Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(14):3182-3196. doi:10.7150/ijms.62219 This issue Cite
Review
Circular RNAs Interaction with MiRNAs: Emerging Roles in Breast Cancer
Department of Medical Oncology, Hangzhou Xiasha Hospital, Hangzhou, Zhejiang Province, China
Received 2021-4-30; Accepted 2021-6-28; Published 2021-7-11
Abstract

Despite significant advances in cancer therapy strategies, breast cancer is one of the most common and lethal malignancies worldwide. Characterization of a new class of RNAs using next-generation sequencing opened new doors toward uncovering etiopathogenesis mechanisms of breast cancer as well as prognostic and diagnostic biomarkers. Circular RNAs (circRNAs) are a novel class of RNA with covalently closed and highly stable structures generated primarily from the back-splicing of precursor mRNAs. Although circRNAs exert their function through various mechanisms, acting as a sponge for miRNAs is their primary mechanism of function. Furthermore, growing evidence has shown that aberrant expression of circRNAs is involved in the various hallmarks of cancers. This paper reviews the biogenesis, characteristics, and mechanism of functions of circRNAs and their deregulation in various cancers. Finally, we focused on the circRNAs roles as a sponge for miRNAs in the development, metastasis, angiogenesis, drug resistance, apoptosis, and immune responses of breast cancer.
Keywords: Breast cancer, Circular RNAs, MiRNAs, Metastasis, Angiogenesis
Introduction
According to the GLOBOCAN 2018 estimates, breast cancer (BC) is the second most common cancer for both sexes (11.6% of the total diagnosed cancers) and the most common cancer among females [1]. Despite advances in diagnostic and treatment, the 5-year survival rate for BC patients is 27.4% [2]. Thus, developing more effective strategies for prophylactic intervention, prediction of prognosis, and therapy evaluation is crucial. BC is a highly heterogeneous malignancy with diverse intratumoral and intertumoral non-uniformity and wide variation in tumors among affected patients [3]. BC has stratified based on the expression of progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and estrogen receptor (ER) into four classes: luminal A (ER+, PR+, HER2-), luminal B (ER-, PR-, HER2±), HER2-positive (ER-, PR-, HER2+), and triple-negative (ER-, PR-, HER2-) [4]. Using sequencing and modern genomic technologies, the molecular profile of BC has been widely deciphered, leading to provide novel information about dysregulated genes and transcripts. Hence, better classification of BC individuals based on the predictive biomarkers at the early stages and the molecular mechanism underlying BC tumorigenesis will help identify and treat BC patients.
Among dysregulated transcripts, non-coding RNAs (ncRNAs), including long-non-coding RNAs (lncRNAs), microRNAs (miRNAs), small nuclear RNAs (snRNAs), and circular RNAs (circRNAs), are associated with cancer development, progression, and treatment [5,6]. The circRNAs, covalently closed RNA structures without 5′ caps or 3′ poly (A)-tails, are insensitive to degradation by RNases R; thus, they are more stable and abundant in the cell [7,8]. It has been revealed that circRNAs are related to the pathogenesis of several human diseases, such as cancer. For instance, circRNAs can be involved in tumorigenesis and tumor progression by promoting self-sufficiency in growth signals, evading cell death, insensitivity anti-growth signals, limitless replication, sustained angiogenesis, and metastasis [9]. Here, we outline the biogenesis and functional roles of circRNAs, describe dysregulation of circRNAs in BC, and the evidence for regulation of BC hallmarks by circRNAs during BC tumorigenesis, circRNAs potential as biomarkers, and their potential therapeutic targets.
Biogenesis, degradation, and characteristics of circRNAs
Initially, Sanger et al. discovered circRNAs in RNA viroid in the 1970s [10]. They were considered as by-products of pre-mRNA processing or the products of mis-splicing, because of the limitations of detection techniques, for a long time. However, abundant numbers of circRNAs in eukaryotes, including fungi, viruses, plants, and animals, have been identified owing to advances in bioinformatics and RNA high-throughput sequencing [11]. As a result, several biochemical tools and techniques have been developed for the identification of circRNAs. Table 1 summarizes the developed tools for circRNAs identification and their advantages and disadvantages.
Advantages and disadvantages of different tools in the identification of circRNAs.
| Tools/Techniques | Advantages | Disadvantages |
|---|---|---|
| Reverse transcription-PCR (RT-PCR) | 1. Simple 2. Rapid | 1. Has biases 2. Has artifactual 3. Requires RNase R to distinguish from linear isoforms 4. Low-throughput |
| Northern blotting | 1. Simple 2. No need to RNase R | 1. Time-consuming 2. Laborious |
| Microarray | 1. High-throughput 2. Sensitive 3. Managable in size | 1. Only detects the known circRNAs 2. Identifies only based on junction sequences 3. Higher noise due to mismatch between probe and target |
| RNA sequencing (RNA-seq) | 1. Detects novel circRNAs 2. More reliable and accurate | 1. Low detection efficiency due to limitation in the detection of head-to-tail junctions 2. Highly expensive |
Following the progress of high-throughput transcriptome analysis techniques, various circRNAs have been predicted and recognized to exist at high levels and stably in body fluids, including serum, plasma, urine, and exosomes. The isolated circRNAs from body fluids and circulating cells contained in body fluids are identified and the analytical results are applied for diagnosis, progression, therapy selection, and therapy monitoring in different diseases, such as cancer [12]. Due to the great variability in the length and genesis from liner RNAs, the isolation of circRNAs is not possible according to their size and/or sequence [13]. Following whole RNA isolation, treatment with exoribonuclease, such as RNase R, is widely used for degrading linear RNAs to enrich circRNAs. Additional exonucleases may be helpful to isolate purer circRNAs. For instance, removing 5′ cap structures in mRNAs with RNA 5′ pyrophosphohydrolase (RppH) or tobacco acid pyrophosphatase (TAP) followed by digestion with an exonuclease degrades RNAs with 5′ cap [14].
Similar to linear RNAs, circRNAs are derived from precursor mRNAs (pre-mRNAs). In contrast, linear RNAs are formed through classical splicing, while circRNAs usually are generated via back-splicing [15]. Although a study revealed that 83% of circRNAs have overlap with exons [16], circRNAs can stem from all regions of the genome, including intronic, untranslational regions (UTRs), antisense, and intergenic [17]. Based on their origin and mechanism of formation, circRNAs can be categorized into three classes: exonic circRNAs (ecircRNAs), circular intron or intronic circRNAs (ciRNAs), and exon-intron circRNAs (eiciRNAs) [18]. Figure 1 shows the mechanism of the formation of various types of circRNAs. EcircRNAs, the most common circRNAs, are generated based on the three models: lariat-driven circularization (or exon skipping model), intron-pairing-driven circularization (or direct back splicing model), and RNA-binding proteins (RBPs)-driven circularization [19]. In the lariat-driven circularization model, the 3′ end of the splice donor covalently joins to the 5′ end of the splice acceptor by exon skipping, leading to an exon-containing lariat structure and a linear mRNA. The introns of the lariat structure are removed by the spliceosome to form an exonic circRNA [12]. In addition to the lariat-driven circularization model, ecircRNAs could be generated through another model called intron-pairing-driven circularization. In this model, the complementary motifs on the flanks of introns form circular and linear structures. In the circular structure, splicing out the introns mediates the formation of ecircRNAs [15,20]. However, during the biogenesis of circRNAs, introns may not be removed completely but are retained between the circulated exons in the circRNA structure. This phenomenon leads to the formation of other types of circRNAs, so-called eiciRNAs [21]. Furthermore, RBPs, including quaking (QKI), nuclear factor 90/110 (NF90/NF110), muscleblind (Mbl), DExH-box helicase 9 (DHX9), and adenosine deaminases that act on RNA 1 (ADAR1), also are involved in the biogenesis of ecircRNAs [22]. The binding of RBPs to the flanking introns mediates circularization of single-stranded RNAs by bringing the flanking introns into the vicinity, leading to the formation of ecircRNAs or eiciRNAs [12,23]. CiRNAs are formed when lariat introns escape from debranching and digestion. Their biogenesis depends on a 7 nt GU-rich element near the 5′ splice site and an 11 nt C-rich element near the branch-point site [24]. The elements cause the intron to generate a circular structure at the branchpoint 2′-5′ junction. Finally, the formed lariats undergo 3′ tail degradation, leading to the formation of ciRNAs [24,25].
