Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(14):3224-3235. doi:10.7150/ijms.60907 This issue Cite
Research Paper
MiR-508-3p promotes proliferation and inhibits apoptosis of middle ear cholesteatoma cells by targeting PTEN/PI3K/AKT pathway
Department of Otolaryngology Head and Neck Surgery, Shengjing Hospital of China Medical University, Liaoning 110004, China
Received 2021-3-26; Accepted 2021-6-21; Published 2021-7-11
Abstract
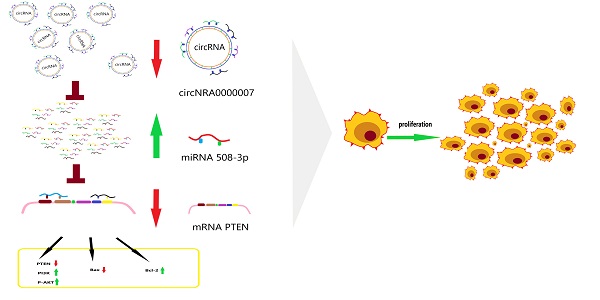
Cholesteatoma of the middle ear is a common disease in otolaryngology, which can lead to serious intracranial and extracranial complications. Recent studies showed that the dysregulation of microRNA may be involved in the formation of middle ear cholesteatoma. This study aimed to explore the regulatory effect of micro ribonucleic acid 508-3p (miR-508-3p) on proliferation and apoptosis of middle ear cholesteatoma cells and excavate its underlying regulatory mechanism. We found miR-508-3p expression was upregulated in tissues and cells of cholesteatoma which was inversely related to the expression of hsa_circ_0000007. Overexpression of miR-508-3p could notably facilitate cholesteatoma cell proliferation. Luciferase reporter assay showed that miR-508-3p bound the 3'-untranslated region of its downstream mRNA PTEN. Gain and loss of functions of miR-508-3p were performed to identify their roles in the biological behaviors of cholesteatoma cells, including proliferation and apoptosis. Rescue assays confirmed that PTEN could reverse the effect of miR-508-3p overexpression on cell proliferation. In a word, this study validated that the development of cholesteatoma may regulated by hsa_circ_0000007/miR-508-3p/ PTEN/ PI3K/Akt axis.
Keywords: hsa_circ_0000007, miR-508-3p, PTEN/ PI3K/Akt pathway, cholesteatoma, proliferation and apoptosis
Introduction
Cholesteatoma is benign collections of keratinized squamous epithelium within the middle ear. There are congenital and acquired middle ear cholesteatomas [1]. Congenital cholesteatoma is formed from remnants of epithelium that get trapped in the temporal bone during development [2]. Acquired cholesteatoma does not result from an embryologic phenomenon, but are the result of pathologic changes that cause the uncontrolled growth of squamous keratinized epithelium in the middle ear [3]. This study is aimed at the acquired cholesteatoma. When the cholesteatoma begins, it can damage temporal bone and nearby structures like ossicles, facial nerve, vestibule, semicircular canal and brain causing many problems like hearing loss, facial paralysis, dizziness, encephalopyosis and so on. Cholesteatoma can be a difficult disease to treat because the underlying cause of the disease, eustachian tube dysfunction, is generally not addressed. This can lead to recurrent disease. Surgical resection of cholesteatoma can also be quite challenging, and residual cholesteatoma is often present after surgery [4]. The pathogenesis of acquired cholesteatoma is not clear. The most popular theory is the keratinocyte of the middle ear becomes hyperproliferative.
Circular RNA (circRNA) was considered as a class of endogenous noncoding RNA (ncRNA) [5]. CircRNA is mainly located in the cytoplasm and is highly stable compared to other ncRNAs [6]. CircRNA is abundantly expressed and evolutionarily conserved across eukaryotic organisms [7] and it plays crucial roles in many diseases, including digestive system neoplasms, cardiovascular disease, and Osteosarcoma [8-10]. It was commonly known that circRNAs regulated cell functions and cancer development by sponging microRNAs (miRNAs) [11-13].
MicroRNAs (miRNAs) are small endogenous RNAs that regulate gene expression post-transcriptionally. MiRNAs are short non-coding RNAs of 19~25 nucleotides that mediate gene silencing by guiding Argonaute (AGO) proteins to target sites in the 3' untranslated region (UTR) of mRNAs. AGOs constitute a large family of proteins that use single-stranded small nucleic acids as guides to complementary sequences in RNA or DNA targeted for silencing [14]. The miRNA-loaded AGO forms the targeting module of the miRNA-induced silencing complex (miRISC), which promotes translation repression and degradation of targeted mRNAs [15]. A single miRNA can target hundreds of mRNAs and influence the expression of many genes often involved in a functional interacting pathway [16].
The PTEN/PI3K/AKT pathway regulates multiple cellular functions, including cell growth, differentiation, proliferation, survival, motility, invasion and intracellular trafficking in various diseases like lung cancer, gastric cancer, breast cancer and so on [17-19]. PTEN, a dual protein and lipid phosphatase, primarily dephosphorylates phosphatidylinositol-3,4,5-trisphosphate (PIP3), which is the product of PI3K and is able to recruit Akt to the membrane, where it is phosphorylated and stimulated [20]. Activated Akt may regulate multiple biological processes, including cell survival, metabolism, cell proliferation and growth, by affecting its downstream substrates [21,22].
Taken together, the current study was designed to explore the role of hsa_circ_0000007 and miR-508-3p in the development of cholesteatoma with the involvement of the PTEN/PI3K/AKT signaling pathway.
Materials and Methods
Patients and samples
The present study was performed using data obtained randomly from 20 patients. All patients were surgically treated at Shengjing Hospital of China Medical University from September 1, 2020 to December 31, 2020. All patients have received pathological diagnosis of middle ear cholesteatoma. We collected and frozen all samples. The lower age limit of these patients was 18 years, and the higher limit was 70 years, with the median age of 53.57±18.67 years, which included 7 women and 13 men. At the same time, 15 cases of posterior auricular normal skin or skin fragments that could not be used during otoplasty were collected as control group. This study was approved by the Institutional Human Ethics Committee of Shengjing Hospital of China Medical University, and prior informed consent obtained from all the patients.
