Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(2):213-224. doi:10.7150/ijms.67116 This issue Cite
Research Paper
Potential mechanism of action of Jing Fang Bai Du San in the treatment of COVID-19 using docking and network pharmacology
1. Department of Pharmacology, Shenyang Pharmaceutical University, 110016, Shenyang, PR China
2. Department of Rehabilitation, Jin Qiu Hospital of Liaoning Province, 110016, Shenyang, PR China
Received 2021-9-14; Accepted 2021-11-2; Published 2022-1-1
Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severely infects people and has rapidly spread worldwide. JingFangBaiDu San (JFBDS) has been used to treat prevalent epidemic pathogens, common cold, headache, cough due to lung-cold, and other symptoms; however, its treatment for COVID-19 is unknown. Molecular docking and network pharmacology were applied to obtain ingredient-protein structures and the herb-ingredient-disease target network model, respectively, to explore the potential mechanism of JFBDS in COVID-19 treatment. Network pharmacology analysis showed that acacetin, wogonin, and isorhamnetin were the main active ingredients of JFBDS, and EGFR, PIK3CA, LCK, MAPK1, MAPK3, MAPK8, STAT3, TNF, IL2, and RELA were speculated to be crucial therapeutic targets. Moreover, the Toll-like receptors, HIF-1, PIK3K/AKT, MAPK, NF-κB and NOD-like receptor signaling pathways were important for JFBDS in COVID-19 treatment. Molecular docking analysis indicated that ingredients of JFBDS could bind to angiotensin converting enzyme II, spike protein, and chymotrypsin like protease (3CLpro), which inhibits virus entry and replication in host cells. This study provides a new perspective for understanding potential therapeutic effects and mechanisms of JFBDS in COVID-19 and may facilitate its clinical application.
Keywords: JingFangBaiDu San, COVID-19, network pharmacology, molecular docking
Introduction
People worldwide have been severely affected by the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) since December 2019. Clinical manifestations of the coronavirus disease 2019 (COVID-19) are fever, cough, shortness of breath, muscle ache, confusion, headache, sore throat, rhinorrhea, chest pain, diarrhea, nausea, and vomiting. Most infected patients developed acute respiratory distress syndrome and eventually died of multiple organ failure [1, 2]. SARS-CoV-2 is deadlier than Middle East respiratory syndrome coronavirus and SARS-CoV. Unlike SARS-CoV, SARS-CoV-2 can generate 3.2 fold more infectious virus particles from infected lung tissues, which indicates its higher infectivity in human lung tissues [3]. Various vaccines, such as virus vectored, protein subunit, genetic, and monoclonal antibody hindrances vaccines have been developed to date [4]. Although vaccines have recently been used in clinical practices, they need more time to gain popularity.
Traditional Chinese medicine (TCM) can protect COVID-19 patients from tissue injury, which is attributed to its anti-inflammatory, antioxidant, and anti-apoptotic effects [5]. TCM has been used to treat acute respiratory tract infections, acute bronchitis, influenza, measles, sore throat, common cold and SARS [6]. According to the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) in China [7], several Chinese herbal formulas, including Qing Fei Pai Du Decoction (QFPD), Lian Hua Qing Wen capsule (LHQW), Shen fu decoction (SF), and Xuebijing injection (XBJ) have been shown to be beneficial for COVID-19 treatment. Among multiple prescriptions for epidemic pathogens in ancient China, JingFangBaiDu San (JFBDS), an old prescription documented in “She Sheng Zhong Miao Fang” written by Shiche Zhang during the Ming dynasty deserves attention. JFBDS is used for exogenous febrile disease, prevalent epidemic pathogens, common cold, headache, dizziness, restlessness, body pain, cough due to lung-cold and other symptoms in clinical practice. The indications for JFBDS include cold dampness pathogenic factors, epidemic diseases, dysentery, sores, common cold and influenza in clinical practice [8], which are consistent with symptoms of COVID-19.
Network pharmacology has been widely applied in TCM studies in recent years [9-13]. This could transform TCM from an experience-based to an evidence-based medicine system and hasten drug discovery [14]. The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) is widely applied as a unique pharmacology platform for Chinese herbal medicines that captures relationships between drugs, targets, and diseases from TCM researches done using network pharmacology. Many studies have indicated that angiotensin-converting enzyme II (ACE2), spike protein (S1), and chymotrypsin like protease (3CLpro) play an important role in virus infection and replication. However, mechanisms of action of JFBDS against COVID-19 are unknown and ingredients interactions with COVID-19 targets have not been reported.
In this study, we constructed an herb-ingredient-disease target network model using network pharmacology to elucidate the possible effect of JFBDS in the treatment of COVID-19. Ingredients of JFBDS and target of COVID-19 were entered into Cytoscape to present herb-ingredient-disease target network model. Moreover, the intersectant targets (named ingredient-disease-targets) made from intersecting ingredient-associated targets and COVID-19-associated disease targets were analyzed using DAVID database. Finally, to verify the validity of the predicted results, molecular docking was used to analyze the interaction of ingredients with ACE2, S1, and 3CLpro.
Methods and Materials
Experimental design
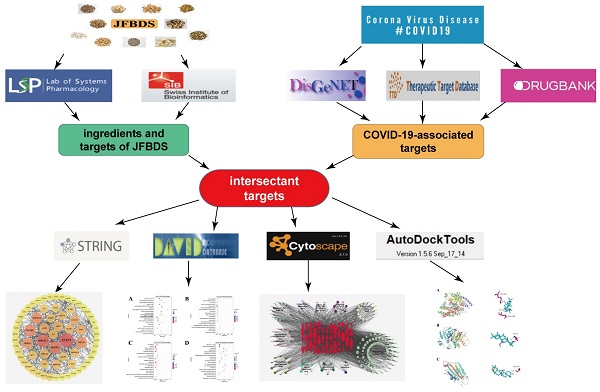
The experimental design is shown in Figure 1. Ingredients of JFBDS were acquired using TCMSP and targets of these ingredients were predicted by Swiss Target Prediction (http://swisstargetprediction.ch/). COVID-19 associated disease targets were mined using DisGeNet (https://www.disgenet.org), TTD (http://db.idrblab.net/ttd), and Drug Bank (https://www.drugbank.ca). Subsequently, ingredient-disease-targets were analyzed using DAVID database. These data were entered into Cytoscape to present the herb-ingredient-disease target network model.
The experiment design of JFBDS on COVID-19 by network pharmacology and molecular docking
Collection of ingredients in JFBDS
JFBDS containes eleven herbs, including Bupleurum chinense DC. (Chaihu), Ligusticum chuanxiong Hort. (Chuanxiong), Angelica pubescens Maxim. f. biserrata Shan et Yuan (Duhuo), Saposhnikovia divaricata (Turcz.) Schischk. (Fangfeng), Poria cocos (Schw.) Wolf (Fuling), Glycyrrhiza uralensis Fisch. (Gancao), Schizonepeta tenuifolia Briq. (Jingjie), Platycodon grandiflorum (Jacq.) A. DC. (Jiegeng), Peucedanum praeruptorum Dunn (Qianhu), Notopterygium inchum Ting ex H.T. Chang (Qianghuo) and Citrus aurantium L. (Zhiqiao). The quantity of Gancao in this prescription was 1.5 g and that of other herbs was 4.5 g. The instructions in TCMSP, conditions of oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 were limitations to obtaining the potential ingredients.