As mentioned later, one of the mechanisms that circRNAs exert their regulatory functions is their interaction with miRNAs in the cytoplasm [26,27]. Although exosomal circRNAs also regulate cellular function by interacting with miRNAs, exosomes act as intercellular carriers of circRNAs and their regulatory functions occur in the cytoplasm of receptor cells [28,29]. Several databases and software have been developed to predict circRNAs/miRNAs interaction and target mRNAs. For instance, the cancer-specific circRNA database (CSCD) at https://gb.whu.edu.cn/CSCD predicts RNA binding protein (RBP) and miRNA response element (MRE) sites [30]. Also, the circular RNA Interactome (CircInteractome) web maps RBP and MRE sites on human circRNAs and provides binding sites on circRNAs [31].
The mechanism of the formation of various types of circRNAs.
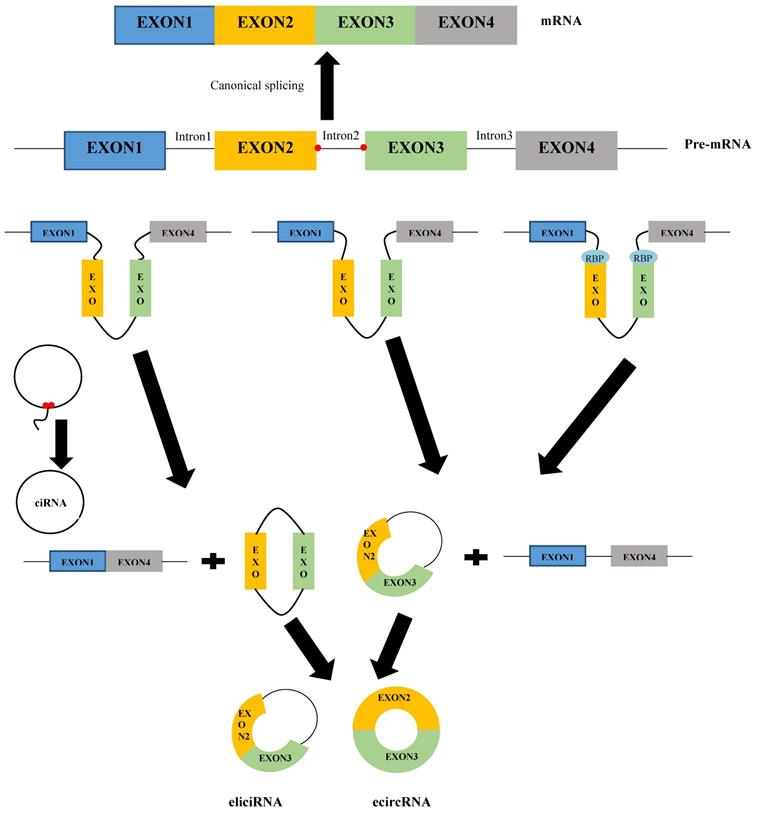
To exert their function, the turnover of circRNAs and how they are degraded are crucial. Owing to 3′ poly (A)-tail and 5′ 7- methylguanosine cap in their structures, mRNAs are degraded by exonucleases shortening their length in 3′→5′ and 5′→3′ directions. Under certain conditions, some mRNAs are cleaved by endonucleases and then are degraded by the mentioned exonucleases. [32]. Due to their circular structure and lack of 3′ poly (A)-tail and 5′ cap, circRNAs are cleaved by endonucleases. It has been demonstrated that circRNAs have a tendency to form imperfect duplexes and inhibit double-stranded RNA-activated protein kinase (PKR). Upon viral infection, RNase L endonuclease degrades circRNAs, leading to PKR activation and immune response induction [33]. Under normal conditions, RBPs, UPF1, and G3BP1 selectively bind to highly-structured circRNAs regulate their decay [34]. Furthermore, circRNAs carrying N6-methyladenosine (m6A) modification undergo endoribonucleolytic cleavage via ribonuclease P (RNase P)/mitochondrial RNA processing (MRP) complex in interaction with YTHDF2, which identifies m6A modification. In this cleavage system, heat-responsive protein 12 (HRSP12) acts as a bridge between the RNase P/MRP complex and YTHDF2 [35].
In addition to their higher stability, there are several particular characteristics of circRNAs: (1) they have a length ranging from below 100 nts to larger than 1000 nts [36,37]; (2) although most of the circRNAs are located in the cytoplasm (ecircRNAs), a small number resides in the nucleus (ciRNAs and eiciRNAs) [17,38]; (3) they are broadly expressed in eukaryotic cells and more than one million circRNAs are detected in human [39]; (4) the expression pattern of circRNAs has been considered as developmental stage-specific and cell type-specific [40]; (5) the sequence of most of the circRNAs are conserved between different species [41]; and (6) they play regulatory roles at both transcriptional and posttranscriptional levels [17].
Functions of circRNA
Figure 2 represents the functional mechanisms of circRNAs in cells.
Interaction with RBPs
Since RBPs are involved in various cellular processes, including proliferation, apoptosis, differentiation, senescence, migration, and oxidative stress responses, through posttranscriptional regulation of RNAs (transportation, splicing, and translation) [42], binding of RBPs to circRNAs and the formation of RNA-protein complexes (RPCs) can mediate several cellular functions. For instance, Zhu et al. found that circZKSCAN1 inhibits cancer stem cells in hepatocellular carcinoma via binding to the RBP fragile X mental retardation protein (FMRP). They showed that cell cycle and apoptosis regulator 1 (CCAR1), a coactivator of the Wnt/β-catenin pathway, is a downstream target of FMRP, suggesting this pathway as a potential therapeutic target [43]. On the other hand, as mentioned above, RBPs mediate circRNAs formation. The RBP trinucleotide repeat-containing 6A (TNRC6A) regulates circ0006916 formation via binding to the intron regions [44].
Regulation of transcription or splicing
It has been suggested that circRNAs, especially eiciRNAs and ciRNAs which are located in the nucleus, are involved in the regulation of gene transcription. For example, two eiciRNAs, circ-PAIP2 and circ-EIF3J, in association with the U1 snRNP can interact with RNA polymerase II to promote their parental genes' expression in HEK293 and HeLa cells, whereas their knockdown with either RNaseH-based antisense oligonucleotides or siRNAs decreases their parental gene expression [21]. In another study, Zhang et al. indicated that the knockdown of ci-ankrd52, a ciRNA, with synthetic antisense targeting the pre-mRNA intron, suppresses mRNA levels of its parental gene [24].
Translation into proteins
It has been reported that some ncRNAs contain small open reading frames (smORFs) for encoding small peptides which contribute to diverse cellular functions and processes, such as pathological conditions [45]. Peptides or proteins encoded by ncRNA play crucial roles in cancer development and progression, including tumor cell metabolism and metastasis [46]. Due to lacking internal ribosome entry sites (IRES), poly (A)-tail, and 5′-3′ polarity, circRNAs were primarily considered as non-coding RNAs [47,48]. Besides their non-coding functions, convincing evidence revealed that circRNAs could be translated into proteins. For example, it has been shown that circ-ZNF609 has an open reading frame (ORF) and could translate into a protein that controls myoblast proliferation [49]. Circ-FBXW7 is another circRNA that carries IRES and codes FBXW7-185aa protein. This protein acts as a tumor suppressor in glioma, which inhibits the proliferation of tumor cells [50]. Mechanistically, the presence of the most common modified base of RNA structures within circRNA sequences, N6- methyladenosine(m6A), recruits YTHDF3 and eIF4G2, leading to translation initiation [51]. However, only a small number of circRNAs have been identified to be involved in the translation.
The function mechanisms of circRNAs. CircRNAs modulate cellular functions by (A) interacting with RBPs, (B) regulating splicing, (C) translation into proteins, and (D) sponging miRNAs.