Data source
The microarray data analyzed in this study were obtained from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE102715, published on Apr 27, 2020. GEO is a public functional genomics data repository supporting MIAME-compliant data submissions. Array- and sequence-based data are accepted. Tools are provided to help users query and download experiments and curated gene expression profiles [23,24]. This dataset GSE102715 profiled the differences in circRNA expression between 4 pairs of cholesteatoma (GSM2743683, GSM2743685, GSM2743687, GSM2743689) and matched normal skin samples (GSM2743684, GSM2743686, GSM2743688, GSM2743690). All specimens were obtained from 2 female and 2 male patients aged 18-year-old to 32-year-old who received unilateral middle ear cholesteatoma surgeries. The post-auricular skins were taken as control samples from the same patients. GSE102715 was based on the Agilent GPL21825 platform (Arraystar Human CircRNA microarray V2). All of the data were freely available online.
Data processing and differential expression analysis
After getting raw expression data, the volcano figure was created using GraphPad Prism 7.0 software. Differentially expressed genes (DEGs) analysis between cholesteatoma and normal samples was performed using the online analysis tool GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE102715) and NetworkAnalyst 3.0 (www.networkanalyst.ca/NetworkAnalyst/home.xhtml). The intersecting part between the two analysis was identified using the Venn diagram webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/). The adjusted P value and |logFC| were calculated. Genes that met the cutoff criteria, adjusted P value<0.05 and |logFC|≥2.0, were considered as DEGs. The heatmap for the DEGs was created using GraphPad Prism 7.0 software.
Functional enrichment analysis
GO analysis is a common useful method for large scale functional enrichment research; The GO analysis included 3 categories, namely, biological process (BP), cellular component (CC) and molecular function (MF), which were used to predict protein functions [25]. Pathway functional analysis was performed on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [26]. GO annotation analysis and KEGG pathway enrichment analysis of DEGs in this study was performed using the clusterProfiler of Limma R package available on Bioconductor (http://bioconductor.org/packages/release/bioc/html/limma.html). P<0.05 and gene counts≥2 were considered statistically significant.
Prediction of circRNA-miRNA-mRNA target gene associations
We predicted circRNA/miRNA target genes using online tools Circular RNA Interactome (https://circinteractome.irp.nia.nih.gov/mirna_target_sites.html), and predicted the interactive relationships between miRNA and target mRNA using TargetScan Human 7.2 (http://www.targetscan.org/vert_72/).
Cell culture and transfection
After extraction of cholesteatoma tissues from middle ear, the tissues were washed 3 times with pre-cooled phosphate buffer solution (PBS) and cut into 1×1 mm3 blocks using a surgical scissor with high temperature sterilization. Digestion of them with 0.25% pancreatin at 37℃ for 3h after centrifugation was terminated by adding culture medium. The filtrate filtered by a 200mesh cell sieve was collected and centrifuged at 1500 rpm for 10 min, then the supernatant was discarded, and the complete medium was added to re-suspend cell precipitate for subsequent experiments. The passage cells were re-inoculated in a 6-well plate. After the cell fusion degree reached about 80%, miR-508-3p NC, miR-508-3p mimic and miR-508-3p inhibitor GenePharma (Shanghai, China) were transfected into middle ear cholesteatoma cells respectively according to the instructions of LipofectamineTM2000 (Invitrogen, Carlsbad, CA, USA) transfection reagent, and cultured in a CO2 incubator at 37°C. After 48 hours, ELISA, EdU staining and TUNEL staining were explored, and the transfection efficiency was evaluated using RT-qPCR. Experimental line of human immortalized keratinocytes (HaCaT) was obtained from shanghai Zhong Qiao Xin Zhou Biotechnology Co.,Ltd. Cells were cultured in high-glucose Dulbecco's Modified Eagle Media (DMEM) (HyClone, Thermo Fisher, Shanghai, China) with 10% fetal bovine serum (FBS) (Corning, Thermo Fisher, Waltham, MA). HaCaT cells were grown under sterile, humidified conditions at 37℃ and 5% CO2.
RT-qPCR
Total RNAs were separated by using trizol reagent (Takara, Otsu, Japan). And Prime Script RT Reagent Kit (HaoranBio, Xuhui, Shanghai, China) and TaqManTM Advanced miRNA cDNA Synthesis Kit (Waltham, MA, USA) were then respectively applied to synthesize complementary DNA. Subsequently, the SYBR Green Master Mix (Takara, Dalian, China) was utilized to conduct the RT-qPCR on ABI 7500 System (Applied iosystems, Carlsbad, California). GAPDH and U6 served as internal controls. Relative expression of RNAs was calculated by using the 2-ΔΔCt method.
CCK-8
Cell counting kit-8 (CCK-8) reagent (Beyotime Institute of Biotechnology, Shanghai, China) was used to perform CCK-8 assay in accordance with the manufacturer's suggestions. Transfected cells (1 × 103) were seeded into the 96-well plates and cultured for 0, 24, 48, 72 and 96 h. Then each well was added with CCK-8 reagent. After 4 h incubation, the optical density was measured using a microplate reader at a wavelength of 450 nm to detect cell proliferation at each time points. The detection was repeated 3 times.
ELISA
The ELISA kit was taken out from a low-temperature refrigerator, and left at room temperature, and then standard substances and diluents were added into blank holes according to instructions of the ELISA kit, and standard substances with different concentrations were added into the rest holes to draw standard curves. Then diluted enzyme conjugate was added, incubated at 37℃ for 30 min and washed for 5 times, and 100μL of chromogenic substrate was added, and then incubated in dark for 15 min. Finally, the reaction termination solution was added dropwise, and absorbance was detected with a microplate reader.
5-ethynyl-2'-deoxyuridine (EdU) staining
Each well was added with 100μL of penetrant, incubated for 15 min, washed with PBS, then added with 100μL of EdU staining solution diluted with culture medium, and incubated for 30 min. The culture medium was discarded and the cells were decolorized with PBS for 3 times, each time for 5 min. The staining was observed under a fluorescence microscope.