Targets prediction of ingredients in JFBDS
Targets of ingredients in JFBDS were obtained by entering the SMILES numbers of each ingredient into the Swiss Target Prediction basement, which predicted targets of the ingredients based on 2D and 3D structures of the ingredient.
Collection of COVID-19-associated targets
DisGeNet, TTD and Drug Bank were used to collect COVID-19-associated targets by searching novel coronavirus, Pneumonia and cytokine-related in above three databases.
Construction of network and bioinformatics analysis
Intersectant targets were obtained by intersecting the ingredient-associated targets and COVID-19-associated disease targets. The ingredient-disease-targets were entered into STRING to obtain the protein-protein interaction (PPI) network. To display the relationship between herb, ingredient, and ingredient-disease-target, the herb-ingredient-disease target network model was created using Cytoscape. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using DAVID (https://david.ncifcrf.gov/). The results are presented as bubble charts drawn using OmicShare platform (https://www.omicshare.com/tools/).
Ingredient-protein molecular docking
3D structures of the ACE2 (PDB ID: 6M2N), S1 (PDB ID:7BZ5) and 3CLpro (PDB ID: 6LU7) were downloaded from the RCSB PDB database (https://www.rcsb.org/). AutoDock Tools 1.5.6 software was used to process protein structures, including removing water molecules, adding hydrogen, calculating Gasteiger charges. The PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was used to download the two-dimensional (2D) structures of the top ten compounds. The 2D structure was processed and converted into the PDB format using Chem3D. Structures of the three proteins (ACE2, S1, and 3CLpro) and their ligands were obtained using PyMol 2.4. Ten ingredients and three proteins were produced using molecular docking. Finally, the three proteins were produced with their original ligands using molecular docking. Binding energies were obtained and visualized using PyMol 2.4.
Results
Collection of chemical ingredients from JFBDS and predicting of potential targets
Chemical ingredients of these herbs were collected using TCMSP, which contained 220 active ingredients. The representative active ingredients are shown in Table 1, and detailed chemical ingredient information is provided in the supplementary materials. The structural information of 25 chemical ingredients was not available; therefore, their targets could not be predicted. Potential targets of the remaining 195 chemical ingredients were predicted using by Swiss Target Prediction, and 1073 potential targets were collected. The representative potential targets are shown in Table 2, and detailed target information is provided in the supplementary materials.
Collection of COVID-19-associated targets
DisGeNet, TTD, and Drug Bank were used to search for COVID-19 targets. Keywords including COVID-19, pneumonia, and cytokine-related syndrome were used to obtain 747 targets. The information on partial COVID-19 associated targets is shown in Table 3, while detailed target information is provided in the supplementary materials.
Collection of ingredient-disease-targets and analysis of PPI network
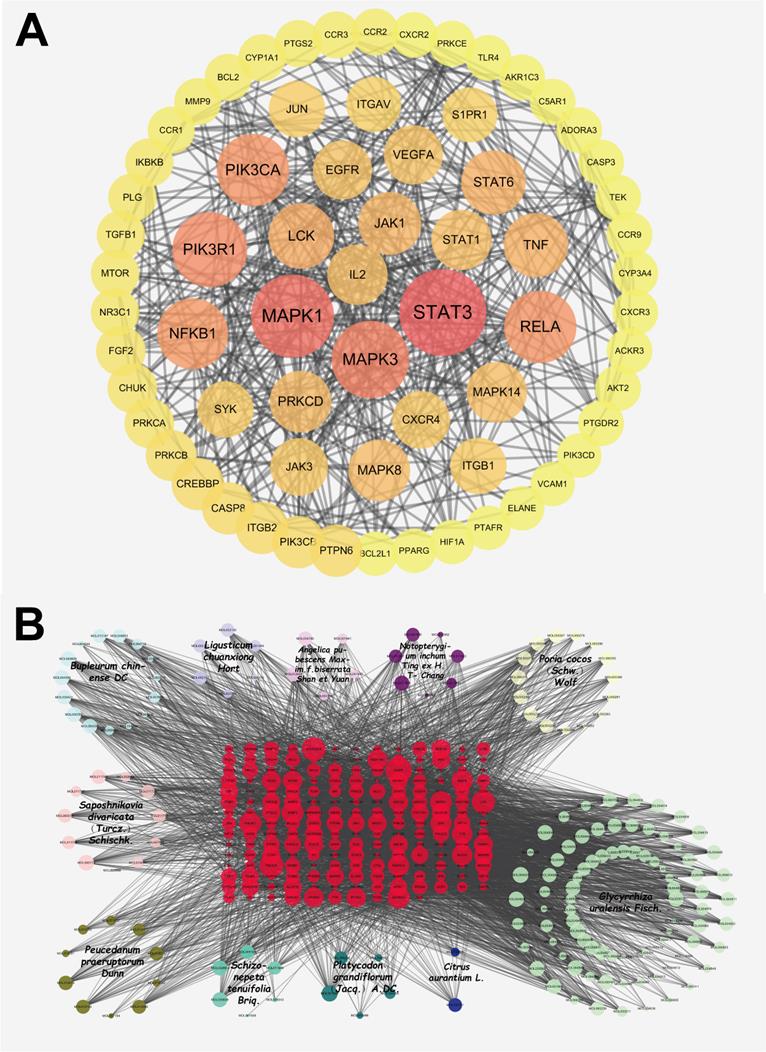
According to the method, 192 ingredient-disease-targets were obtained. Thereafter, the STRING database was used to obtain a PPI network of 192 interactive targets by limiting the highest degree (degree ≥ 0.9) of confidence level and eliminating independent target proteins. This network finally contained 192 nodes and 772 edges (Figure 2A), in which nodes represent the target, edges represent the relationship between the targets, and different colors represent different cluster interactions. To display the PPI network intuitively, the degree values from 0 to 9 were hidden (Figure 2A). The top ten targets of the degree values are displayed in the supplementary materials.