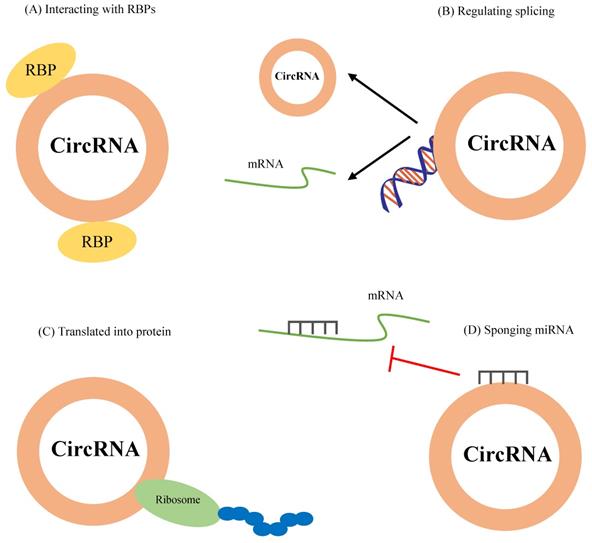
As miRNA sponges
Several studies have been demonstrated that circRNAs can act as competitiveendogenous RNAs (ceRNAs) via binding to miRNAs to suppress their function. MiRNAs are 21-25 nts in length non-coding RNAs that negatively regulate mRNA expression by binding to their 3′‐untranslated region [5]. The most well-known circRNA in interaction with miRNAs is ciRS-7, also called CDR1as, which contains 63 conserved regions for binding to miR-7 [17]. Also, circSry acts as a sponge for miR-138 with having 16 conserved regions for binding to miR-138 [40]. Table 2 summarizes some circRNAs dysregulation in various cancers and their interaction with miRNAs, leading to upregulation of miRNA target mRNAs.
CircRNAs as diagnostic and prognostic biomarkers in cancer
Owing to their stable structure, the long half-life, and the abundant presence in tissues and fluids, including plasma, blood, uterine, and exosomes, as well as expression in a developmental stage- and tissue-specific manner, circRNAs have been considered as promising biomarkers in the diagnosis and prognosis of various cancers [70].
CircRNAs act as sponges for miRNAs in various cancers.
| CircRNA | Up/Downregulated | Cancer | Target | Ref |
|---|---|---|---|---|
| circDLGAP4 | Upregulated | Lung | miR-143/CDK1 | [52] |
| circ-0006282 | Upregulated | Gastric | miR-155/ FBXO22 | [53] |
| circBCRC-3 | Downregulated | Bladder | miR-182-5p/p27 | [54] |
| cSMARCA5 | Upregulated | Cervical | miR‑432/ERK | [55] |
| circZFR | Upregulated | PTC | miR-1261/C8orf4 | [56] |
| circMCTP2 | Downregulated | Gastric | miR-99a-5p/MTMR3 | [57] |
| circATRNL1 | Downregulated | OSCC | miR-23a-3p/PTEN | [58] |
| circ-CPA4 | Upregulated | Lung | let-7 miRNA/PD-L1 | [59] |
| circ-ITCH | Downregulated | Cervical | miR-93-5p/FOXK2 | [60] |
| circPVT1 | Upregulated | HCC | miR‐3666/SIRT7 | [61] |
| circFAM114A2 | Downregulated | Bladder | miR-762/∆NP63 | [62] |
| hsa_circ_0053277 | Upregulated | CRC | miR‐2467‐3p/MMP14 | [63] |
| circEXOC6B | Downregulated | Ovarian | miR-421/RSU1 | [64] |
| circ_SFMBT2 | Downregulated | Glioma | mir-182-5p/Mtss1 | [65] |
| hsa_circ_0000370 | Upregulated | AML | miR-1299/ S100A7A | [66] |
| circMTO1 | Downregulated | HCC | miR‐9/p21 | [67] |
| circ_0020710 | Upregulated | Melanoma | miR-370-3p/CXCL12 | [68] |
| circ-001971 | Upregulated | CRC | miR-29c-3p/VEGFA | [69] |
CDK1, cyclin-dependent kinase 1; FBXO22, f-box protein 22; PTC, papillary thyroid cancer; MTMR3, myotubularin-related protein 3; OSCC, oral squamous cell carcinoma; PD-L1, programmed cell death ligand 1; FOXK2, forkhead box K2; HCC, hepatocellular carcinoma; SIRT7, Sirtuin 7; CRC, colorectal cancer; MMP14, matrix metalloproteinase 14; RSU1, ras suppressor-1; Mtss1, metastasis suppressor 1; AML, acute myeloid leukemia; VEGFA, vascular endothelial growth factor A.
The analysis of circSNAP47 expression in 83 tissue samples with qRT-PCR uncovered that circSNAP47 significantly upregulated in lung cancer patients. The higher expression levels of circSNAP47 were correlated with decreased overall survival (OS), advanced metastasis, and adverse prognosis [71]. Similarly, upregulation of circ-ZKSCAN1 was closely related to poor prognosis, malignant characteristics, tumor stage, and tumor size in patients with lung cancer [72]. In contrast, microarray and qRT-PCR analysis revealed that circBCAR3 upregulation in lung adenocarcinoma is correlated with improved OS, whereas its downregulation is associated with lymph node metastasis and advanced stage [73]. It has been shown that higher expression of CDR1as is associated with poor prognosis and diagnosis of various cancers, including esophageal cancer, CRC, hepatocellular carcinoma, and gastric cancer [74-77]. The analysis of circRNA expression in exosomes isolated from the serum of 170 CRC patients indicated that hsa-circ-0004771 distinguishes patients with stage I/II CRC and patients with benign intestinal diseases [78]. In gastric cancer, the upregulation of circPRMT5, circLMTK2, and circHIPK3 is notably correlated with worse OS [79-81]. Zou et al. identified circLARP4 as a prognosis biomarker in ovarian cancer which its lower expression was associated with poor prognosis in patients with ovarian cancer [82]. Serum circRNA analysis revealed that the levels of circSETDB1 act as a predicting biomarker for response to chemotherapy in ovarian cancer [83]. The summarize of circRNAs as prognostic and diagnostic biomarkers in various cancers is listed in Table 3.
CircRNAs regulate the hallmarks of breast cancer in interaction with miRNAs
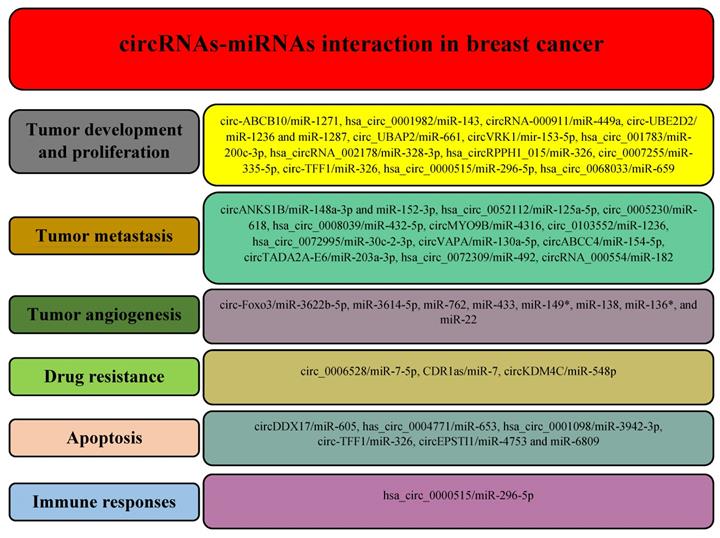
Based on the mentioned techniques, a growing number of circRNAs have been identified with a potential role in the different processes during breast cancer, including carcinogenesis, metastasis, angiogenesis, response to treatments, apoptosis, and immune responses (Figure 3).
Different circRNAs regulate various hallmarks of breast cancer by modulating miRNAs.
CircRNAs as prognostic and diagnostic biomarkers in various cancers.
| CircRNA | Cancer | Prognostic/Diagnostic | Source | Method | Ref |
|---|---|---|---|---|---|
| circ-ITCH | Lung | Diagnostic | Tissue | RT | [84] |
| circMTO1 | HCC | Diagnostic | Tissue | MA/RT | [67] |
| circPVT1 | Gastric | Prognostic/Diagnostic | Tissue | NGS/RT | [85] |
| circ_0034642 | Glioma | Prognostic | Tissue | RT | [86] |
| hsa_circ_0014717 | CRC | Prognostic/Diagnostic | Tissue | RT | [87] |
| hsa_circ_002059 | Gastric | Prognostic/Diagnostic | Tissue/Plasma | RT | [41] |
| has_circ_0067934 | ESCC | Prognostic | Tissue | RT | [88] |
| circ-LDLRAD3 | Pancreatic | Diagnostic | Tissue/Plasma | RT | [89] |
| hsa_circ_0126897 | CRC | Diagnostic | Tissue | MA | [90] |
| circPVT1 | OS | Diagnostic | Tissue/Serum | RT | [91] |
| circFARSA | Lung | Diagnostic | Tissue/Exosome | NGS/RT | [92] |
| circ-PDE8A | Pancreatic | Prognostic | Exosome | MA | [29] |
| CDR1as | HCC | Diagnostic | Tissue | RT | [93] |
| circLPAR1 | Bladder | Prognostic | Tissue | RT | [94] |
RT, qRT-PCR; HCC, hepatocellular carcinoma; MA, microarray; NGS, next-generation sequencing (RNAseq); CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; OS, osteosarcoma.