TUNEL staining
Cells in each group were washed with PBS, fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.3% TritonX-100 solution for 15 min, added with 50μL of TUNEL test solution per well, incubated at 37℃ in dark for 60 min, washed with PBS, added with anti-fluorescence quencher, and observed via TUNEL staining under a microscope.
Western blot
The cells from each group were collected and lysed with RIPA lysis buffer, and the protein concentrations in each group were determined by Braford method. At room temperature, proteins were separated by 10% SDS-PAGE at constant pressure, transferred to a PVDF membrane, and sealed with 5% skimmed milk powder for 1 h. After that, the membrane was incubated with primary anti-antibodies PTEN (1:1000), PI3K (1:1000) and p-Akt (1:1000) overnight at 4℃ and then the HRP labeled secondary antibody (1: 2000) for 2 h. The bands were developed using DAB color development method, and the absorbance of each band was analyzed by Image J software.
Luciferase reporter assay
Reporter plasmids were obtained by inserting PTEN 3'-UTR sequence into pmirGLO vector (Promega, Madison, WI, USA). For the luciferase assay, miR-508-3p mimics and reporter plasmids were co-transfected into 239T cells using Lipofectamine2000. After culturing for 48 h, firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, Sunnyvale, CA, USA) according to the manufacturer's instructions.
Statistical analysis
The statistical analysis was performed using the SPSS 20.0 and the data were visualized using the GraphPad 7. Data has been displayed as the mean± standard deviation (SD). The one-way ANOVA or student's t-test was utilized for the comparisons among groups. Pearson analysis was used to observe the correlation between hsa_circ_0000007 and miR-508-3p in tissue samples. Each experiment of this study was performed in triplicate. Any value of p < 0.05 was thought to be of statistical significance.
Results
Identification of different expression circRNA
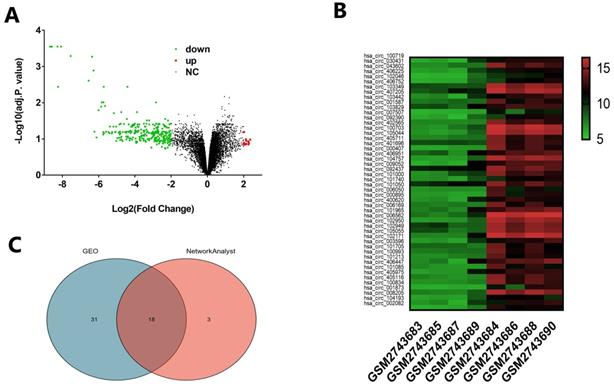
We downloaded the microarray expression dataset GSE102715 from Gene Expression Omnibus (GEO) and analyzed the different circRNAs between cholesteatoma and normal skin using the online analysis tool GEO2R and NetworkAnalyst 3.0. In total, there are 13247 raw circRNAs in dataset GSE102715. Based on the criteria of |logFC|≥2, GEO2R identified 16 upregulated and 283 downregulated circRNAs showed in Volcano (Fig. 1A). Based on the criteria of adjusted p. value <0.05 and |logFC|≥2, We obtained top 49 different circRNAs to show in heatmap (Fig. 1B). NetworkAnalyst 3.0 identified 21 dysregulated circRNAs on the criteria of P<0.05 and |logFC|≥2. Subsequently, Venn analysis was performed to get the intersection of the dysregulated circRNAs between GEO2R and NetworkAnalyst 3.0 result (Fig. 1C). As showed in Venn diagram, there are 18 common candidates of dysregulated circRNAs.
Distribution of differentially expressed circRNAs between cholesteatoma and normal skin. (A) Volcano plot representing log2(fold change) and -log10 (adjusted. p. value). Red stands for upregulations, green stands for downregulations and black stands for normal expression in volcanoes. Each point represents a gene. (B) Heatmap of differentially expressed circRNAs in cholesteatoma and normal skin with the criteria |log2FoldChange|>2 and adjusted p. value < 0.05. The horizontal axis represents the names of the samples. GSM2743683, GSM2743685, GSM2743687and GSM2743689 are the samples of cholesteatoma. GSM2743684, GSM2743686, GSM2743688 and GSM2743690 represent normal skin. The vertical axis represents differentially expressed circRNAs. Similar with Volcano, red stands for upregulations, green stands for downregulations and black stands for normal expression. (C) Venn diagram showing the intersection of significative dysregulated circRNA between GEO2R and NetworkAnalyst 3.0.
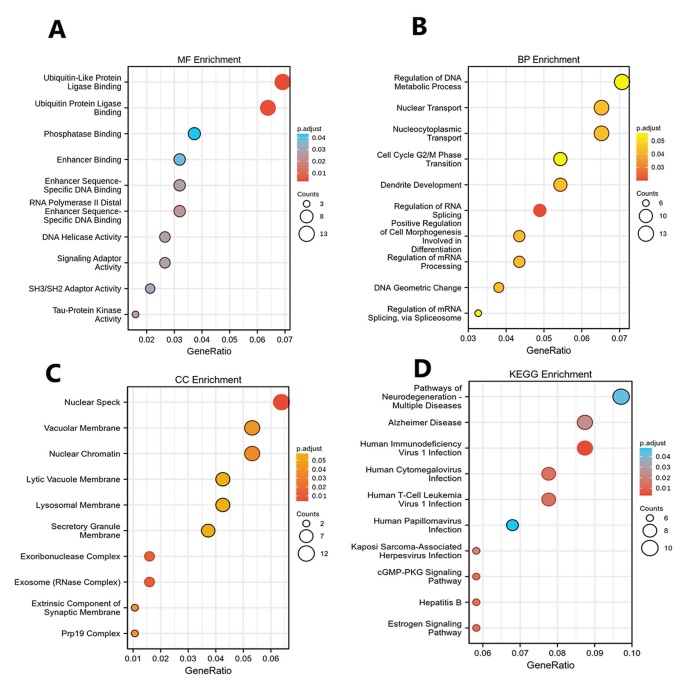
GO and KEGG analysis. (A) Top 10 significant biological process (BP) GO functional terms; (B) Top 10 significant cellular component (CC) GO functional terms; (C) Top 10 significant molecular function (MF) GO functional terms in cholesteatoma compared to the normal skin, respectively. (D) Top 10 enriched pathways identified by KEGG analysis. The X-axis represents the gene ratio for each of the differentially expressed genes in each pathway. The Y-axis gives the name of the enriched term. The size of each node indicates the number of significant genes in each category. The adjust p value are indicated by changing colors.