The information of partial active ingredients in JFBDS
| Herb | MOL ID | Ingredient | OB% | DL |
|---|---|---|---|---|
| Bupleurum chinense DC. (Chaihu) | MOL001645 | Linoleyl acetate | 42.1 | 0.2 |
| MOL000449 | Stigmasterol | 43.83 | 0.76 | |
| MOL000354 | isorhamnetin | 49.6 | 0.31 | |
| MOL001645 | Linoleyl acetate | 42.1 | 0.2 | |
| Ligusticum chuanxiong Hort. (Chuanxiong) | MOL001494 | Mandenol | 42 | 0.19 |
| MOL002135 | Myricanone | 40.6 | 0.51 | |
| MOL002140 | Perlolyrine | 65.95 | 0.27 | |
| MOL002151 | senkyunone | 47.66 | 0.24 | |
| Angelica pubescens Maxim.f.biserrata Shan et Yuan (Duhuo) | MOL001941 | Ammidin | 34.55 | 0.22 |
| MOL001942 | isoimperatorin | 45.46 | 0.23 | |
| MOL000358 | beta-sitosterol | 36.91 | 0.75 | |
| MOL003608 | O-Acetylcolumbianetin | 60.04 | 0.26 | |
Saposhnikovia divaricata (Turcz.) Schischk. (Fangfeng) | MOL000011 | (2R,3R)-3-(4-hydroxy-3-methoxy-phenyl)-5-methoxy-2-methylol-2,3-dihydropyrano[5,6-h] [1,4] benzodioxin-9-one | 68.83 | 0.66 |
| MOL011730 | 11-hydroxy-sec-o-beta-d-glucosylhamaudol_qt | 50.24 | 0.27 | |
| MOL011732 | anomalin | 59.65 | 0.66 | |
| MOL011737 | divaricatacid | 87 | 0.32 | |
Poria cocos (Schw.) Wolf (Fuling) | MOL000273 | (2R)-2-[(3S,5R,10S,13R,14R,16R,17R)-3,16-dihydroxy-4,4,10,13,14-pentamethyl-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta[a]phenanthren-17-yl]-6-methylhept-5-enoic acid | 30.93 | 0.81 |
| MOL000275 | trametenolic acid | 38.71 | 0.8 | |
| MOL000276 | 7,9(11)-dehydropachymic acid | 35.11 | 0.81 | |
| MOL000279 | Cerevisterol | 37.96 | 0.77 | |
| Glycyrrhiza uralensis Fisch. (Gancao) | MOL001484 | Inermine | 75.18 | 0.54 |
| MOL001792 | DFV | 32.76 | 0.18 | |
| MOL000211 | Mairin | 55.38 | 0.78 | |
| MOL002311 | Glycyrol | 90.78 | 0.67 | |
| Schizonepeta tenuifolia Briq. (Jingjie) | MOL011849 | Schizonepetoside B | 31.02 | 0.28 |
| MOL011856 | Schkuhrin I | 54.45 | 0.52 | |
| MOL002881 | Diosmetin | 31.14 | 0.27 | |
| MOL000359 | sitosterol | 36.91 | 0.75 | |
| Platycodon grandiflorum (Jacq.) A. DC. (Jiegeng) | MOL001689 | acacetin | 34.97 | 0.24 |
| MOL004355 | Spinasterol | 42.98 | 0.76 | |
| MOL004580 | cis-Dihydroquercetin | 66.44 | 0.27 | |
| MOL005996 | 2-O-methyl-3-O-β-D-glucopyranosyl platycogenate A | 45.15 | 0.25 | |
| Peucedanum praeruptorum Dunn (Qianhu) | MOL013076 | (8S,9R)-9-hydroxy-8-(2-hydroxypropan-2-yl)-8,9-dihydrofuro[2,3-h] chromen-2-one | 37.3 | 0.2 |
| MOL013077 | Decursin | 39.27 | 0.38 | |
| MOL013078 | praeruptorin E | 51.22 | 0.66 | |
| MOL013079 | dl-praeruptorin a | 46.46 | 0.53 | |
| Notopterygium inchum Ting ex H.T. Chang (Qianghuo) | MOL001941 | Ammidin | 34.55 | 0.22 |
| MOL011962 | 6'-Feruloylnodakenin | 32.02 | 0.67 | |
| MOL011963 | 8-geranoxy-5-methoxypsoralen | 40.97 | 0.5 | |
| MOL011968 | coumarin,glycoside | 33.07 | 0.78 | |
| Citrus aurantium L. (Zhiqiao) | MOL013381 | Marmin | 38.23 | 0.31 |
| MOL002341 | Hesperetin | 70.31 | 0.27 | |
| MOL000358 | beta-sitosterol | 36.91 | 0.75 | |
| MOL004328 | naringenin | 59.29 | 0.21 |
The information of partial potential targets in JFBDS
| Gene Symbol | UniProt ID | Gene Symbol | UniProt ID |
|---|---|---|---|
| ALOX12 | P18054 | MMP3 | P08254 |
| BTK | Q06187 | MMP9 | P14780 |
| CD81 | P60033 | NFKB1 | P19838 |
| CTSK | P43235 | NLRP3 | Q96P20 |
| NOD2 | Q9HC29 | CASP1 | P29466 |
| RELA | Q04206 | CASP8 | Q14790 |
| ICAM1 | P05362 | MAPK3 | P27361 |
| IKBKB | O14920 | JUN | P05412 |
| IL2 | P60568 | PIK3CA | P42336 |
| IRAK4 | Q9NWZ3 | PIK3R1 | P27986 |
| TLR4 | O00206 | STAT1 | P42224 |
| TLR9 | Q9NR96 | STAT3 | P40763 |
| TNF | P01375 | TBK1 | Q9UHD2 |
| VCAM1 | P19320 | SYK | P43405 |
| MAPK1 | P28482 | JAK1 | P23458 |
The information of partial COVID-19-associated targets
| Gene Symbol | UniProt ID | Gene Symbol | UniProt ID |
|---|---|---|---|
| CD19 | P15391 | TNF | P01375 |
| CRP | P02741 | IL13 | P35225 |
| IFNG | P01579 | CSF2 | P04141 |
| IL6 | P05231 | IL4 | P05112 |
| TWIST1 | Q15672 | CXCR2 | P25025 |
| CARTPT | Q16568 | IL33 | O95760 |
| IL1B | P01584 | CXCL5 | P42830 |
| CXCL1 | P09341 | HMOX1 | P09601 |
| CCL2 | P13500 | MYD88 | Q99836 |
| CCL11 | P51671 | TGFB1 | P01137 |
| IL18 | Q14116 | CCL24 | O00175 |
| CCL17 | Q92583 | CASP1 | P29466 |
| ITGB2 | P05107 | SIRT1 | Q96EB6 |
| TLR4 | O00206 | IL6ST | P40189 |
| IL17A | Q16552 | CD40LG | P29965 |
Creation of herb-ingredient-disease targets network in JFBDS
As shown in Figure 2B, the network of herb-ingredient-disease targets was built using Cytoscape. 360 nodes and 3901 edges were obtained in the herb-ingredient-disease targets network. According to the degree in network, the top ten ingredients were MOL001689 (acacetin), MOL004835 (glypallichalcone), MOL000173 (wogonin), MOL004905 (glyuranolide), MOL004598 (3',4',5',3,5,6,7-Heptamethoxy flavone), MOL004828 (glepidotin A), MOL004856 (gancaonin A), MOL005013 (18α-hydroxy glycyrrhetic acid), MOL000354 (Isorhamnetin) and MOL002565 (medicarpin). The top targets were mainly EGFR, PIK3CA, LCK, MAPK1, MAPK3, MAPK8, STAT3, TNF, IL2, and RELA. This suggests that JFBDS has therapeutic effects on COVID-19 probably through the interaction between the ingredients and targets. Detailed information on the top ten ingredients and targets are shown in Tables 4 and 5.
