3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(1):35-49. doi:10.7150/ijms.75625 This issue Cite
Research Paper
Glutamine synthetase regulates the immune microenvironment and cancer development through the inflammatory pathway
1. Graduate Institute of Cancer Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei 11031, Taiwan.
2. Department of Neurosurgery, Taipei Medical University Hospital, Taipei 11031, Taiwan.
3. Department of Surgery, School of Medicine, College of Medicine, Taipei Medical University, Taipei 11031, Taiwan.
4. Research Center for Cancer Biology, China Medical University, Taichung 40676, Taiwan.
5. Department of Surgery, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan 70101, Taiwan.
6. Ph.D. Program for Cancer Molecular Biology and Drug Discovery, College of Medical Science, Taipei Medical University, Taipei 11031, Taiwan.
7. Department of Statistics, Faculty of Science and Technology, PGRI Adi Buana University, East Java, Surabaya 60234, Indonesia.
8. TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei 11031, Taiwan.
*Equal contributors to this work.
Abstract
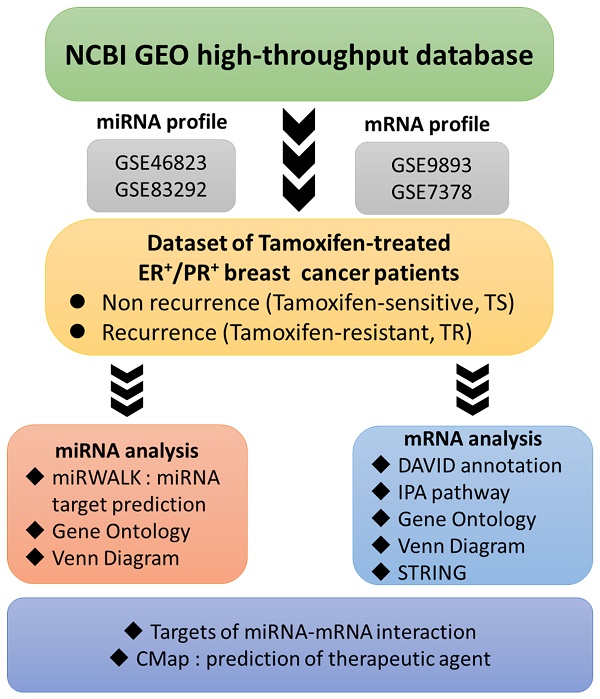
Although adjuvant tamoxifen therapy is beneficial to estrogen receptor-positive (ER+) breast cancer patients, a significant number of patients still develop metastasis or undergo recurrence. Therefore, identifying novel diagnostic and prognostic biomarkers for these patients is urgently needed. Predictive markers and therapeutic strategies for tamoxifen-resistant ER+ breast cancer are not clear, and micro (mi)RNAs have recently become a focal research point in cancer studies owing to their regulation of gene expressions, metabolism, and many other physiological processes. Therefore, systematic investigation is required to understand the modulation of gene expression in tamoxifen-resistant patients. High-throughput technology uses a holistic approach to observe differences among expression profiles of thousands of genes, which provides a comprehensive level to extensively investigate functional genomics and biological processes. Through a bioinformatics analysis, we revealed that glutamine synthetase/glutamate-ammonia ligase (GLUL) might play essential roles in the recurrence of tamoxifen-resistant ER+ patients. GLUL increases intracellular glutamine usage via glutaminolysis, and further active metabolism-related downstream molecules in cancer cell. However, how GLUL regulates the tumor microenvironment for tamoxifen-resistant ER+ breast cancer remains unexplored. Analysis of MetaCore pathway database demonstrated that GLUL is involved in the cell cycle, immune response, interleukin (IL)-4-induced regulators of cell growth, differentiation, and metabolism-related pathways. Experimental data also confirmed that the knockdown of GLUL in breast cancer cell lines decreased cell proliferation and influenced expressions of specific downstream molecules. Through a Connectivity Map (CMap) analysis, we revealed that certain drugs/molecules, including omeprazole, methacholine chloride, ioversol, fulvestrant, difenidol, cycloserine, and MK-801, may serve as potential treatments for tamoxifen-resistant breast cancer patients. These drugs may be tested in combination with current therapies in tamoxifen-resistant breast cancer patients. Collectively, our study demonstrated the crucial roles of GLUL, which provide new targets for the treatment of tamoxifen-resistant breast cancer patients.
Keywords: breast cancer, tamoxifen, glutamine synthetase (GLUL), immune microenvironment, bioinformatics

 Global reach, higher impact
Global reach, higher impact