Functional enrichment analysis of DEGs
GO function and KEGG pathway enrichment analysis for DEGs were showed in (Figs. 2A-D). The enriched GO terms were divided into CC, BP, and MF ontologies. The results of GO analysis indicated that DEGs were mainly enriched in BPs, including regulation of DNA metabolic process (GO:0051052), nucleocytoplasmic transport (GO:0006913), nuclear transport (GO:0051169), regulation of chromosome organization (GO:0033044), ncRNA processing (GO:0034470) and so on. MF analysis showed that the DEGs were significantly enriched in ubiquitin-like protein ligase binding (GO:0044389), ubiquitin protein ligase binding (GO:0031625), protein serine/threonine kinase activity (GO:0004674), catalytic activity, acting on RNA (GO:0140098), phospholipid binding (GO:0005543) and so on. For the cell component, the DEGs were enriched in nuclear speck (GO:0016607), nuclear chromatin (GO:0000790), vacuolar membrane (GO:0005774), lysosomal membrane (GO:0005765), lytic vacuole membrane (GO:0098852) and so on. In addition, KEGG pathway analysis (Fig. 2D) showed that upregulated genes were mainly enriched in Pathways of neurodegeneration - multiple diseases (KEGG ID: hsa05022), Human immunodeficiency virus 1 infection (KEGG ID: hsa05170), Alzheimer disease (KEGG ID: hsa05010), Human T-cell leukemia virus 1 infection (KEGG ID: hsa05166), Human cytomegalovirus infection (KEGG ID: hsa05163) and so on. The top ten of P < 0.05 were chosen in each enrichment.
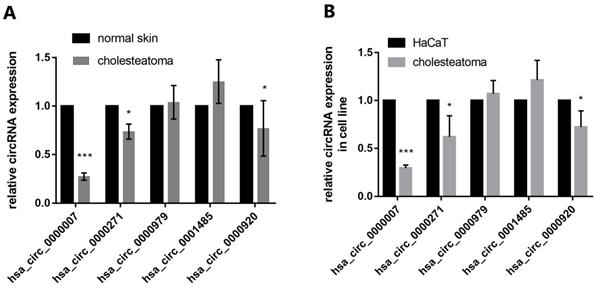
The expression of 5 candidate circRNAs in both specimens and cell lines. (A) RT-qPCR detection show the expression of 5 candidate circRNAs (hsa_circ_0000007, hsa_circ_0000271, hsa_circ_0000979, hsa_circ_0001485, hsa_circ_0000920) in both cholesteatoma (n=10) and normal skin specimens (n=10). (B) RT-qPCR assay was conducted to detect the expression of the 5 candidate circRNAs in cholesteatoma cells and HaCaT cell line. Data are presented as mean ± SD. *p < 0.05, **p < 0.01. ***p < 0.001. Repetition=3
Identification of genes of interest
We identified five candidates of circRNAs (hsa_circ_0000007, hsa_circ_0000271, hsa_circ_0000979, hsa_circ_0001485, hsa_circ_0000920) which are markedly downregulated in cholesteatoma to be further studied. RT-qPCR assay depicted that hsa_circ_0000007 expression was most significantly downregulated in comparison with other 4 circRNAs both in tissue and cells (Fig. 3A and B).
Prediction and verification of potential target microRNA and mRNA
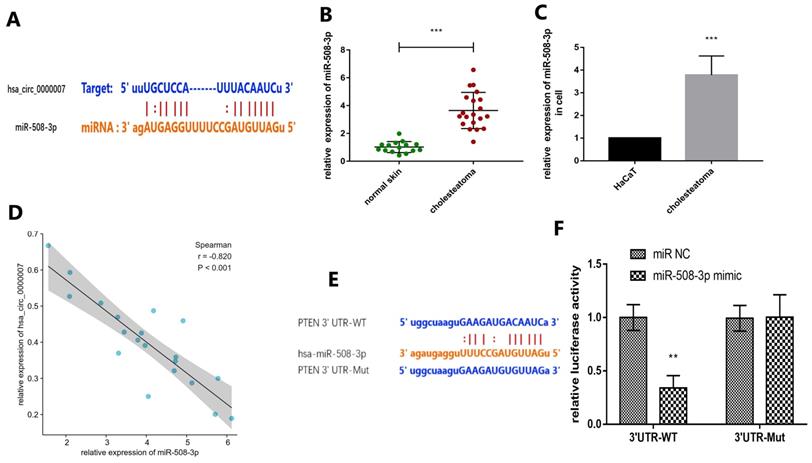
We intended to explore the molecular mechanism of hsa_circ_0000007 in cholesteatoma. We used online prediction tool Circular RNA Interactome to predict potential miRNA which could possibly bind with hsa_circ_0000007. The prediction results showed four miRNAs, including miR-492, miR-508-3p, miR-665 and miR-876-3p. We chose miR-508-3p because its context+ score was minimum of all (Fig. 4A). In this study, RT-qPCR assay was applied to examine the expression of miR-508-3p both in cholesteatoma tissue and cells (Fig. 4B and C). The results demonstrated that the expression of miR-508-3p was notably higher in cholesteatoma tissue and cells than that in normal skin and HaCaT cells (p<0.001). Moreover, miR-508-3p expression was negatively correlated with hsa_circ_0000007 expression (Fig. 4D) (p<0.001). Next, bioinformatics analysis tool Targetscan (http://www.targetscan.org/) showed that PTEN was a potential miR-508-3p target mRNA. PTEN was also found to have a binding site for miR-508-3p through searching starBase (Fig. 4E). To confirm that PTEN was a miR-508-3p target, we cloned mutant and wild-type PTEN sequences to construct mutant vectors and reporter plasmids respectively. The results showed that the reporter plasmid and miR-508-3p mimic co-transfections visibly suppressed luciferase activity and mutated PTEN vectors, but miR-508-3p mimic co-transfection had no significant effect on luciferase activity. These results proved that miR-508-3p directly targeted PTEN (Fig. 4F).