The PPI network of ingredient-disease-targets (A). The interaction network of herb-ingredient-disease targets (B). Among this, red color nodes represented ingredient-disease-targets; light blue nodes represented Bupleurum chinense DC. (Chaihu), dark bule nodes represented Citrus aurantium L. (Zhiqiao), light purple nodes represented Ligusticum chuanxiong Hort. (Chuanxiong), dark purple nodes represented Notopterygium inchum Ting ex H.T. Chang (Qianghuo), light pink nodes represented Saposhnikovia divaricata (Turcz.) Schischk. (Fangfeng), dark pink nodes represented Angelica pubescens Maxim.f.biserrata Shan et Yuan (Duhuo), light yellow nodes represented Poria cocos (Schw.) Wolf (Fuling), dark yellow nodes represented Peucedanum praeruptorum Dunn (Qianhu), light green nodes represented Glycyrrhiza uralensis Fisch. (Gancao), dark green nodes represented Platycodon grandiflorum (Jacq.) A. DC. (Jiegeng) and bright green nodes represented Schizonepeta tenuifolia Briq. (Jingjie).
The information of top ten ingredients of degree values from analysis cytoscape
| MOL ID | Molecule Name | Herb |
|---|---|---|
| MOL001689 | Acacetin | Platycodon grandiflorum (Jacq.) A. DC. (Jiegeng) |
| MOL004835 | Glypallichalcone | Glycyrrhiza uralensis Fisch. (Gancao) |
| MOL000173 | Wogonin | Saposhnikovia divaricata (Turcz.) Schischk. (Fangfeng) |
| MOL004905 | Glyuranolide | Glycyrrhiza uralensis Fisch. (Gancao) |
| MOL004598 | 3',4',5',3,5,6,7-Heptamethoxy flavone | Bupleurum chinense DC. (Chaihu) |
| MOL004828 | Glepidotin A | Glycyrrhiza uralensis Fisch. (Gancao) |
| MOL004856 | Gancaonin A | Glycyrrhiza uralensis Fisch. (Gancao) |
| MOL005013 | 18α-hydroxy glycyrrhetic acid | Glycyrrhiza uralensis Fisch. (Gancao) |
| MOL000354 | Isorhamnetin | Glycyrrhiza uralensis Fisch. (Gancao) and Bupleurum chinense DC. (Chaihu) |
| MOL002565 | Medicarpin | Glycyrrhiza uralensis Fisch. (Gancao) |
The information of top 50 targets of degree values from analysis cytoscape
| Name of targets | Degree | Name of targets | Degree |
|---|---|---|---|
| EGFR | 85 | PTGS2 | 47 |
| PIK3CA | 73 | ALOX12 | 46 |
| ADORA2A | 71 | JAK3 | 46 |
| LCK | 69 | ALOX15 | 45 |
| ADORA3 | 64 | CDK4 | 45 |
| PIK3R1 | 64 | TNF | 45 |
| MMP3 | 63 | ADAM17 | 44 |
| MAPK1 | 62 | MAPK3 | 44 |
| MAPK14 | 60 | PIK3CB | 43 |
| MMP9 | 60 | PLA2G1B | 43 |
| SYK | 60 | ADORA2B | 42 |
| ALOX5 | 57 | CYP17A1 | 42 |
| F10 | 57 | SHBG | 42 |
| HSD11B1 | 53 | ERN1 | 41 |
| MTOR | 52 | IL2 | 40 |
| PPARG | 52 | MMP8 | 40 |
| ABCG2 | 51 | JAK1 | 39 |
| MMP13 | 51 | PIK3CG | 39 |
| PTGS1 | 51 | PTPN6 | 38 |
| NOS2 | 49 | RELA | 38 |
| PARP1 | 49 | AKR1B1 | 37 |
| NR3C1 | 48 | ITGB1 | 37 |
| STAT3 | 48 | S1PR1 | 37 |
| ABCB1 | 47 | PIK3CD | 36 |
| MAPK8 | 47 | AKR1C3 | 35 |
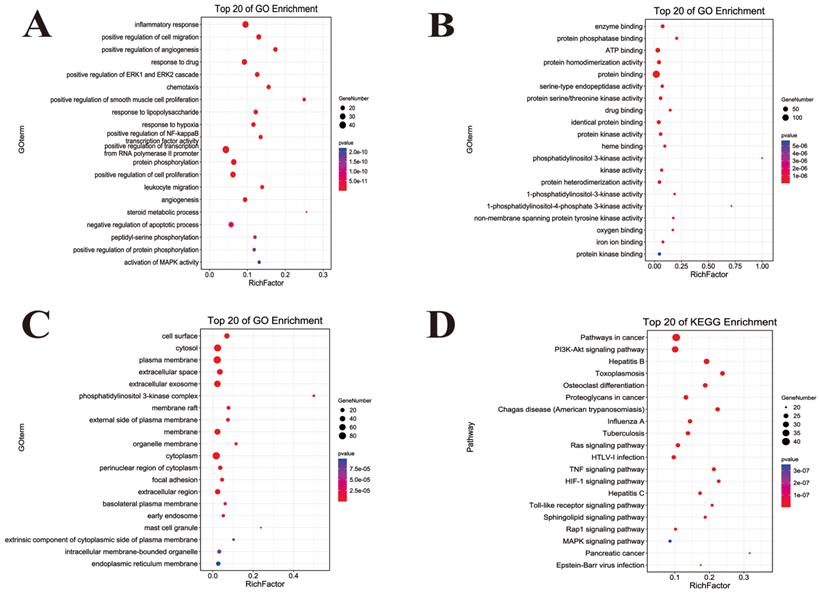
GO and KEGG functional annotation analysis of ingredient-disease-targets in JFBDS for treating COVID-19. GO enrichment of putative targets was divided into biological process (A), molecular function (B) and cellular component (C). The pathway of putative targets was predicted by KEGG enrichment (D).
Bioinformatics of ingredient-disease-target
The GO function enrichment analysis of ingredient-disease-targets was performed using DAVID in the categories of biological process (BP), molecular function (MF) and cellular component (CC) are shown in Figure 3. In BP term, the results showed that inflammatory response, chemotaxis, response to lipopolysaccharide (LPS) positive regulation of NF-κB transcription factor activity, and leukocyte migration played important roles in the treatment of COVID-19. In MF terms, the results demonstrated that ATP and protein binding were two important molecular functions. In CC terms, the cytosol, plasma membrane and cytoplasm were the main sites of the targets. To uncover the potential therapeutic mechanisms of JFBDS in COVID-19, the ingredient-disease-targets were analyzed using KEGG. The top 20 pathways of KEGG enrichment analysis are shown in Figure 4D. Among these, the most significant were the Toll-like receptors, HIF-1, PIK3K/AKT, MAPK, NF-κB and NOD-like receptor signaling pathways.