CircRNAs and tumor development and proliferation
There is increasing evidence that circRNAs are involved in breast cancer development and proliferation. For example, circ-ABCB10 is significantly overexpressed in breast cancer cells and increases breast cancer cell proliferation and progression via directly targeting miR-1271 [95]. Similarly, hsa_circ_0001982 enhances carcinogenesis of breast cancer cells by negatively regulating miR-143, whereas knockdown of hsa_circ_0001982 inhibits cell proliferation and invasion and induces apoptosis [96]. Microarray analysis manifested downregulation circRNA-000911 in breast cancer, whereas its upregulation promotes carcinogenesis via targeting miR‑449a, leading to elevation of Notch1 expression [97]. In another study, Wang et al. demonstrated the upregulation of circ-UBE2D2 in breast cancer tissues was related to poor prognosis. They indicated that circ-UBE2D2 could promote breast cancer progression via acting as a sponge for two miRNAs, miR-1236 and miR-1287. They showed upregulation of MTA2, AFP, ZEB1, HOXB7, and KLF8, and downregulation of p21 expression as target genes of miR-1236, whereas the expression of CD105, PIK3CB, EGFR, GAGE1, ANGPT1, and ATF6α overexpressed as target genes of miR-1287. Therapeutically, intratumoral administration cholesterol-conjugated si-circ-UBE2D2 in a xenograft tumor model notably delayed tumor growth [98]. circ_UBAP2 upregulation significantly promoted the progression of TNBC by modulating the miR-661/MTA1 pathway [99]. Yan et al. found that circVRK1 is a tumor suppressor circRNA that suppresses the expansion and stemness of breast cancer cells through binding to mir-153-5p [100]. It has been shown that mir-153 is involved in the maintenance of stemness of TNBC cells by targeting KLF5 [101]. There is evidence that cancer stem cells (CSCs) are the initiator cells of cancers and are responsible for cancer treatment failure owing to their pluripotency and self-renewal characteristics, leading to tumor recurrence [102,103]. Other circRNAs also mediate breast cancer cell development and progression, including hsa_circ_001783, hsa_circRNA_002178, hsa_circRPPH1_015, circ_0007255, circ‐TFF1, hsa_circ_0000515, and hsa_circ_0068033, via sponging miR-200c-3p, miR-328-3p, miR-326, miR-335-5p, miR-326, miR-296-5p, and miR-659, respectively [104-110].
CircRNAs and tumor metastasis
It has been demonstrated that metastasis is responsible for 90% of cancer-related deaths and is a primary determining factor in response to treatments [111]. During the metastasis process, cancerous cells are dissociated from the primary site and systemically migrate through the vascular or lymphatic system to the secondary site. The migrated cells proliferate and survive in the secondary niche with the help of the components and cells within the tissue microenvironment [112]. Epithelial to mesenchymal transition (EMT) plays an essential role in tumor metastasis in which non-motile epithelial cells are transformed into motile mesenchymal cells containing invasive properties [113].
There is growing evidence that circRNAs can mediate cancer metastasis. For example, circMTO1, hsa_circ_000984, circPIP5K1A, and circ-SMAD7 are associated with metastasis in bladder, colon, lung, and ovarian cancers [114-117]. Yuan et al. found that circSCYL2 is downregulated in breast cancer cells, whereas its upregulation could inhibit the migration, invasion, and EMT progression in breast cancer cells [118]. RNA analysis of TNBC tissues and cell lines revealed that circANKS1B is overexpressed in TNBC, leading to the promotion of breast cancer cell migration, invasion, and metastasis both in vitro and in vivo via inducing EMT. Mechanistically, circANKS1B acts as a sponge for two miRNAs, miR-148a-3p and miR-152-3p, which leads to upregulation of their target, upstream transcription factor 1 gene (USF1). Upregulation of the USF1 transcription factor transcriptionally increases the expression of TGF-β1, resulting in activation of the TGF-β1/Smad pathway to stimulate EMT [119]. In another study, Zhang et al. found that hsa_circ_0052112 could promote migration and invasion of MCF-7 cells via directly sponging miR-125a-5p and regulating ZNF83 expression, whereas upregulation of miR-125a-5p notably reduces cell migration and invasion [120]. Circ_0005230 is another pro-metastatic circRNA in breast cancer that promotes cell invasion by sponging miR-618 and regulating the expression of chromobox protein homolog 8 (CBX8) [121]. It has been reported that CBX8 is an oncogenic protein that promotes the migratory properties of several cancer cells [122]. Using microarray and qRT-PCR analyses, Liu et al. identified the overexpression of hsa_circ_0008039 in breast cancer samples which increases cell proliferation and migration via sponging miR-432-5p and elevating E2F3 expression [123]. E2F3 is an oncogene that enhances tumor growth and metastasis in various cancers [124-126]. Wang et al. indicated that circMYO9B is upregulated in breast cancer tissues, whereas its knockdown remarkably inhibits cell proliferation and invasion in vitro and tumor growth in vivo. Mechanistically, circMYO9B enhances the expression of FOXP4 through directly sponging miR-4316 [127]. By microarray, Du et al. found that circSKA3 is highly expressed in breast cancer cells and promoted cell invasion. The pro-metastatic property of circSKA3 is correlated with its binding to Tks5 and integrin β1, which promotes invadopodia formation [128]. Invadopodia are actin, integrins, and TKS5 rich structures on the cancer cell membrane's outer surface that facilitate cancer cell movement and invasiveness [129,130]. In addition to the mentioned circRNAs, there are other ones that promote migratory properties and metastasis of breast cancer cells, including circ_0103552, hsa_circ_0072995, circVAPA, and circABCC4, which act as a sponge for miR‐1236, miR-30c-2-3p, miR-130a-5p, and miR-154-5p, respectively [131-134].
There are also circRNAs that act as suppressors and negatively regulate the metastasis of breast cancer. For instance, Xu et al. indicated that two circTADA2As, circTADA2A-E6 and circTADA2A-E5/E6, are suppressors of progression and metastasis of breast cancer. The expression levels of these two circRNAs were significantly reduced in breast cancer patients and their downregulation was associated with poor survival of patients with TNBC. They showed that circTADA2A-E6 could exert their inhibitory effects on metastasis via targeting miR-203a-3p and restoring SOCS3 expression [135]. It has been demonstrated that SOCS3 has anti-proliferative and anti-metastatic properties in breast cancer [136,137]. In another study, Yan et al. identified hsa_circ_0072309 as another suppressor circRNA that inhibits breast cancer cell proliferation and invasion via inhibiting miR-492 [138]. In silico and RT‐qPCR analyses demonstrated that circRNA_000554 is notably reduced in breast cancer tissues and cell lines. Transfection of breast cancer cells with circRNA_000554 suppresses cell migration and invasion via reversing the EMT process and sponging miR‐182 [139]. Zinc finger protein 36 (ZFP36) is the direct target of miR-182 [139] that acts as a tumor suppressor gene by inhibiting tumor cell proliferation and growth and inducing cell cycle arrest [140]. Hou et al. reported that circASS1 expression is downregulated in MDA-MB-231, whereas its upregulation inhibits the invasiveness of breast cancer cells [141].
CircRNAs and tumor angiogenesis
To proliferate and survive, tumor cells need blood vessel formation to supply sufficient nutrients and oxygen, so-called angiogenesis [142,143]. This process requires pro-angiogenic factors, including vascular endothelial growth factors (VEGFs), platelet-derived growth factors (PDGFs), fibroblast growth factors (FGFs), hypoxia-inducible factors (HIFs), transforming growth factors (TGFs), and some chemokines [144]. VEGF is the most potent promoter among the angiogenic agents, which exerts its functions through VEGF receptors (VEGFRs) [145]. Mechanistically, the binding of pro-angiogenic factors to their cognate receptors leads to the production of proteases, such as matrix metalloproteinases (MMPs), which degrade extracellular matrix components. Then, endothelial cells (ECs) proliferate and migrate into the degraded area to form primary sprouts. The lamination of primary sprouts results in vascular tube formation and subsequent maturation to blood vessels [146].