Effect of miR-508-3p on the biological phenotype of cholesteatoma cells
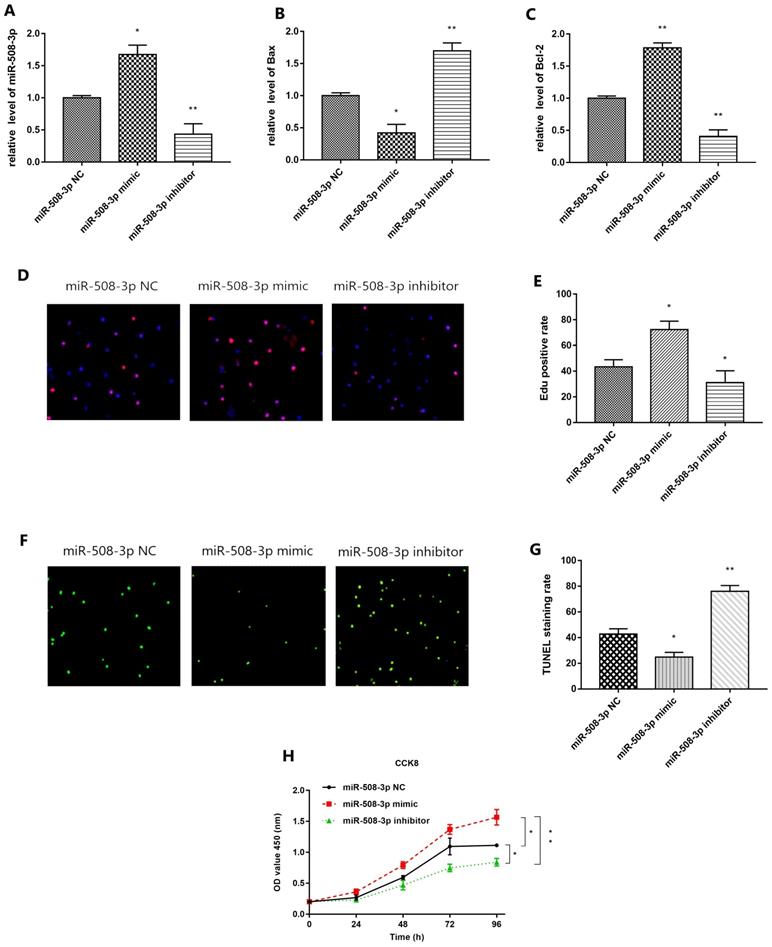
QRT-PCR results are shown in Fig. 5A. Compared with that in miR-508-3p NC group, miR-508-3p level in middle ear cholesteatoma cells overtly increased in miR-508-3p mimic group (p<0.05) and notably decreased in miR-508-3p inhibitor group (p<0.01). The results of ELISA are shown in Fig. 5B and 5C. Compared with those in miR-508-3p NC group, Bax level in middle ear cholesteatoma cells was decreased in miR-508-3p mimic group(p<0.05) and enhanced in miR-508-3p inhibitor group (p<0.01) respectively, while Bcl-2 level was elevated in miR-508-3p mimic group (p<0.01) and declined in miR-508-3p inhibitor group (p<0.01) respectively. Fig. 5D presents the results of EdU staining. Compared with that in miR-508-3p NC group, the proliferation rate of middle ear cholesteatoma cells was increased in miR-508-3p mimic group(p<0.05) and decreased in miR-508-3p inhibitor group (p<0.05), as shown in Fig. 5E. Fig. 5F shows TUNEL staining results. Compared with that in miR-508-3p NC group, the apoptosis rate of middle ear cholesteatoma cells was lowered in miR-508-3p mimic group (p<0.05) and elevated in miR-508-3p inhibitor group (p<0.01), as shown in Fig. 5G. The CCK-8 assay results suggested that over-expression of miR-508-3p significantly promoted the cholesteatoma cell proliferation 96 h after transfection, while down-expression of miR-508-3p reduced proliferation (all p< 0.05) (Fig. 5H). In conclusion, miR-508-3p can promote proliferation and inhibit apoptosis in cholesteatoma cells.
Prediction and verification of potential target miRNA and mRNA. (A) Prediction of the binding of miR-508-3p and hsa_circ_0000007 performed via an online website (https://circinteractome.irp.nia.nih.gov/). (B) MiR-508-3p levels in 20 cholesteatoma tissues and 15 normal skin tissues were measured by RT-qPCR. (C) Expression of miR-508-3p in cholesteatoma cells and HaCaT cell line were measured by RT-qPCR. (D) The negative correlation between the expressions of miR-508-3p and hsa_circ_0000007 was verified by Spearman test (r = -0.820, p< 0.001). (E) Prediction of the binding of miR-508-3p and PTEN performed by an online website (http://www.targetscan.org). (F) Binding relation of miR-508-3p and PTEN identified using dual luciferase reporter gene assay. *p < 0.05, **p < 0.01. ***p < 0.001.