Molecular docking of 18α-hydroxy glycyrrhetic acid bound with ACE2 (A). Molecular docking of Glyuranolide bound with 3CLpro and S1(B, C).
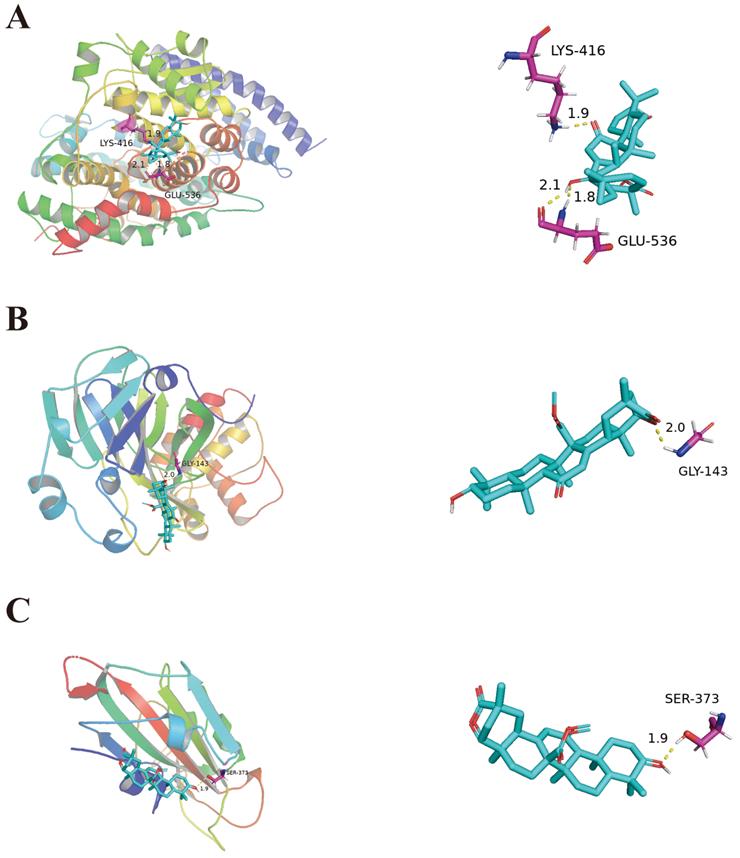
Results of molecular docking
Molecular docking was used to verify the impact of top ten ingredients on ACE2, S1, and 3CLpro. The binding energies of ACE2, S1, and 3CLpro with their original ligands were -2.51 kJ/mol, -4.6 kJ/mol and -5.44 kJ/mol, respectively. These results indicate that the top ten ingredients have a strong affinity for ACE2, S1, and 3CLpro. For instance, 18α-hydroxy glycyrrhetic acid was strongly bound to ACE2. Glyuranolide was strongly associated with S1 and 3CLpro. As shown in Figure 4, 18α-hydroxy glycyrrhetic acid was bound to ACE2 through three hydrogen bonds with the amino acid residues Lys416 and Glu536. Glyuranolide was bound to S1 and 3CLpro by one hydrogen bond with the amino acid residues Gly143 and Ser373, respectively. The binding energies of various compounds are shown in Table 6.
Molecular docking results of aim proteins and the ten ingredients of top degree
| MOL ID | Molecule Name | ACE2 binding energy (kJ/moL) | S1 binding energy (kJ/moL) | 3CL binding energy (kJ/moL) |
|---|---|---|---|---|
| MOL001689 | Acacetin | -4.1 | -6.14 | -5.81 |
| MOL004835 | Glypallichalcone | -3.84 | -6.1 | -6.24 |
| MOL000173 | Wogonin | -4.1 | -4.59 | -6.6 |
| MOL004905 | Glyuranolide | -5.66 | -6.97 | -7.09 |
| MOL004598 | 3',4',5',3,5,6,7-Heptamethoxy flavone | -3.03 | -3.05 | -4.29 |
| MOL004828 | Glepidotin A | -4.92 | -4.33 | -4.53 |
| MOL004856 | Gancaonin A | -4.23 | -5.42 | -6.44 |
| MOL005013 | 18α-hydroxy glycyrrhetic acid | -5.73 | -6.42 | -6.85 |
| MOL000354 | Isorhamnetin | -4.5 | -5.52 | -6.61 |
| MOL002565 | Medicarpin | -5.39 | -5.56 | -5.91 |
Discussion
COVID-19 is a clinical syndrome caused by SARS-CoV-2 that has been sweeping the globe since December 2019. COVID-19 may result in cytokine storm syndrome and acute respiratory distress syndrome [15]. Cytokine storms contain several disorders of immune dysregulation characterized by systemic inflammation and multiple organ dysfunction [16], which results in the clinical deterioration of COVID-19 patients. Many cytokines played a crucial role in COVID-19, such as IL-2, IL-6, IL-7, IL-8, IL-1β, IL10 and TNF-α [2, 17]. Moreover, many signaling pathways, including the toll-like receptors, NF-κB, MAPK, and TNF, are involved in COVID-19 [18, 19]. Overcoming this epidemic has become the toughest issue worldwide. Currently, antiviral therapy is widely used in clinical practices, including alpha-interferon, lopinavir/ritonavir, ribavirin, chloroquine phosphate, and arbidol [20]. However, these medicines have some side effects, and the combination of these medicines is not recommended [21]. Despite the partial use of vaccines, effective drug development remains an urgent task.
The TCM treatment approach has been widely used to treat COVID-19 in China. Many studies have reported that TCM could resist viral pneumonia by modulating cytokine production, controlling viral replication, and regulating immunological function [22]. According to the COVID-19 seventh edition diagnostic criteria, four prescriptions, QFPD, LHQW, SF, and XBJ, achieved satisfactory results [13, 19, 23, 24]. Among them, QFPD was reported to exhibit immune regulation, anti-infection, anti-inflammation and multi-organ protection by regulating toll-like receptors, NF-κB, and MAPK signaling pathways, which subsequently inhibits the release of pro-inflammatory factors [18, 23]. SF and XBJ inhibit inflammation and regulate immunity. XBJ also alleviated pneumonia-induced multi-organ damage [19, 24]. LHQW significantly inhibited SARS-CoV-2 replication and diminished the cytokine release [13]. These data indicate that TCM plays an irreplaceable role in immune regulation, anti-infection, anti-inflammation, and multi-organ protection. JFBDS contains eleven herbs that have been used to treat cough, influenza, and respiratory tract infections for nearly four hundred years. According to the indications of JFBDS, it could treat epidemic pathogens and lung diseases, improve digestive tract symptoms, and relieve pain, which is consistent with most symptoms of COVID-19.