It has been demonstrated that various circRNAs are associated with tumor angiogenesis in different cancers, including circ-001971, circRNA-MYLK, circ-RanGAP1, and circSMARCA5 in the colorectal, bladder, and gastric cancers, and glioblastoma, respectively [69,147-149]. In a study, Yang et al. investigated the effect of two non-coding RNAs, Foxo3 pseudogene (Foxo3P) and circ-Foxo3, on the proliferation and angiogenesis of breast cancer cells. They revealed that non-cancerous cell lines express high levels of Foxo3, Foxo3P, and circ-Foxo3 compared to cancer cells, whereas transfection of Foxo3, Foxo3P, and circ-Foxo3 into MDA-MB-231 cells, triple-negative breast cancer (TNBC) cells, significantly decreased proliferation and survival of cancer cells. Furthermore, subcutaneous injection of circ-Foxo3-, Foxo3-, Foxo3P-transfected MDA-MB-231 cells into nude mice indicated that tumor growth in mice receiving transfected cells was notably slower than those received the control cells. They demonstrated that the inhibitory effects of Foxo3, Foxo3P, and circ-Foxo3 on tumor growth were due to decreased blood vessel formation and nutrition supply. Also, they showed that breast cancer cells transfected with Foxo3, Foxo3P, and circ-Foxo3 underwent extensive apoptosis. To exert their anti-tumor effects, both Foxo3P and circ-Foxo3 acted as a sponge for eight miRNAs, including miR-3622b-5p, miR-3614-5p, miR-762, miR-433, miR-149*, miR-138, miR-136*, and miR-22. Sponging these miRNAs increased free Foxo3 mRNAs and promoted their translation [150]. It has been shown that Foxo3 upregulation increases apoptosis and inhibits angiogenesis by downregulating an antiapoptotic protein, FLIP, and decreasing EC proliferation, migration, and tube formation [151-153].
CircRNAs and drug resistance
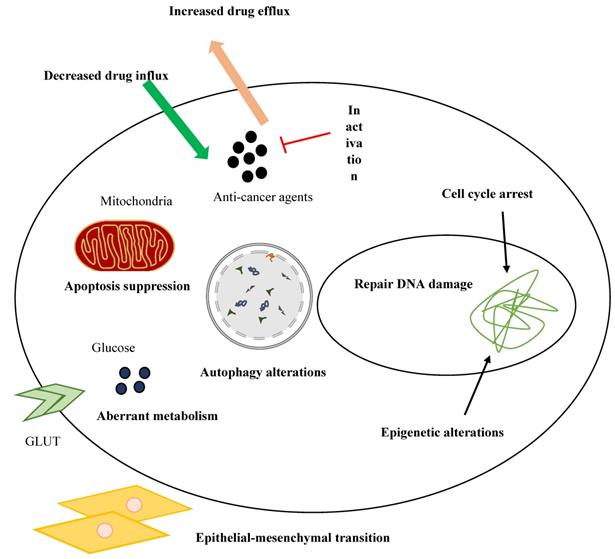
Despite significant advances in the therapeutic strategies of breast cancer, resistance to treatments is a major obstacle to successful cancer therapy. Several mechanisms have been identified in drug resistance of breast cancer, including dysregulation of ATP binding cassette (ABC) transporters, modifications of signaling pathways, EMT and CSCs, cell cycle arrest, autophagy, and apoptosis [154]. Figure 4 represents drug resistance mechanisms in cancer cells.
Drug resistance mechanisms in cancer cells.
It has been shown that circRNAs are involved in resistance to chemotherapy. For example, circPVT1 upregulation mediates resistance of osteosarcoma cells to cisplatin and doxorubicin via regulating ABCB1, whereas knockdown of circPVT1 enhances chemoresistance of the cancer cells [91]. Moreover, a number of circRNAs are involved in the resistance of breast cancer cells to therapeutic agents. The comparison of circRNA expression between monastrol resistant and non-resistance cells with microarray analysis by Liu et al. revealed that 398 circRNAs were dysregulated between the cell types. Further analyses demonstrated that the expression levels of hsa‑circRNA-007874 (circRNA‑MTO1) were decreased in monastrol-resistant breast cancer cells, whereas its upregulation enhanced the sensitivity of the cells to monastrol and inhibited cell viability. CircRNA‑MTO1 exerts its anti-tumor functions by targeting Eg5 protein without influencing Eg5 at the mRNA level. Mechanistically, circRNA‑MTO1 acts as a ceRNA for tumor necrosis factor receptor-associated factor 4 (TRAF4) which inhibits its interaction with the Eg5 gene, resulting in suppression of Eg5 at the protein level [155]. TRAF4 is an oncogene that promotes translation without affecting the mRNA level via binding to the 3′ UTR of the target gene [156]. In another study, Ma et al. identified circAMOTL1 as a mediator of paclitaxel resistance in MDA-MB-231 cells. On the other hand, siRNA against circAMOTL1 reversed the resistance in breast cancer cells by regulating the AKT pathway [157]. Gao et al. revealed that among 3093 detected circRNAs, 18 circRNAs are differentially expressed between adriamycin (ADM) resistant MCF-7 cells (MCF-7/ADM) and parental MCF-7 cells. They identified that circ_0006528 is highly expressed in ADM-resistant tissues and cell lines compared to ADM-sensitive ones, whereas circ_0006528 knockdown using a siRNA increased the sensitivity of breast cancer cells to ADM. Mechanistically, circ_0006528 targets miR-7-5p which leads to overexpression of Raf1 [158]. It has been shown that Raf1 protein is involved in the drug resistance of breast cancer cells via regulating the ERK pathway [159]. In another study, Yang et al. investigated the expression and role of CDR1as in the resistance of breast cancer cells to chemotherapy with 5‐fluorouracil (5‐FU). They uncovered the overexpression of CDR1as and downregulation of miR-7 in 5‐FU-resistant breast cancer cells, whereas knockdown of CDR1as or ectopic overexpression of miR-7 increased chemosensitivity of the cells by promoting apoptosis [160]. circKDM4C is another circRNA that plays a role in the chemoresistance of breast cancer. The expression levels of circKDM4C are decreased in breast cancer and its low levels are associated with metastasis and poor prognosis. CircKDM4C could attenuate doxorubicin resistance of breast cancer cells via targeting the miR-548p/ phenazine biosynthesis-like domain-containing protein (PBLD) axis [161].
CircRNAs and apoptosis
Apoptosis, programmed cell death, is a highly selective and crucial process that balances cell survival and death to prevent various diseases, including cancer [162]. Generally, there are two pathways of apoptosis: extrinsic (death receptor) and intrinsic (mitochondrial). In the extrinsic pathway, binding of extracellular ligands, such as Fas ligand (Fas-L), tumor necrosis factor (TNF), and TNF-related apoptosis-inducing ligand (TRAIL) to their cognate receptors initiates death signals, whereas the intrinsic pathway is activated after an increase of mitochondrial permeability and release of pro-apoptotic proteins [163]. In both of the pathways, cysteine proteases called caspases are activated and cleaved the proteins.
Recently, Peng et al. revealed that circDDX17 levels were decreased in breast cancer, whereas its upregulation promoted cell apoptosis and reduced cell proliferation and colony formation through the regulation of p21 and CDK genes via directly binding to miR-605 [164]. High-throughput RNA sequencing revealed that 49 circRNAs were differentially expressed (30 were upregulated, and 19 were downregulated) between breast cancer tissues and nontumorous ones. Further analyses indicated that has_circ_0004771 expression was higher than other circRNAs in breast cancer tissues. Has_circ_0004771 upregulated the expression of Zinc finger E-box binding homeobox 2 (ZEB2) by binding to miR-653. Has_circ_0004771 and ZEB2 knockdown could induce apoptosis and inhibition of cell growth [165]. In another study, Zhao et al. found that hsa_circ_0001098 (circRNA_BARD1) overexpression could promote breast cancer cell apoptosis by regulating the miR-3942-3p/BARD1 axis [166]. It has been demonstrated that BARD1could promote apoptosis through binding to, phosphorylating, and stabilizing p53 in DNA double-strand break repair [167]. In addition to the mentioned circRNAs, it has been shown that circ‐TFF1 (hsa_circ_0061825) and circEPSTI1 also are involved in the apoptosis of breast cancer cells through sponging miRNAs. Silencing circ‐TFF1 promotes apoptosis by miR‐326/TFF1 axis [108], while circEPSTI1 knockdown induces apoptosis via regulating the miR-4753 and miR-6809/ BCL11A pathway [168].