MiR-508-3p facilitates cell proliferation and inhibits apoptosis in cholesteatoma cell through PTEN/PI3K/Akt signal pathway
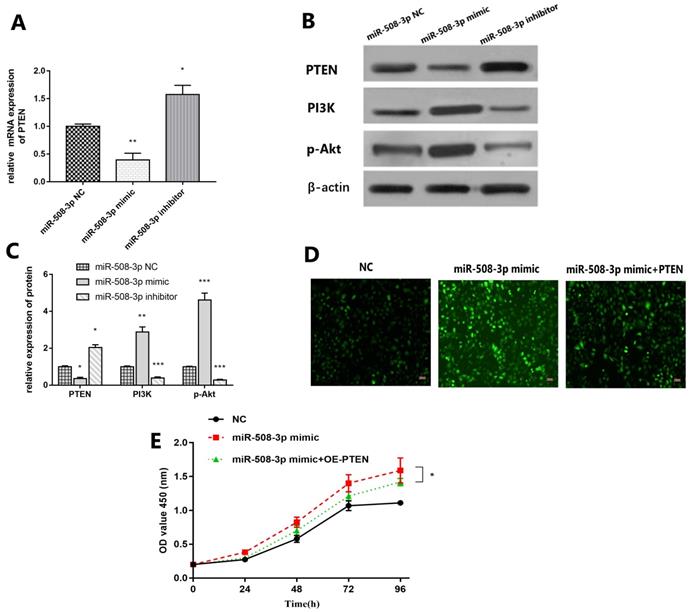
QRT-PCR results are shown in Fig. 6A. Compared with that in miR-508-3p NC group, PTEN level in middle ear cholesteatoma cells notably decreased in miR-508-3p mimic group (p<0.01) and obviously increased in miR-508-3p inhibitor group (p<0.05). The Western blotting results are revealed in Fig. 6B and 6C. Compared with those in miR-508-3p NC group, the level of PTEN protein in middle ear cholesteatoma cells decreased in miR-508-3p mimic group and increased in miR-508-3p inhibitor group (p<0.05), while the levels of PI3K and p-Akt proteins raised in miR-508-3p mimic group (p<0.01, p<0.001) and lowered in miR-508-3p inhibitor group (p<0.001, p<0.001). Fig. 6D and 6E suggested that over-expression of miR-508-3p visibly facilitated the cholesteatoma cell proliferation after transfection. Co-transfection with miR-508-3p mimic and oe-PTEN led to significantly reduced cell proliferation compared to miR-508-3p mimic transfection alone (p < 0.05). This rescue assays were performed to prove overexpression PTEN can reverse the trendy of proliferation after upregulation of miR-508-3p.
Discussion
In recent years, with the gradual maturity of biochip and sequencing technology, a variety of biological databases can provide more reliable data for researchers [23,27]. Subsequently, Non-coding RNA (ncRNA) has been increasingly studied in various diseases [28]. Studies have shown that ncRNAs play important roles in various biological processes [29,30]. NcRNAs are commonly employed for RNA that does not encode a protein, but can regulate biological transcription and translation. NcRNAs include miRNAs, circRNAs and so on [31].
MicroRNAs (miRNAs), widely distributed, small regulatory RNA genes, target both messenger RNA (mRNA) degradation and suppression of protein translation based on sequence complementarity between the miRNA and its targeted mRNA [32]. MiRNAs are involved in human health and disease as endogenous suppressors of the translation of coding genes. Specific cognate mRNA targets for miRNA are the key to the regulation of mRNA [33]. MiRNAs have been reported and studied in various diseases. For instance, microRNA is a potential blood-based epigenetic biomarker for Alzheimer's disease [34]. Epigenetic abnormalities in meningiomas include abnormal microRNA expression [35]. Many studies have found that miRNA is most closely related to proliferation and apoptosis. For example, Xia MM et al. [36] summarized many microRNAs can regulate Sertoli cell proliferation and adhesion. Zhu ZJ et al. [37] found overexpression of microRNA-181a (miR-181a) promoted the proliferation and inhibited the apoptosis of osteosarcoma cells.
Effect of miR-508-3p on proliferation and apoptosis in cholesteatoma. (A) RT-qPCR detection show the expression of miR-508-3p in miR-508-3p NC group, miR-508-3p mimic group and miR-508-3p inhibitor group. (B, C) Level of Bax and Bcl-2 in miR-508-3p NC group, miR-508-3p mimic group and miR-508-3p inhibitor group of cholesteatoma cells via ELISA. (D) EdU staining of transfected cells. (E) EdU positive rate of miR-508-3p NC group, miR-508-3p mimic group and miR-508-3p inhibitor group. (F) TUNEL staining of transfected cells. (G) TUNEL staining rate of miR-508-3p NC group, miR-508-3p mimic group and miR-508-3p inhibitor group. (H) The proliferative capability of transfected cells was evaluated by CCK-8 assay. *p < 0.05, **p < 0.01. ***p < 0.001
Upregulation of PTEN reverses the effect of miR-508-3p on cholesteatoma. (A) The expression of PTEN was detected by RT-qPCR in miR-508-3p NC group, miR-508-3p mimic group and miR-508-3p inhibitor group. (B) Western blot analysis showed the protein level of PTEN, PI3K and p-Akt in miR-508-3p NC group, miR-508-3p mimic group and miR-508-3p inhibitor group in vitro. (C) Relative protein level of PTEN, PI3K and p-Akt in vitro. (D) The transfected cells growth situation of miR-508-3p NC, miR-508-3p mimic and miR-508-3p+PTEN. (E) The proliferative capability of transfected cells was evaluated by CCK-8 assay. *p < 0.05, **p < 0.01. ***p < 0.001
Circular RNAs (circRNAs), a novel class of long noncoding RNAs, are characterized by a covalently closed continuous loop without 5′ or 3′ polarities structure and have been widely found in thousands of lives including plants, animals and human beings. Utilizing the high-throughput RNA sequencing (RNA-seq) technology, recent findings have indicated that a great deal of circRNAs, exhibit cell type-specific, tissue-specific or developmental-stage-specific expression. Evidences are arising that some circRNAs might regulate microRNA (miRNA) function as microRNA sponges and play a significant role in transcriptional control. CircRNAs associate with related miRNAs and the circRNA-miRNA axes are involved in a serious of disease pathways such as apoptosis, vascularization, invasion and metastasis [38]. The aberrant expression of circRNAs has been reported in many human diseases including gastric cancer [39], colorectal cancer [40], papillary thyroid cancer [41], lung adenocarcinoma [42] and so on. Function of circRNA-miRNAs-mRNA axis is increasingly studied in human diseases, but has not been reported in middle ear cholesteatoma. In this experiment, we carried out a study on hsa_circ_0000007-miR-508-3p-PTEN axis.