According to the degree value of the targets, several potential active ingredients that played an important role in the treatment of COVID-19, including acacetin, glypallichalcone, wogonin, gancaonin A, and isorhamnetin were screened in JFBDS. Acacetin was obtained from Schizonepeta tenuifolia Briq. Glypallichalcone and Gancaonin A were obtained from Glycyrrhiza uralensis Fisch. Wogonin was obtained from Saposhnikovia divaricata (Turcz.) Schischk. Isorhamnetin was obtained from both Glycyrrhiza uralensis Fisch. and Bupleurum chinense DC. Acacetin is a flavone that has anti-inflammatory effects [25, 26]. It has been reported to decrease the activity of cyclooxygenase-2 (COX-2) and nitric oxide synthase (NOS) [27], suppress levels of proinflammatory cytokines and chemokines, reduced the adhesion of eosinophils to tracheal epithelial cells [28], and inhibit sepsis-induced acute lung injury [26, 29]. Wogonin is also a flavonoid with anti-inflammatory, anti-fibrotic, and anti-cancer effects [30]. Some studies have shown that the possible mechanism of action of wogonin on inflammation was associated with the reduction of phosphorylation and activation of transcription factors STAT1 or STAT3 [31]. Moreover, wogonin inhibits the expression of COX-2 and HIF-1α and reverses hypoxia resistance by regulating the PI3K/Akt signaling pathway [21, 32]. As one of the major metabolites of quercetin, isorhamnetin has anti-inflammatory activity and reduces the expression of inflammatory genes in macrophages stimulated by LPS [33]. It also significantly inhibits the inflammation, proliferation and migration of BEAS-2B cells by regulating the MAPK and NF-κB signaling pathways [34]. Therefore, it is reasonable to speculate that acacetin, wogonin, and isorhamnetin are the main ingredients of JFBDS that treat COVID-19 by regulating PI3K/AKT, STAT3, MAPK, and NF-κB signaling pathways.
According to the results of cytoscape, the targets of JFBDS for treating COVID-19 mainly contained EGFR, PIK3CA, lymphocyte-specific protein tyrosine kinase (LCK), mitogen-activated protein kinase 1 (MAPK1), MAPK3, MAPK8, STAT3, TNF, IL2 and RELA. These targets were mainly involved in toll-like receptors, HIF-1, PIK3K/AKT, NF-κB and NOD-like receptor signaling pathways, which have important effects on cytokines. For example, EGFR is related to pulmonary inflammation and its phosphorylation can regulate the expression of pro-inflammatory factors, such as IL-8 and CXCL1 [35, 36]. EGFR also activates multiple signaling pathways, such as PI3K/Akt/mTOR, JAK/STAT and NF-κB, which are closely related to inflammation [37]. LCK, a cytoplasmic tyrosine kinase expressed in T cells and natural killer cells, is a necessary step to activate T cell. Therefore, selective inhibition of LCK can produce immunosuppressive effects [38]. As an important pathway for extracellular signal transduction into the nucleus, MAPK cascade plays an important role in cell proliferation, differentiation and apoptosis [39]. MAPK1, MAPK3, MAPK8 and MAPK14 can be activated by stimulating phosphorylation during changes in the internal environment, such as osmotic pressure changes, oxidative stress, inflammatory factors and viral infection. By suppressing MAPK signalling pathways, flavonoids inhibit the production of inflammatory cytokines in LPS-stimulated macrophages [40]. IL-2, with multiple effects on the immune system, was considered a key cytokine because of its regulatory effects on T cells. IL-2 performs crucial functions during immune homeostasis, which can optimize and regulate lymphocyte responses [41]. IL-2 can also activate the STAT, PI3K/AKT and MAPK signaling pathways [42]. In GO terms analysis, it was found that inflammatory response, chemotaxis, response to LPS, positive regulation of NF-κB transcription factor activity and leukocyte migration might play an important role in the treatment of COVID-19. KEGG pathway analysis revealed that the toll-like receptors, HIF-1, PIK3K/AKT, MAPK, NF-κB, and NOD-like receptor signaling pathways had significant effects on COVID-19 therapy. The toll-like receptors and PI3K/Akt signaling pathways have already been identified as important for anti-inflammatory effects on COVID-19 [18, 24]. The activation of PI3K/Akt/mTOR pathway causes pulmonary fibrosis and lung injury by increasing lung fibroblasts and lung epithelial cells [13]. This indicates that it might be closely related to the development and treatment of COVID-19 [43].
Some studies have reported targets closely related to COVID-19. ACE2 mainly exists among he epithelial cells of alveoli, trachea, bronchus, serous bronchial glands, alveolar monocytes, and macrophages of the lower respiratory tract. It is the cellular receptor of COVID-19 [44]. The entry of SARS-CoV-2 into cells requires S1 binding to the cell surface receptor. S1 also facilitates the attachment of target cells to the viral surface of the S protein [45]. 3CLpro is present in the multi-protein ORF1Ab of SARS-CoV-2 and is an attractive target against SARS-CoV-2 because of its importance in the replication of the virus[46]. Therefore, the main ingredients of high degree were docked with ACE2, 3CLpro, and S1 proteins. In the molecular docking process, lower the binding energy, more effective is the molecular association. The ingredient-protein interaction with docking energy less than 5 kJ/mol signified effective docking [47]. According to the results of molecular docking, the sectional ingredients showed a strong binding affinity to the three proteins. In particular, 18α-hydroxy glycyrrhetic acid and glyuranolide showed high affinity, indicating that both were important active ingredients of JFBDS for COVID-19. Glypallichalcone, glyuranolide, glepidotin A, gancaonin A, 18α-hydroxy glycyrrhetic acid, medicarpin, and isorhamnetin were seven of the top ten ingredients with high binding energy that were derived from Glycyrrhiza uralensis Fisch. However, isorhamnetin was also derived from Bupleurum chinense DC. Therefore, Glycyrrhiza uralensis Fisch. took an irreplaceable position in JFBDS [48]. The ingredients, targets, and signaling pathways of JFBDS in COVID-19 are shown in Figure 5. However, exact ingredients and therapeutic targets of JFBDS need to be studied further and confirmed.
The ingredients, targets and signalling pathway of JFBDS on COVID-19.
