3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(7):1670-1679. doi:10.7150/ijms.54460 This issue Cite
Research Paper
Screening, identification and interaction analysis of key MicroRNAs and genes in Asthenozoospermia
1. Center for Translational Medicine, Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, West China Second University Hospital, Sichuan University, Chengdu, China.
2. Department of Cardiovascular Surgery, Zhongnan Hospital of Wuhan University, Wuhan, China.
Abstract
Background: Asthenozoospermia, one of the most common causes of male infertility, is a complicate multifactorial pathological condition that genetic factors are involved in. However, the epigenetic signature and mechanism of asthenozoospermia still remain limited. Our study aimed to confirm the key microRNAs (miRNAs) and genes in asthenozoospermia and demonstrate the underlying epigenetic regulatory mechanisms.
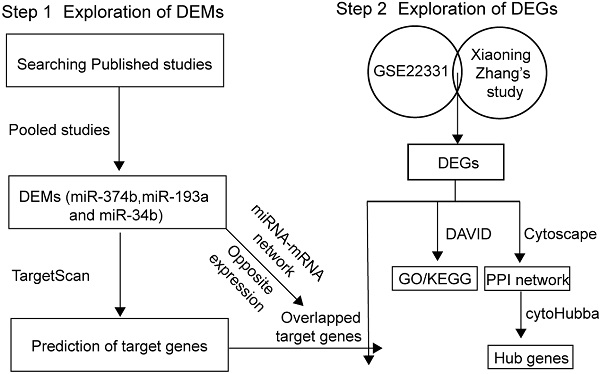
Methods: We screened out and pooled previous studies to extracted potential differentially expressed miRNAs (DEMs). GSE22331 and a published profile dataset were integrated to identify differentially expressed genes (DEGs). Pathway and gene ontology analysis were performed using DAVID. A protein-protein network (PPI) was constructed using STRING. The target genes of DEMs were predicted using TargetScan and the miRNA-mRNA network was built.
Results: We reported 3 DEMs and 423 DEGs by pooling included dataset and published studies. Pathway analysis showed that these DEGs might participate in signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway and Notch signaling pathway. 25 hub genes were identified, and the most significant gene was BDNF. We screened out the overlapped DEGs between the predicted target genes of 3 DEMs and the 423 DEGs. Finally, a potential miRNA-mRNA regulatory network was constructed.
Conclusion: This study firstly pooled several published studies and a GEO dataset to determine the significance of potential miRNAs and genes, such as miR-374b, miR-193a, miR-34b, BDNF, NTRK2, HNRNPD and EFTUD2 in regulating asthenozoospermia and underscore their interactions in the pathophysiological mechanism. Our results provided theoretical basis and new clues for potential therapeutic treatment in asthenozoospermia. Validations in vivo and in vitro are required in future studies.
Keywords: Asthenozoospermia, miRNAs, genes

 Global reach, higher impact
Global reach, higher impact