Cholesteatoma is a noncancerous cystic lesion derived from an abnormal growth of keratinizing squamous epithelium in the temporal bone [43]. Although not malignant, cholesteatoma can destroy temporal bone and nearby structures like ossicles, facial nerve, vestibule, semicircular canal and brain causing many problems like hearing loss, facial paralysis, dizziness, encephalopyosis and so on. Cholesteatoma is a serious disease in otolaryngology. Most of the cholesteatoma mechanisms that have been proposed to explain the pathogenesis of acquired cholesteatoma can be divided into four categories: (1) invagination theory (retraction pocket theory), (2) the theory of epithelial invasion or migration (immigration theory), (3) the theory of squamous metaplasia, and (4) basal cell hyperplasia theory (papillary ingrowth theory) [44]. Ultimately, the accumulation of epithelial keratinocytes with over-proliferation and inhibited apoptosis in a deepening retraction pocket leads to the formation of cholesteatoma.
With the increasing research on ncRNA in recent years, many RNA microarray databases [45] can be used for free. In this study, we mined GSE102715 in GEO database. By obtaining the raw data, we found hsa_circ_0000007 through two analysis software (GEO2R and Network Analyst 3.0). Then we detected hsa_circ_0000007 with RT-PCR and found that the expression of hsa_circ_0000007 was significantly lower in cholesteatoma than normal skin. As mentioned above, circRNAs may regulate miRNAs and act as microRNA sponges. Therefore, we found miR-508-3p that was the targeted miRNA downstream of hsa_circ_0000007 by using biological prediction software. Subsequently, we observed by RT-PCR that the expression of miR-508-3p in cholesteatoma was significantly higher than that in normal skin, and there was a statistically negative correlation with hsa_circ_0000007. Our experiment also demonstrated that the changes of miR-508-3p expression could affect the phenotypes of proliferation and apoptosis in cholesteatoma cells. In this study, cells with miR-508-3p NC, high miR-508-3p expression and low miR-508-3p expression were successfully obtained. The levels of Bax and Bcl-2 in cells of each group were determined by the ELISA kit. Bax and Bcl-2 are pro-apoptotic and anti-apoptotic proteins respectively, which play important roles in apoptosis [46]. The experimental results revealed that compared with miR-508-3p NC group, miR-508-3p inhibitor can overtly elevate the level of pro-apoptotic factor Bax and lower the level of anti-apoptotic factor Bcl-2. On the contrary, miR-508-3p mimic can raise the level of anti-apoptotic factor Bcl-2 and lower the level of pro-apoptotic factor Bax. Then, EdU and TUNEL staining methods were adopted to detect the effect of miR-508-3p on the proliferation and apoptosis of cholesteatoma cells, respectively. The results suggest that miR-508-3p mimic can significantly promote the proliferation of middle ear cholesteatoma cells and suppress their apoptosis.
MiR-508-3p has also been reported in other diseases, and its biological functions are related to proliferation, apoptosis and invasion. For instance, Lin C et al. [47] find that overexpressing miR-508 promotes, while silencing miR-508 impairs, the aggressive phenotype of oesophageal squamous cell carcinoma both in vitro and in vivo. Another study demonstrated the functional role of miR-508-3p in promoting the proliferation, invasion and migration of ESCC cells. They also identified a PCAT-1/miR-508-3p/ANXA10 axis in mediating the promoting role of miR-508-3p as a potential therapeutic target of ESCC [48]. But its mechanism of action in cholesteatoma has not been clarified.
We found a targeting relationship between miR-508-3p and PTEN through the TargetScan database analysis. Some CLIP-seq experiments also verified the targeted regulatory relationship between PTEN and miR-508-3p [49,50]. In this study, we verified the targeting relationship between miR-508-3p and PTEN by Luciferase Reporter Assay. In addition, the expression of PTEN decreased significantly in miR-508-3p mimic group and increased significantly in miR-508-3p inhibitor group. This proves the targeted regulation of miR-508-3p on PTEN at the transcriptional level further. Moreover, the rescue experiment also proved that PTEN could reverse the proliferation trend of miR-508-3p mimic group cells. Therefore, we infer that miR-508-3p has an effect on the phenotype of cholesteatoma through PTEN.
PTEN (phosphatase and tens in homolog deleted on chromosome 10) (also named MMAC1/TEP1) was discovered in 1997 independently by three laboratories as a tumor suppressor of which the expression is often lost in tumors [51]. Later studies established that PTEN is a negative regulator of a major cell growth and survival signaling pathway, namely the phosphatidylinositol-3-kinase (PI3K)/AKT signaling pathway [52]. Phosphatidylinositol‑4,5‑bisphosphate 3‑kinase (PI3K) is activated and leads to protein kinase B (Akt) phosphorylated with the help of phosphoinositide‑dependent kinase, in the PI3K/Akt signal transduction pathway [53]. Activated Akt may regulate multiple biological processes, including cell survival, metabolism, cell proliferation and growth, by affecting its downstream substrates [21]. In order to investigate the regulatory mechanism of miR-508-3p in cholesteatoma, the protein expression levels of PTEN, PI3K and p-Akt in cholesteatoma cells were detected via Western Blotting in this study. As results presented in Fig. 6, compared with miR-508-3p NC group, miR-508-3p inhibitor can enhance PTEN protein level and impede PI3K and Akt protein expressions. On the contrary, miR-508-3p mimic can decrease the expression of PTN and increase the expression of PI3K and p-AKT.
To sum up, we concluded that miR-508-3p played a key role in the formation of cholesteatoma by regulating the PTEN/PI3K/Akt signaling pathway. While the overexpression of miR-508-3p in cholesteatoma is probably mediated by the regulation of upstream hsa_circ_0000007.
Acknowledgements
We acknowledge the Gene Expression Omnibus for generating GEO datasets.
Funding
This work was supported by the Education Funding Project of Liaoning Province (grant no. ZF2019015).
Author Contributions
Xiulan Ma designed the study, made a critical review and improvement scheme; Dongliang Liu collected the samples, performed molecular analysis and wrote the manuscript; all authors read and approved the final version.
