell Signal. 2025;127:111572
143. Diomede F, Marconi GD, Fonticoli L, Pizzicanella J, Merciaro I, Bramanti P. et al. Functional Relationship between Osteogenesis and Angiogenesis in Tissue Regeneration. Int J Mol Sci. 2020 21
144. Tuckermann J, Adams RH. The endothelium-bone axis in development, homeostasis and bone and joint disease. Nat Rev Rheumatol. 2021;17:608-20
145. Jiang W, Zhu P, Huang F, Zhao Z, Zhang T, An X. et al. The RNA Methyltransferase METTL3 Promotes Endothelial Progenitor Cell Angiogenesis in Mandibular Distraction Osteogenesis via the PI3K/AKT Pathway. Front Cell Dev Biol. 2021;9:720925
146. Khandelwal S, Lane NE. Osteoporosis: Review of Etiology, Mechanisms, and Approach to Management in the Aging Population. Endocrinol Metab Clin North Am. 2023;52:259-75
147. Clynes MA, Harvey NC, Curtis EM, Fuggle NR, Dennison EM, Cooper C. The epidemiology of osteoporosis. Br Med Bull. 2020;133:105-17
148. Li J, Chen X, Lu L, Yu X. The relationship between bone marrow adipose tissue and bone metabolism in postmenopausal osteoporosis. Cytokine Growth Factor Rev. 2020;52:88-98
149. Liang J, Yi Q, Liu Y, Li J, Yang Z, Sun W. et al. Recent advances of m6A methylation in skeletal system disease. J Transl Med. 2024;22:153
150. Wang Y, Chen Y, Xiao H, Liu Z, Liu X, Feng Z. et al. METTL3-mediated m6A modification increases Hspa1a stability to inhibit osteoblast aging. Cell Death Discov. 2024;10:155
151. Song J, Wang Y, Zhu Z, Wang W, Yang H, Shan Z. Negative Regulation of LINC01013 by METTL3 and YTHDF2 Enhances the Osteogenic Differentiation of Senescent Pre-Osteoblast Cells Induced by Hydrogen Peroxide. Adv Biol (Weinh). 2024;8:e2300642
152. Liu XW, Xu HW, Yi YY, Zhang SB, Chang SJ, Pan W. et al. Inhibition of Mettl3 ameliorates osteoblastic senescence by mitigating m6A modifications on Slc1a5 via Igf2bp2-dependent mechanisms. Biochim Biophys Acta Mol Basis Dis. 2024;1870:167273
153. Huang Y, Wang S, Hu D, Zhang L, Shi S. ALKBH5 regulates etoposide-induced cellular senescence and osteogenic differentiation in osteoporosis through mediating the m(6)A modification of VDAC3. Sci Rep. 2024;14:23461
154. Mo XB, Zhang YH, Lei SF. Genome-wide identification of m(6)A-associated SNPs as potential functional variants for bone mineral density. Osteoporos Int. 2018;29:2029-39
155. Han L, Wu J, Wang M, Zhang Z, Hua D, Lei S. et al. RNA Modification-Related Genetic Variants in Genomic Loci Associated with Bone Mineral Density and Fracture. Genes (Basel). 2022 13
156. Tran B, Nguyen ND, Center JR, Eisman JA, Nguyen TV. Association between fat-mass-and-obesity-associated (FTO) gene and hip fracture susceptibility. Clin Endocrinol (Oxf). 2014;81:210-7
157. Guo Y, Liu H, Yang TL, Li SM, Li SK, Tian Q. et al. The fat mass and obesity associated gene, FTO, is also associated with osteoporosis phenotypes. PLoS One. 2011;6:e27312
158. Usategui-Martín R, Pérez-Castrillón JL, Briongos-Figuero L, Abadía-Otero J, Lara-Hernandez F, García-Sorribes S. et al. Genetic variants in obesity-related genes and the risk of osteoporotic fracture. The Hortega Follow-up Study. Front Biosci (Landmark Ed). 2022;27:32
159. Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H. et al. m(6)A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol Ther Nucleic Acids. 2020;19:421-36
160. Dong X, Liao B, Zhao J, Li X, Yan K, Ren K. et al. METTL14 mediates m(6)a modification on osteogenic proliferation and differentiation of bone marrow mesenchymal stem cells by regulating the processing of pri-miR-873. Mol Med Rep. 2023 28
161. Yang J, Wu J. Discovery of potential biomarkers for osteoporosis diagnosis by individual omics and multi-omics technologies. Expert Rev Mol Diagn. 2023;23:505-20
162. Bai Q, Shi M, Sun X, Lou Q, Peng H, Qu Z. et al. Comprehensive analysis of the m6A-related molecular patterns and diagnostic biomarkers in osteoporosis. Front Endocrinol (Lausanne). 2022;13:957742
163. Qiao Y, Li J, Liu D, Zhang C, Liu Y, Zheng S. Identification and experimental validation of key m6A modification regulators as potential biomarkers of osteoporosis. Front Genet. 2022;13:1072948