CircRNAs and immune responses
According to the cancer immunosurveillance concept, tumor cells are identified, eradicated, and repressed by host immune systems [169]. However, tumor cells can escape from immunosurveillance and immune control, resulting in tumor proliferation and clinical emergence [170]. Among immune cells, myeloid-derived suppressor cells (MDSCs), regulatory T (Treg) cells, and tumor-associated macrophages (TAMs) act as immunosuppressive cells, whereas cytotoxic T cells (CTLs), T helper (Th) lymphocytes, and natural killer (NK) cells fight against cancer [171]. Furthermore, inhibitory proteins, including programmed death receptor-1 (PD-1) and cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4), also mediate immune evasion of cancer cells. Thus, blocking the inhibitory proteins has been developed as a promising strategy in breast cancer therapy [172,173].
It has been shown that various circRNAs are involved in the immune regulation of cancers [174]. For instance, Zhang et al. found that upregulation of hsa_circ_0020397 in colorectal cancer mediates cell viability and invasion via targeting miR‐138, leading to overexpression of PD‐L1 and telomerase reverse transcriptase (TERT) [175]. Circ‐UBAP2 is another circRNA that promotes cancer progression by enhancing immune evasion. It was demonstrated that the higher levels of circ‐UBAP2 modulate the ZEB1 and CXCR4 expression in pancreatic adenocarcinoma, which is correlated with exhausted T cells, Tregs, and M2 macrophages as well as higher levels of PD-1 and CTLA-4 [176]. Cai et al. indicated that upregulation of hsa_circ_0000515 in breast cancer was associated with poor prognosis, whereas its silencing impaired cell proliferation, the progression of the cell cycle, and invasion, reduced pro-angiogenic potential, and decreased inflammatory response of breast cancer. Mechanistically, hsa_circ_0000515 acts as a sponge for miR‐296‐5p to inhibit CXCL10 repression [109]. The binding of CXCL10 to the CXCR3 receptor activates and initiates G protein‐mediated signaling, resulting in Th1 cell generation and their aggregation at the inflammatory niche [177]. Moreover, CXCL10 acts as an oncogene that is associated with breast cancer progression [177].
Conclusions and perspectives
There is growing evidence that the circRNAs expression can affect breast cancer development and progression and may show great potential as a biomarker in diagnosing, prognosis, and treatment of breast cancer. Compared to other RNAs, circRNAs contain a stable structure and are broadly expressed in body fluids such as blood and uterine, suggesting circRNAs as promising and reliable biomarkers. Moreover, circRNAs detection using qRT-PCR and RNA sequencing is more accurate and convenient and needs a shorter time and smaller amounts of samples than the detection of proteins. However, their biological functions, formation, maturation, and transport from the nucleus to the cytoplasm need further investigation. Due to our incomplete understanding of circRNAs, some issues need to be addressed for their application as diagnostic and therapeutic biomarkers as well as therapeutic agents in clinical practice. First, it is necessary to assess the specificity and sensitivity of circRNAs as potential biomarkers. Second, targeting a circRNA as a therapeutic strategy for cancer therapy has potential risks for off-target effects on other tissues because a circRNA participates in the function of many tissues. Third, selecting an efficient delivery system, including viral vectors, liposomes, nanoparticles, and exosomes, based on the target cells is essential for the delivery of large amounts of circRNA. Another strategy to enhance their efficacy is engineering artificial circRNAs containing a large number of binding sites for miRNAs. Thus, circRNAs can be used in combination with other prognostic and diagnostic biomarkers for breast cancer, and their clinical application still requires more analysis and optimization.
Abbreviations
BC: breast cancer; HER2: human epidermal growth factor receptor 2; miRNAs: microRNAs; circRNAs: circular RNAs; ecircRNAs: exonic circRNAs; ciRNAs: circular intron or intronic circRNAs; eiciRNAs: exon-intron circRNAs; RBPs: RNA-binding proteins; ceRNAs: competitive endogenous RNAs; OS: overall survival; EMT: epithelial to mesenchymal transition; TNBC: triple-negative breast cancer; PD-1: programmed death receptor-1; CTLA-4: cytotoxic T-lymphocyte-associated antigen-4.
Author Contributions
LG provided suggestions and edited the manuscript. XYZ and JS drafted the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Quinn KM. Innovative approaches to immunotherapy in breast cancer. J Thorac Dis. 2020;12:4536
3. Turashvili G, Brogi E. Tumor heterogeneity in breast cancer. Front Med. 2017;4:227
4. Li X-X, Wang L-J, Hou J, Liu H-Y, Wang R, Wang C. et al. Identification of Long Noncoding RNAs as Predictors of Survival in Triple-Negative Breast Cancer Based on Network Analysis. Biomed Res Int. 2020. 2020
5. Goradel NH, Mohammadi N, Haghi-Aminjan H, Farhood B, Negahdari B, Sahebkar A. Regulation of tumor angiogenesis by microRNAs: State of the art. J Cell Physiol. 2019;234:1099-110
6. Grillone K, Riillo C, Scionti F, Rocca R, Tradigo G, Guzzi PH, et al. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter.” J Exp Clin Cancer Res. 2020;39:1-19
7. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q. et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588
8. Drula R, Braicu C, Harangus A, Nabavi SM, Trif M, Slaby O. et al. Critical function of circular RNAs in lung cancer. Wiley Interdiscip Rev RNA. 2020 e1592
9. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y. et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90
10. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci. 1976;73:3852-6
11. Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10:258
12. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267-74
13. Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y. et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116-e116
14. Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5′→ 3′ exoribonuclease Xrn1. Nat Struct Mol Biol. 2011;18:270
15. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141-57
16. Ye C, Chen L, Liu C, Zhu Q, Fan L. Widespread noncoding circular RNA s in plants. New Phytol. 2015;208:88-95
17. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
18. Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18:547-57
19. Zhang C, Ma L, Niu Y, Wang Z, Xu X, Li Y. et al. Circular RNA in Lung Cancer Research: Biogenesis, Functions, and Roles. Int J Biol Sci. 2020;16:803
20. Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
21. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256
22. Guo Y, Yang J, Huang Q, Hsueh C, Zheng J, Wu C. et al. Circular RNAs and their roles in head and neck cancers. Mol Cancer. 2019;18:44
23. Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829-42
24. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
25. Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. Rna. 2014;20:1476-87
26. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Acids. 2020;22:285-97
27. Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M. et al. A novel circular RNA, circFAT1 (e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222-32
28. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A. et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11:8182
29. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237-50
30. Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F. et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925-9
31. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34-42
32. Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246-59
33. Liu C-X, Li X, Nan F, Jiang S, Gao X, Guo S-K. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865-80
34. Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell. 2020.
35. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494-507
36. Shao J, Wang L, Liu X, Yang M, Chen H, Wu B. et al. Identification and characterization of circular RNAs in Ganoderma lucidum. Sci Rep. 2019;9:1-13
37. Guria A, Velayudha Vimala Kumar K, Srikakulam N, Krishnamma A, Chanda S, Sharma S. et al. Circular RNA profiling by illumina sequencing via template-dependent multiple displacement amplification. Biomed Res Int. 2019. 2019
38. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-61
39. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984-92
40. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409
41. Li P, Chen S, Chen H, Mo X, Li T, Shao Y. et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim acta. 2015;444:132-6
42. Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243-55
43. Zhu Y-J, Zheng B, Luo G-J, Ma X-K, Lu X-Y, Lin X-M. et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526
44. Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z. et al. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39:981-92
45. Dragomir MP, Manyam GC, Ott LF, Berland L, Knutsen E, Ivan C. et al. FuncPEP: A Database of Functional Peptides Encoded by Non-Coding RNAs. Non-coding RNA. 2020;6:41
46. Wang J, Zhu S, Meng N, He Y, Lu R, Yan G-R. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27:1718-25
47. Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. 2018;37:206
48. Liu D, Mewalal R, Hu R, Tuskan GA, Yang X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic Res. 2017;4:1-8
49. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22-37
50. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F. et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI J Natl Cancer Inst. 2018;110:304-15
51. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017;27:626-41
52. Wang B, Hua P, Zhao B, Li J, Zhang Y. Circular RNA circDLGAP4 is involved in lung cancer development through modulating microRNA-143/CDK1 axis. Cell Cycle. 2020;19:2007-17
53. He Y, Wang Y, Liu L, Liu S, Liang L, Chen Y. et al. Circular RNA circ_0006282 contributes to the progression of gastric cancer by sponging miR-155 to upregulate the expression of FBXO22. Onco Targets Ther. 2020;13:1001
54. Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F. et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17:144
55. Huang P, Qi B, Yao H, Zhang L, Li Y, Li Q. Circular RNA cSMARCA5 regulates the progression of cervical cancer by acting as a microRNA-432 sponge. Mol Med Rep. 2020;21:1217-23
56. Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503:56-61
57. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X. et al. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. 2020;39:1-17
58. Chen G, Li Y, He Y, Zeng B, Yi C, Wang C. et al. Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol Ther Acids. 2020;19:961-73
59. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39:1-19
60. Li J, Guo R, Liu Q, Sun J, Wang H. Circular RNA Circ-ITCH inhibits the malignant behaviors of cervical cancer by microRNA-93-5p/FOXK2 Axis. Reprod Sci. 2020:1-9
61. Li Y, Shi H, Yuan J, Qiao L, Dong L, Wang Y. Downregulation of circular RNA circPVT1 restricts cell growth of hepatocellular carcinoma through downregulation of Sirtuin 7 via microRNA-3666. Clin Exp Pharmacol Physiol. 2020
62. Liu T, Lu Q, Liu J, Xie S, Feng B, Zhu W. et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating∆ NP63 by sponging miR-762. Cell Death Dis. 2020;11:1-14
63. Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 2020;235:2881-90
64. Wang Z, Zhang W, Fang J, Xie P, Miao M, Yang H. Circular RNA circEXOC6B Inhibits the Progression of Ovarian Cancer by Sponging miR-421 and Regulating RUS1 Expression. Onco Targets Ther. 2020;13:8233
65. Zhang S, Qin W, Yang S, Guan N, Sui X, Guo W. Circular RNA SFMBT2 Inhibits the Proliferation and Metastasis of Glioma Cells Through Mir-182-5p/Mtss1 Pathway. Technol Cancer Res Treat. 2020;19:1533033820945799
66. Zhang L, Bu Z, Shen J, Shang L, Chen Y, Wang Y. A novel circular RNA (hsa_circ_0000370) increases cell viability and inhibits apoptosis of FLT3-ITD-positive acute myeloid leukemia cells by regulating miR-1299 and S100A7A. Biomed Pharmacother. 2020;122:109619
67. Han D, Li J, Wang H, Su X, Hou J, Gu Y. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
68. Wei C-Y, Zhu M-X, Lu N-H, Liu J-Q, Yang Y-W, Zhang Y. et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol Cancer. 2020;19:1-14
69. Chen C, Huang Z, Mo X, Song Y, Li X, Li X. et al. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res. 2020;39:1-15
70. Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019 p. 90-9
71. Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X. et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun. 2018;503:2089-94
72. Wang Y, Xu R, Zhang D, Lu T, Yu W, Wo Y. et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl Lung Cancer Res. 2019;8:862
73. Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2019;508:37-45
74. Sang M, Meng L, Sang Y, Liu S, Ding P, Ju Y. et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018;426:37-46
75. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T. et al. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918-28
76. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17-27
77. Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z. et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119:440-6
78. Pan B, Qin J, Liu X, He B, Wang X, Pan Y. et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096
79. Du W, Li D, Guo X, Li P, Li X, Tong S. et al. Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif cells, nanomedicine, Biotechnol. 2019;47:4120-30
80. Wang S, Tang D, Wang W, Yang Y, Wu X, Wang L. et al. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol Cancer. 2019;18:162
81. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/beta-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23:7905-12
82. Zou T, Wang PL, Gao Y, Liang WT. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22:7178-82
83. Wang W, Wang J, Zhang X, Liu G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:7451
84. Wan L, Zhang L, Fan K, Cheng Z-X, Sun Q-C, Wang J-J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016. 2016
85. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208-19
86. Yang M, Li G, Fan L, Zhang G, Xu J, Zhang J. Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem Biophys Res Commun. 2019;508:980-5
87. Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775-82
88. Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J. et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576
89. Yang F, Liu D-Y, Guo J-T, Ge N, Zhu P, Liu X. et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345
90. Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499-512
91. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14:321
92. Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G. et al. A novel plasma circular RNA circ FARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783-91
93. Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res. 2017;16:3891-902
94. Lin G, Sheng H, Xie H, Zheng Q, Shen Y, Shi G. et al. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol Lett. 2019;17:3537-47
95. Liang H-F, Zhang X-Z, Liu B-G, Jia G-T, Li W-L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566
96. Tang Y-Y, Zhao P, Zou T-N, Duan J-J, Zhi R, Yang S-Y. et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901-8
97. Wang H, Xiao YI, Wu LI, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743-54
98. Wang Y, Li J, Du C, Zhang L, Zhang Y, Zhang J. et al. Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl Oncol. 2019;12:1305-13
99. Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y. et al. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun. 2018;505:996-1002
100. Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L. et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704
101. Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W. et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6:533
102. Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717-28
103. Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells—what challenges do they pose? Nat Rev Drug Discov. 2014;13:497-512
104. Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L. et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:1-14
105. Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 2020;24:2189-201
106. Zhao C, Li L, Li Z, Xu J, Yang Q, Shi P. et al. A Novel Circular RNA hsa_circRPPH1_015 Exerts an Oncogenic Role in Breast Cancer by Impairing miRNA-326-Mediated ELK1 Inhibition. Front Oncol. 2020;10:906
107. Jia Q, Ye L, Xu S, Xiao H, Xu S, Shi Z. et al. Circular RNA 0007255 regulates the progression of breast cancer through miR-335-5p/SIX2 axis. Thorac cancer. 2020;11:619-30
108. Pan G, Mao A, Liu J, Lu J, Ding J, Liu W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53:e12720
109. Cai F, Fu W, Tang L, Tang J, Sun J, Fu G. et al. Hsa_circ_0000515 is a novel circular RNA implicated in the development of breast cancer through its regulation of the microRNA-296-5p/CXCL10 axis. FEBS J. 2020.
110. Yuan P, Lei L, Dong S, Liu D. Circular RNA hsa_circ_0068033 Acts as a Diagnostic Biomarker and Suppresses the Progression of Breast Cancer Through Sponging miR-659. Onco Targets Ther. 2020;13:1921
111. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670-91
112. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5:1-17
113. Mondal P, Meeran SM. Long non-coding RNAs in breast cancer metastasis. Non-coding RNA Res. 2020.
114. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508:991-6
115. Xu X-W, Zheng B-A, Hu Z-M, Qian Z-Y, Huang C-J, Liu X-Q. et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674
116. Chi Y, Luo Q, Song Y, Yang F, Wang Y, Jin M. et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem. 2019;120:19019-30
117. Zhao Y, Qin XP, Lang YP, Kou D, Shao ZW. Circular RNA circ-SMAD7 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur Rev Med Pharmacol Sci. 2019;23:5603-10
118. Yuan C, Luo X, Zhan X, Zeng H, Duan S. EMT related circular RNA expression profiles identify circSCYL2 as a novel molecule in breast tumor metastasis. Int J Mol Med. 2020;45:1697-710
119. Zeng K, He B, Yang BB, Xu T, Chen X, Xu MU. et al. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17:1-19
120. Jiang L, Hou J, Zhong S, Zhou S, Zhu L, Li J. et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342-53
121. Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y. et al. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell Physiol Biochem. 2018;51:1710-22
122. Jia Y, Wang Y, Zhang C, Chen MY. Upregulated CBX8 promotes cancer metastasis via the WNK2/MMP2 pathway. Mol Ther. 2020;19:188-96
123. Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502:358-63
124. Zhang Y, Zhang Z, Li Z, Gong D, Zhan B, Man X. et al. MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. Oncol Rep. 2020;43:1032
125. Yang H, Wang L, Tang X, Bai W. miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncol Lett. 2017;14:7687-90
126. Yang J, Zhang HF, Qin CF. MicroRNA-217 functions as a prognosis predictor and inhibits pancreatic cancer cell proliferation and invasion via targeting E2F3. Eur Rev Med Pharmacol Sci. 2017;21:4050-7
127. Wang N, Gu Y, Li L, Wang F, Lv P, Xiong Y. et al. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR-4316. Arch Biochem Biophys. 2018;653:63-70
128. Du WW, Yang W, Li X, Fang L, Wu N, Li F. et al. The Circular RNA circSKA3 Binds Integrin β1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Mol Ther. 2020;28:1287-98
129. Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS. Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol. 2017;27:595-607
130. Stylli SS, Stacey TT, Verhagen AM, Pass I, Courtneidge SA, Lock P. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J Cell Sci. 2009;122:2727-40
131. Yang L, Song C, Chen Y, Jing G, Sun J. Circular RNA circ_0103552 forecasts dismal prognosis and promotes breast cancer cell proliferation and invasion by sponging miR-1236. J Cell Biochem. 2019;120:15553-60
132. Zhang H-D, Jiang L-H, Hou J-C, Zhou S-Y, Zhong S-L, Zhu L-P. et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10:1229-42
133. Zhou S, Chen W, Yang S, Li J, Zhang J, Zhang H. et al. Circular RNA circVAPA regulates breast cancer cell migration and invasion via sponging miR-130a-5p. Epigenomics. 2020;12:303-17
134. Jiang J, Cheng X. Circular RNA circABCC4 acts as a ceRNA of miR-154-5p to improve cell viability, migration and invasion of breast cancer cells in vitro. Cell Cycle. 2020;19:2653-61
135. Xu J-Z, Shao C-C, Wang X-J, Zhao X, Chen J-Q, Ouyang Y-X. et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:1-16
136. Barclay JL, Anderson ST, Waters MJ, Curlewis JD. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int J cancer. 2009;124:1756-66
137. Kim G, Ouzounova M, Quraishi AA, Davis A, Tawakkol N, Clouthier SG. et al. SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene. 2015;34:671-80
138. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033
139. Mao Y, Lv M, Cao W, Liu X, Cui J, Wang Y. et al. Circular RNA 000554 represses epithelial-mesenchymal transition in breast cancer by regulating microRNA-182/ZFP36 axis. FASEB J. 2020;34:11405-20
140. Xu L, Ning H, Gu L, Wang Q, Lu W, Peng H. et al. Tristetraprolin induces cell cycle arrest in breast tumor cells through targeting AP-1/c-Jun and NF-κB pathway. Oncotarget. 2015;6:41679
141. Hou J, Xu Z, Zhong S, Zhang H, Jiang L, Chen X. et al. Circular RNA circASS1 is downregulated in breast cancer cells MDA-MB-231 and suppressed invasion and migration. Epigenomics. 2019;11:199-213
142. Hashemi Goradel N, Ghiyami-Hour F, Jahangiri S, Negahdari B, Sahebkar A, Masoudifar A. et al. Nanoparticles as new tools for inhibition of cancer angiogenesis. J Cell Physiol. 2018;233:2902-10
143. Goradel NH, Asghari MH, Moloudizargari M, Negahdari B, Haghi-Aminjan H, Abdollahi M. Melatonin as an angiogenesis inhibitor to combat cancer: Mechanistic evidence. Toxicol Appl Pharmacol. 2017;335:56-63
144. Saman H, Raza SS, Uddin S, Rasul K. Inducing Angiogenesis, a Key Step in Cancer Vascularization, and Treatment Approaches. Cancers (Basel). 2020;12:1172
145. Rajabi M, Mousa SA. The role of angiogenesis in cancer treatment. Biomedicines. 2017;5:34
146. Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell Mol Life Sci. 2020;77:1745-70
147. Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L. et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305-17
148. Lu J, Wang Y, Yoon C, Huang X, Xu Y, Xie J. et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38-48
149. Barbagallo D, Caponnetto A, Barbagallo C, Battaglia R, Mirabella F, Brex D. et al. The GAUGAA Motif Is Responsible for the Binding between circSMARCA5 and SRSF1 and Related Downstream Effects on Glioblastoma Multiforme Cell Migration and Angiogenic Potential. Int J Mol Sci. 2021;22:1678
150. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-31
151. Skurk C, Maatz H, Kim H-S, Yang J, Abid MR, Aird WC. et al. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J Biol Chem. 2004;279:1513-25
152. Potente M, Fisslthaler B, Busse R, Fleming I. 11, 12-Epoxyeicosatrienoic acid-induced inhibition of FOXO factors promotes endothelial proliferation by down-regulating p27Kip1. J Biol Chem. 2003;278:29619-25
153. Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A. et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382-92
154. Hu W, Tan C, He Y, Zhang G, Xu Y, Tang J. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther. 2018;11:1529
155. Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53:1752-62
156. Zhang L, Zhou F, de Vinuesa AG, de Kruijf EM, Mesker WE, Hui L. et al. TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol Cell. 2013;51:559-72
157. Ma J, Fang L, Yang Q, Hibberd S, Du WW, Wu N. et al. Posttranscriptional regulation of AKT by circular RNA angiomotin-like 1 mediates chemoresistance against paclitaxel in breast cancer cells. Aging (Albany NY). 2019;11:11369
158. Gao D, Zhang X, Liu B, Meng D, Fang K, Guo Z. et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175-88
159. Lee M, Koh WS, Han SS. Down-regulation of Raf-1 kinase is associated with paclitaxel resistance in human breast cancer MCF-7/Adr cells. Cancer Lett. 2003;193:57-64
160. Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou D. et al. Inhibition of circular RNA CDR1as increases chemosensitivity of 5-FU-resistant BC cells through up-regulating miR-7. J Cell Mol Med. 2019;23:3166-77
161. Liang Y, Song X, Li Y, Su P, Han D, Ma T. et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene. 2019;38:6850-66
162. Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014. 2014
163. Jan R. Understanding apoptosis and apoptotic pathways targeted cancer therapeutics. Adv Pharm Bull. 2019;9:205
164. Peng HH, Wen YG. CircDDX17 acts as a competing endogenous RNA for miR-605 in breast cancer progression. Eur Rev Med Pharmacol Sci. 2020;24:6794-801
165. Xie R, Tang J, Zhu X, Jiang H. Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR-653 by targeting ZEB2 signaling pathway. Biosci Rep. 2019 39
166. Zhao J, Zou H, Han C, Ma J, Zhao J, Tang J. Circlular RNA BARD1 (Hsa_circ_0001098) overexpression in breast cancer cells with TCDD treatment could promote cell apoptosis via miR-3942/BARD1 axis. Cell Cycle. 2018;17:2731-44
167. Feki A, Jefford CE, Berardi P, Wu J-Y, Cartier L, Krause K-H. et al. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene. 2005;24:3726-36
168. Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D. et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003
169. Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137-48
170. Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases—elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16-25
171. Nicolini A, Ferrari P, Rossi G, Carpi A. Tumour growth and immune evasion as targets for a new strategy in advanced cancer. Endocr Relat Cancer. 2018;25:R577-604
172. Zou Y, Zou X, Zheng S, Tang H, Zhang L, Liu P. et al. Efficacy and predictive factors of immune checkpoint inhibitors in metastatic breast cancer: a systematic review and meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920940928
173. Swoboda A, Nanda R. Immune checkpoint blockade for breast cancer. Optim Breast Cancer Manag. 2018 p. 155-65
174. Li W, Liu J, Chen M, Xu J, Zhu D. Circular RNA in cancer development and immune regulation. J Cell Mol Med. 2020.
175. Zhang X, Xu L, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056-64
176. Zhao R, Ni J, Lu S, Jiang S, You L, Liu H. et al. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging (Albany NY). 2019;11:8484
177. Smit MJ, Verdijk P, van der Raaij-Helmer EMH, Navis M, Hensbergen PJ, Leurs R. et al. CXCR3-mediated chemotaxis of human T cells is regulated by a Gi-and phospholipase C-dependent pathway and not via activation of MEK/p44/p42 MAPK nor Akt/PI-3 kinase. Blood. 2003;102:1959-65
Author contact
![]() Corresponding author: Liu Gong, Department of Medical Oncology, Hangzhou Xiasha Hospital, No. 3 Qingchun East Road, Hangzhou, China. Tel: +86-571- 85893117, Fax: +86-571-85893117, Email: 3x036edu.cn
Corresponding author: Liu Gong, Department of Medical Oncology, Hangzhou Xiasha Hospital, No. 3 Qingchun East Road, Hangzhou, China. Tel: +86-571- 85893117, Fax: +86-571-85893117, Email: 3x036edu.cn

 Global reach, higher impact
Global reach, higher impact