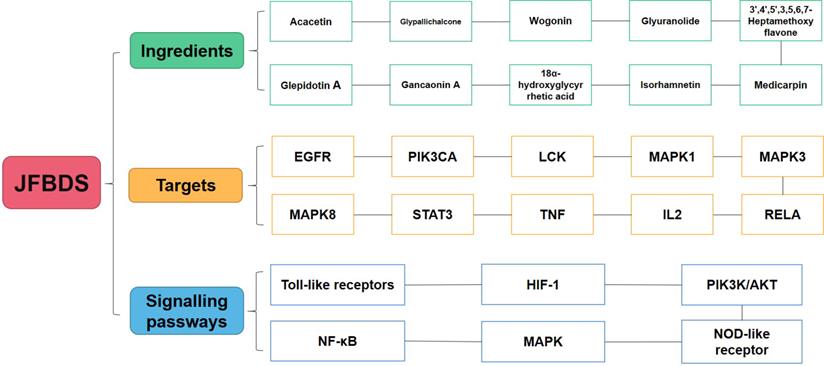
Conclusion
In this study, 220 active ingredients of JFBDS were collected using TCMSP and 192 ingredient-disease-targets were collected from Swiss Target Prediction database, DisGeNet, TTD, and DrugBank. Bioinformatics analysis revealed that acacetin, wogonin, and isorhamnetin were main active ingredients of JFBDS for the treatment of COVID-19. EGFR, PIK3CA, LCK, MAPK1, MAPK3, MAPK8, STAT3, TNF, IL2, and RELA were crucial therapeutic targets for the treatment of COVID-19 using JFBDS. Moreover, the toll-like receptors, HIF-1, PIK3K/AKT, MAPK, NF-κB, and NOD-like receptor signaling pathways were involved in the treatment of COVID-19. The partially predicted ingredients of JFBDS were verified using molecular docking. This study provides a new perspective for understanding the potential therapeutic effect and mechanism of action of JFBDS on COVID-19 and facilitates its clinical application.
Abbreviations
JingFangBaiDu San, JFBDS; coronavirus disease 2019, COVID-19; severe acute respiratory syndrome corona virus 2, SARS-CoV-2; traditional Chinese medicine, TCM; Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform, TCMSP; severe acute respiratory syndrome coronavirus, SARS-CoV; Qing Fei Pai Du Decoction, QFPD; Lian Hua Qing Wen Capsule, LHQW; Shen fu Decoction, SF; Xuebijing injection, XBJ; Therapeutic Target Database, TTD; oral bioavailability, OB; drug-likeness, DL; protein-protein interaction, PPI; Gene Ontology, GO; Kyoto Encyclopedia of Genes and Genomes, KEGG; angiotensin converting enzyme II, ACE2.
Supplementary Material
Supplementary information.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Author Contributions
Experimental operation, data acquisition and validation and manuscript writing: Jiaojiao Li, Kuo Zhang. Review and editing: Kuo Zhang, Jingyu Yang and Chunfu Wu. Design of the project: Chunfu Wu, Jimin Bao. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395:507-13
2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497-506
3. Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y. et al. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study With Implications for the Pathogenesis of COVID-19. Clin Infect Dis. 2020;71:1400-9
4. Kaur SP, Gupta V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020;288:198114
5. Zhang JL LW, Li Y, Wong MS, Wang YJ, Zhang Y. Therapeutic options of TCM for organ injuries associated with COVID-19 and the underlying mechanism. Phytomedicine. 2020 153297
6. Wu T, Yang X, Zeng X, Poole P. Traditional Chinese medicine in the treatment of acute respiratory tract infections. Respir Med. 2008;102:1093-8
7. Wang H, Li X, Li T, Zhang S, Wang L, Wu X. et al. The genetic sequence, origin, and diagnosis of SARS-CoV-2. European Journal of Clinical Microbiology & Infectious Diseases. 2020;39:1629-35
8. Li L, Yu AL, Wang ZL, Chen K, Zheng W, Zhou JJ. et al. Chaihu-Shugan-San and absorbed meranzin hydrate induce anti-atherosclerosis and behavioral improvements in high-fat diet ApoE(-/-) mice via anti-inflammatory and BDNF-TrkB pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;115:108893
9. Zhang M, Yuan Y, Zhou W, Qin Y, Xu K, Men J. et al. Network pharmacology analysis of Chaihu Lizhong Tang treating non-alcoholic fatty liver disease. Computational Biology and Chemistry. 2020 86
10. Huang S, Zhang Z, Li W, Kong F, Yi P, Huang J. et al. Network Pharmacology-Based Prediction and Verification of the Active Ingredients and Potential Targets of Zuojinwan for Treating Colorectal Cancer. Drug Design, Development and Therapy. 2020 Volume 14: 2725-40
11. Xuan X, Sun Z, Yu C, Chen J, Chen M, Wang Q. et al. Network pharmacology-based study of the protective mechanism of conciliatory anti-allergic decoction on asthma. Allergologia et Immunopathologia. 2020;48:441-9
12. Xu T, Wang Q, Liu M. A Network Pharmacology Approach to Explore the Potential Mechanisms of Huangqin-Baishao Herb Pair in Treatment of Cancer. Medical science monitor: international medical journal of experimental and clinical research. 2020;26:e923199
13. Xia QD, Xun Y, Lu JL, Lu YC, Yang YY, Zhou P. et al. Network pharmacology and molecular docking analyses on Lianhua Qingwen capsule indicate Akt1 is a potential target to treat and prevent COVID-19. Cell Prolif. 2020;53:e12949
14. Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines. 2013;11:110-20
15. Soy M, Keser G, Atagunduz P, Tabak F, Atagunduz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085-94
16. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med. 2020;383:2255-73
17. Chen X, Yin YH, Zhang MY, Liu JY, Li R, Qu YQ. Investigating the mechanism of ShuFeng JieDu capsule for the treatment of novel Coronavirus pneumonia (COVID-19) based on network pharmacology. Int J Med Sci. 2020;17:2511-30
18. Yang R, Liu H, Bai C, Wang Y, Zhang X, Guo R. et al. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): In silico and experimental study. Pharmacol Res. 2020;157:104820
19. Li X, Lin H, Wang Q, Cui L, Luo H, Luo L. Chemical composition and pharmacological mechanism of Shenfu decoction in the treatment of novel coronavirus Pneumonia (COVID-19). Drug Dev Ind Pharm. 2020:1-32
20. Trivedi N, Verma A, Kumar D. Possible treatment and strategies for COVID-19: review and assessment. European review for medical and pharmacological sciences. 2020;24:12593-608
21. Wang H, Zhao L, Zhu LT, Wang Y, Pan D, Yao J. et al. Wogonin reverses hypoxia resistance of human colon cancer HCT116 cells via downregulation of HIF-1α and glycolysis, by inhibiting PI3K/Akt signaling pathway. Mol Carcinog. 2014;53(Suppl 1):E107-18
22. Zhang DH, Wu KL, Zhang X, Deng SQ, Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. 2020;18:152-8
23. Zhao J, Tian S, Lu D, Yang J, Zeng H, Zhang F. et al. Systems pharmacological study illustrates the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanism of Qing-Fei-Pai-Du decoction in the treatment of COVID-19. Phytomedicine. 2020: 153315.