164. Zhang P, Chen H, Xie B, Zhao W, Shang Q, He J. et al. Bioinformatics identification and experimental validation of m6A-related diagnostic biomarkers in the subtype classification of blood monocytes from postmenopausal osteoporosis patients. Front Endocrinol (Lausanne). 2023;14:990078
165. Ma Y, Xu S, Xu Z, Zhang Y, Lu C, Chen D. et al. Renal safety of zoledronic acid in patients with osteoporosis: a retrospective study. Endocrine. 2024;83:459-65
166. Liu FC, Luk KC, Chen YC. Risk comparison of osteonecrosis of the jaw in osteoporotic patients treated with bisphosphonates vs. denosumab: a multi-institutional retrospective cohort study in Taiwan. Osteoporos Int. 2023;34:1729-37
167. Jin Y, Wu A, Bian S, Teng J. Icariin upregulates methyltransferase-like 14-mediated prolyl 4-hydroxylase beta subunit m6A modification to promote osteogenic differentiation of bone marrow stem cells. Exp Cell Res. 2024;440:114138
168. Tian S, Li YL, Wang J, Dong RC, Wei J, Ma Y. et al. Chinese Ecliptae herba (Eclipta prostrata (L.) L.) extract and its component wedelolactone enhances osteoblastogenesis of bone marrow mesenchymal stem cells via targeting METTL3-mediated m6A RNA methylation. J Ethnopharmacol. 2023;312:116433
169. Wang Y, Yu W, E Y, Rui L, Jia C, Zhu W. Qianggu Decoction Alleviated Osteoporosis by Promoting Osteogenesis of BMSCs through Mettl3-Mediated m(6)A Methylation. Adv Biol (Weinh). 2024: e2400341.
170. Liao M, He Q, Yang J, Gou L, Ma K, Wang H. Study on the Mechanism of Xianling Gubao Capsule Regulating Runt-Related Transcription Factor 2 (RUNX2) and Promoting Osteoblast Differentiation by N6-Methyladenosine (m6A) Methyltransferase-Like 3 (METTL3). Altern Ther Health Med. 2024
171. Wei J, Dong R, Ma Y, Wang J, Tian S, Tu X. et al. Single-cell sequencing reveals that specnuezhenide protects against osteoporosis via activation of METTL3 in LEPR(+) BMSCs. Eur J Pharmacol. 2024;981:176908
172. Cheng BR, Wu RY, Gao QY, Jiang KX, Li SS, Qi SH. et al. Chinese Proprietary Medicine Xianling Gubao Capsule for Osteoporosis: A Systematic Review and Meta-Analysis of Randomized Clinical Trials. Front Endocrinol (Lausanne). 2022;13:870277
173. Zhang G, Qin L, Shi Y. Epimedium-derived phytoestrogen flavonoids exert beneficial effect on preventing bone loss in late postmenopausal women: a 24-month randomized, double-blind and placebo-controlled trial. J Bone Miner Res. 2007;22:1072-9
174. Yong EL, Cheong WF, Huang Z, Thu WPP, Cazenave-Gassiot A, Seng KY. et al. Randomized, double-blind, placebo-controlled trial to examine the safety, pharmacokinetics and effects of Epimedium prenylflavonoids, on bone specific alkaline phosphatase and the osteoclast adaptor protein TRAF6 in post-menopausal women. Phytomedicine. 2021;91:153680
175. Li D, Li K, Zhang W, Yang KW, Mu DA, Jiang GJ. et al. The m6A/m5C/m1A Regulated Gene Signature Predicts the Prognosis and Correlates With the Immune Status of Hepatocellular Carcinoma. Front Immunol. 2022;13:918140
176. Wang L, Li J, Mei N, Chen H, Niu L, He J. et al. Identifying subtypes and developing prognostic models based on N6-methyladenosine and immune microenvironment related genes in breast cancer. Sci Rep. 2024;14:16586
177. Jiang J, Qu H, Zhan X, Liu D, Liang T, Chen L. et al. Identification of osteosarcoma m6A-related prognostic biomarkers using artificial intelligence: RBM15. Sci Rep. 2023;13:5255
Author contact
![]() Corresponding author: Xijie Yu, Laboratory of Endocrinology and Metabolism/Department of Endocrinology and Metabolism, Rare Disease Center, West China Hospital, Sichuan University, Chengdu 610041, China. Tel: +86 2885422362, Fax: +86 2885423459, E-mail: xijieyuedu.cn
Corresponding author: Xijie Yu, Laboratory of Endocrinology and Metabolism/Department of Endocrinology and Metabolism, Rare Disease Center, West China Hospital, Sichuan University, Chengdu 610041, China. Tel: +86 2885422362, Fax: +86 2885423459, E-mail: xijieyuedu.cn

 Global reach, higher impact
Global reach, higher impact