Ethics approval and consent to participate
The Ethics Committee of the Shengjing Hospital of China Medical University approved this study (2018PS268K). All patients involved in the study provided written informed consent.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rutkowska J, Özgirgin N, Olszewska E. Cholesteatoma Definition and Classification: A Literature Review. J Int Adv Otol. 2017;13:266-271
2. Kazahaya K, Potsic WP. Congenital cholesteatoma. Curr Opin Otolaryngol Head Neck Surg. 2004;12:398-403
3. Kennedy KL, Singh AK. Middle Ear Cholesteatoma. 2020 Jul 10. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. 2021 Jan-
4. Keeler JA, Kaylie DM. Cholesteatoma: Is a second stage necessary? Laryngoscope. 2016;126:1499-500
5. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440-1
6. Danan M, Schwartz S, Edelheit S. et al. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131-42
7. Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423-37
8. Sun Z, Chen C, Su Y. et al. Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms. J Cancer. 2019;10:2885-2891
9. Li JJ, Wang W, Wang XQ. et al. A novel strategy of identifying circRNA biomarkers in cardiovascular disease by meta-analysis. J Cell Physiol. 2019;234:21601-21612
10. Liu J, Yang L, Fu Q. et al. Emerging Roles and Potential Biological Value of CircRNA in Osteosarcoma. Front Oncol. 2020;10:552236
11. Dori M, Bicciato S. Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations. Genes (Basel). 2019;10:642
12. Liang ZZ, Guo C, Zou MM. et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20:173
13. Lin F, Chen HW, Zhao GA. et al. Advances in Research on the circRNA-miRNA-mRNA Network in Coronary Heart Disease Treated with Traditional Chinese Medicine. Evid Based Complement Alternat Med. 2020;2020:8048691
14. Swarts DC, Makarova K, Wang Y. et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 2014;21:743-53
15. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421-33
16. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207
17. Pérez-Ramírez C, Cañadas-Garre M, Molina MÁ. et al. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics. 2015;16:1843-62
18. Hu M, Zhu S, Xiong S. et al. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol Rep. 2019;41:1439-1454
19. Gao X, Qin T, Mao J. et al. PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J Exp Clin Cancer Res. 2019;38:256
20. Dahia PL. PTEN, a unique tumor suppressor gene. Endocr Relat Cancer. 2000;7:115-29
21. Fresno Vara JA, Casado E, de Castro J. et al. PI3K/Akt signaling pathway and cancer. Cancer Treat Rev. 2004;30:193-204
22. Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261-74
23. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207-10
24. Barrett T, Wilhite SE, Ledoux P. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991-5
25. Ashburner M, Ball CA, Blake JA. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-9
26. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30
27. Bono H, Kasukawa T, Hayashizaki Y. et al. READ: RIKEN Expression Array Database. Nucleic Acids Res. 2002;30:211-3
28. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18
29. Panni S, Lovering RC, Porras P. et al. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194417
30. Kaikkonen MU, Adelman K. Emerging Roles of Non-Coding RNA Transcription. Trends Biochem Sci. 2018;43:654-667
31. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17-29
32. Ying SY, Chang DC, Lin SL. The MicroRNA. Methods Mol Biol. 2018;1733:1-25
33. Ioshikhes I, Roy S, Sen CK. Algorithms for mapping of mRNA targets for microRNA. DNA Cell Biol. 2007;26:265-72
34. Fransquet PD, Ryan J. Micro RNA as a potential blood-based epigenetic biomarker for Alzheimer's disease. Clin Biochem. 2018;58:5-14
35. Mitha R, Shamim MS. Significance of micro-RNA expression in patients with meningioma. J Pak Med Assoc. 2020;70:1287-1288
36. Xia MM, Shen XY, Niu CM. et al. MicroRNA regulates Sertoli cell proliferation and adhesion. Yi Chuan. 2018;40:724-732
37. Zhu ZJ, Huang P, Chong YX. et al. MicroRNA-181a promotes proliferation and inhibits apoptosis by suppressing CFIm25 in osteosarcoma. Mol Med Rep. 2016;14:4271-4278
38. Rong D, Sun H, Li Z. et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271-73281
39. Li R, Jiang J, Shi H. et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661-1680
40. Zhang HD, Jiang LH, Sun DW. et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1-7
41. Bi W, Huang J, Nie C. et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37:275
42. Wang J, Zhao X, Wang Y. et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32
43. Isaacson G. Diagnosis of pediatric cholesteatoma. Pediatrics. 2007;120:603-608
44. Kuo CL. Etiopathogenesis of acquired cholesteatoma: prominent theories and recent advances in biomolecular research. Laryngoscope. 2015;125:234-40
45. Li S, Teng S, Xu J. et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20:1420-1433
46. Gaumer S, Guénal I, Brun S. et al. Bcl-2 and Bax mammalian regulators of apoptosis are functional in Drosophila. Cell Death Differ. 2000;7:804-14
47. Lin C, Liu A, Zhu J. et al. miR-508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nat Commun. 2014;5:4620
48. Zang B, Zhao J, Chen C. LncRNA PCAT-1 Promoted ESCC Progression via Regulating ANXA10 Expression by Sponging miR-508-3p. Cancer Manag Res. 2019;11:10841-10849
49. Luna JM, Barajas JM, Teng KY. et al. Argonaute CLIP Defines a Deregulated miR-122-Bound Transcriptome that Correlates with Patient Survival in Human Liver Cancer. Mol Cell. 2017;67:400-410.e7
50. Helwak A, Kudla G, Dudnakova T. et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654-65
51. Li J, Yen C, Liaw D. et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943-7
52. Downes CP, Ross S, Maccario H. et al. Stimulation of PI 3-kinase signaling via inhibition of the tumor suppressor phosphatase, PTEN. Adv Enzyme Regul. 2007;47:184-94
53. Martini M, De Santis MC, Braccini L. et al. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372-83
Author contact
![]() Corresponding author: Xiulan Ma, PhD, Department of Otolaryngology Head and Neck Surgery, Shengjing Hospital of China Medical University, Liaoning 110004, China. E-mail: xiulan_ma1964com
Corresponding author: Xiulan Ma, PhD, Department of Otolaryngology Head and Neck Surgery, Shengjing Hospital of China Medical University, Liaoning 110004, China. E-mail: xiulan_ma1964com

 Global reach, higher impact
Global reach, higher impact