24. Zheng WJ, Yan Q, Ni YS, Zhan SF, Yang LL, Zhuang HF. et al. Examining the effector mechanisms of Xuebijing injection on COVID-19 based on network pharmacology. BioData Min. 2020;13:17
25. Chien S-T, Lin S-S, Wang C-K, Lee Y-B, Chen K-S, Fong Y. et al. Acacetin inhibits the invasion and migration of human non-small cell lung cancer A549 cells by suppressing the p38α MAPK signaling pathway. Molecular and Cellular Biochemistry. 2011;350:135-48
26. Sun LC, Zhang HB, Gu CD, Guo SD, Li G, Lian R. et al. Protective effect of acacetin on sepsis-induced acute lung injury via its anti-inflammatory and antioxidative activity. Arch Pharm Res. 2018;41:1199-210
27. Singh S, Gupta P, Meena A, Luqman S. Acacetin, a flavone with diverse therapeutic potential in cancer, inflammation, infections and other metabolic disorders. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 2020;145:111708
28. Huang WC, Liou CJ. Dietary acacetin reduces airway hyperresponsiveness and eosinophil infiltration by modulating eotaxin-1 and th2 cytokines in a mouse model of asthma. Evid Based Complement Alternat Med. 2012;2012:910520
29. Bhat TA, Nambiar D, Tailor D, Pal A, Agarwal R, Singh RP. Acacetin Inhibits In Vitro and In Vivo Angiogenesis and Downregulates Stat Signaling and VEGF Expression. Cancer Prevention Research. 2013;6:1128-39
30. Khan NM, Haseeb A, Ansari MY, Devarapalli P, Haynie S, Haqqi TM. Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human Osteoarthritis chondrocytes. Free radical biology & medicine. 2017;106:288-301
31. Yeh C-H, Shih H-C, Hong H-M, Lee S-S, Yang M-L, Chen C-J. et al. Protective effect of wogonin on proinflammatory cytokine generation via Jak1/3-STAT1/3 pathway in lipopolysaccharide stimulated BV2 microglial cells. Toxicology and Industrial Health. 2013;31:960-6
32. Zhao L, Sha Y-Y, Zhao Q, Yao J, Zhu B-B, Lu Z-J. et al. Enhanced 5-fluorouracil cytotoxicity in high COX-2 expressing hepatocellular carcinoma cells by wogonin via the PI3K/Akt pathway. Biochemistry and Cell Biology. 2013;91:221-9
33. Boesch-Saadatmandi C, Loboda A, Wagner AE, Stachurska A, Jozkowicz A, Dulak J. et al. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J Nutr Biochem. 2011;22:293-9
34. Ren X, Han L, Li Y, Zhao H, Zhang Z, Zhuang Y. et al. Isorhamnetin attenuates TNF-α-induced inflammation, proliferation, and migration in human bronchial epithelial cells via MAPK and NF-κB pathways. Anat Rec (Hoboken). 2021;304:901-13
35. Tran CT, Garcia M, Garnier M, Burucoa C, Bodet C. Inflammatory signaling pathways induced by Helicobacter pylori in primary human gastric epithelial cells. Innate Immun. 2017;23:165-74
36. Swanson CD, Akama-Garren EH, Stein EA, Petralia JD, Ruiz PJ, Edalati A. et al. Inhibition of epidermal growth factor receptor tyrosine kinase ameliorates collagen-induced arthritis. J Immunol. 2012;188:3513-21
37. Wu Z, Zhao W, Yang Z, Wang YM, Dai Y, Chen LA. Novel Resistance Mechanisms to Osimertinib Analysed by Whole-Exome Sequencing in Non-Small Cell Lung Cancer. Cancer Manag Res. 2021;13:2025-32
38. Vico-Barranco I, Arbulo-Echevarria MM, Serrano-García I, Pérez-Linaza A, Miranda-Sayago JM, Miazek A, et al. A Novel, LAT/Lck Double Deficient T Cell Subline J.CaM1.7 for Combined Analysis of Early TCR Signaling. Cells. 2021; 10
39. Du Y, Tang H, Gu X, Shi Y, Gong P, Yao Y. Radiation Can Regulate the Expression of miRNAs Associated with Osteogenesis and Oxidation in Exosomes from Peripheral Blood Plasma. Oxid Med Cell Longev. 2021;2021:6646323
40. Hong GE, Kim JA, Nagappan A, Yumnam S, Lee HJ, Kim EH. et al. Flavonoids Identified from Korean Scutellaria baicalensis Georgi Inhibit Inflammatory Signaling by Suppressing Activation of NF- kappa B and MAPK in RAW 264.7 Cells. Evid Based Complement Alternat Med. 2013;2013:912031
41. Arenas-Ramirez N, Woytschak J, Boyman O. Interleukin-2: Biology, Design and Application. Trends in immunology. 2015;36:763-77
42. Jiang T, Zhou C, Ren S. Role of IL-2 in cancer immunotherapy. Oncoimmunology. 2016;5:e1163462
43. Wang YX, Ma JR, Wang SQ, Zeng YQ, Zhou CY, Ru YH. et al. Utilizing integrating network pharmacological approaches to investigate the potential mechanism of Ma Xing Shi Gan Decoction in treating COVID-19. European review for medical and pharmacological sciences. 2020;24:3360-84
44. Zhang XY, Huang HJ, Zhuang DL, Nasser MI, Yang MH, Zhu P. et al. Biological, clinical and epidemiological features of COVID-19, SARS and MERS and AutoDock simulation of ACE2. Infect Dis Poverty. 2020;9:99
45. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-80 e8
46. Gimeno A, Mestres-Truyol J, Ojeda-Montes MJ, Macip G, Saldivar-Espinoza B, Cereto-Massague A. et al. Prediction of Novel Inhibitors of the Main Protease (M-pro) of SARS-CoV-2 through Consensus Docking and Drug Reposition. Int J Mol Sci. 2020 21
47. Qian H, Jin Q, Liu Y, Wang N, Chu Y, Liu B. et al. Study on the Multitarget Mechanism of Sanmiao Pill on Gouty Arthritis Based on Network Pharmacology. Evidence-Based Complementary and Alternative Medicine. 2020;2020:1-11
48. Yang R, Yuan BC, Ma YS, Zhou S, Liu Y. The anti-inflammatory activity of licorice, a widely used Chinese herb. Pharm Biol. 2017;55:5-18
Author contact
![]() Corresponding author: Chunfu Wu, Department of Pharmacology, Shenyang Pharmaceutical University, 103 Wenhua Road, 110016, Shenyang, PR China. Tel. & Fax: +86 24 23986340. E-mail address: wucfedu.cn. Kuo Zhang, Department of Pharmacology, Shenyang Pharmaceutical University, 103 Wenhua Road, 110016, Shenyang, PR China. Tel. & Fax: +86 24 23986340. E-mail address: zhangkuoedu.cn
Corresponding author: Chunfu Wu, Department of Pharmacology, Shenyang Pharmaceutical University, 103 Wenhua Road, 110016, Shenyang, PR China. Tel. & Fax: +86 24 23986340. E-mail address: wucfedu.cn. Kuo Zhang, Department of Pharmacology, Shenyang Pharmaceutical University, 103 Wenhua Road, 110016, Shenyang, PR China. Tel. & Fax: +86 24 23986340. E-mail address: zhangkuoedu.cn

 Global reach, higher impact
Global reach, higher impact