Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(7):1670-1679. doi:10.7150/ijms.54460 This issue Cite
Research Paper
Screening, identification and interaction analysis of key MicroRNAs and genes in Asthenozoospermia
1. Center for Translational Medicine, Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, West China Second University Hospital, Sichuan University, Chengdu, China.
2. Department of Cardiovascular Surgery, Zhongnan Hospital of Wuhan University, Wuhan, China.
Received 2020-10-13; Accepted 2021-1-4; Published 2021-2-6
Abstract
Background: Asthenozoospermia, one of the most common causes of male infertility, is a complicate multifactorial pathological condition that genetic factors are involved in. However, the epigenetic signature and mechanism of asthenozoospermia still remain limited. Our study aimed to confirm the key microRNAs (miRNAs) and genes in asthenozoospermia and demonstrate the underlying epigenetic regulatory mechanisms.
Methods: We screened out and pooled previous studies to extracted potential differentially expressed miRNAs (DEMs). GSE22331 and a published profile dataset were integrated to identify differentially expressed genes (DEGs). Pathway and gene ontology analysis were performed using DAVID. A protein-protein network (PPI) was constructed using STRING. The target genes of DEMs were predicted using TargetScan and the miRNA-mRNA network was built.
Results: We reported 3 DEMs and 423 DEGs by pooling included dataset and published studies. Pathway analysis showed that these DEGs might participate in signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway and Notch signaling pathway. 25 hub genes were identified, and the most significant gene was BDNF. We screened out the overlapped DEGs between the predicted target genes of 3 DEMs and the 423 DEGs. Finally, a potential miRNA-mRNA regulatory network was constructed.
Conclusion: This study firstly pooled several published studies and a GEO dataset to determine the significance of potential miRNAs and genes, such as miR-374b, miR-193a, miR-34b, BDNF, NTRK2, HNRNPD and EFTUD2 in regulating asthenozoospermia and underscore their interactions in the pathophysiological mechanism. Our results provided theoretical basis and new clues for potential therapeutic treatment in asthenozoospermia. Validations in vivo and in vitro are required in future studies.
Keywords: Asthenozoospermia, miRNAs, genes
Introduction
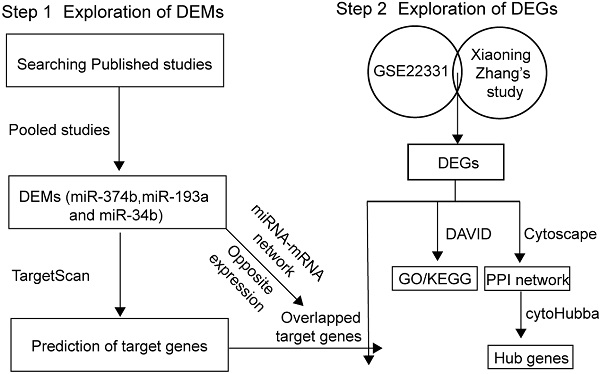
Male infertility is a severe problem in human reproduction health [1, 2]. Abnormal sperm morphology, poor sperm motility and low sperm count are identified as common causes of male infertility. According to the criteria reported by the World Health Organization, asthenozoospermia is characterized by total motility < 40% and progressive motility < 32% in fresh semen samples [3]. Several factors have been confirmed to be associated with asthenozoospermia, as shown in Figure 1. Chromosomal abnormality was correlated with sperm deficiency including asthenozoospermia [4]. The multiple morphological abnormalities of the sperm flagella (MMAF) is one of the most known causes of asthenozoospermia. Mutations in the mitochondrial genome have been reported in MMAF, such as DNAH1, CFAP43, CFAP44, CFAP69, and QRICH2 [5-8], and study have shown compelling evidence that the loss-of-function mutations of these genes could induce MMAF and lead to male infertility. Hormone could also modulate spermatogenesis and impact fertilization. In a clinical trial, most of 786 subfertility patients were characterized as idiopathic oligo or asthenozoospermia who improved their semen quality and concentration by receiving hormonal treatment [9]. Sperm motility was under the control of flagellum [10] and any structural or functional defects in flagellum would lead to reduced motility and induce asthenozoospermia [11]. Moreover, the anti-sperm antibodies played key roles in female and male immunological infertility and sperm-immobilizing antibodies that bound to the surface of ejaculated sperm could impact the sperm motility [12]. There were other exogenous sources could result in asthenozoospermia, for example, drinking alcohol, smoking, toxicant and physical activity also have effect on the quality of sperm [13-17]. However, the epigenetic signatures which affect asthenozoospermia are not well understood and require to be further investigated.
Currently, increasing genetic factors contributing to male infertility were reported. Alterations were observed between fertile men and asthenozoospermia patients' transcriptomes, including pathways and genes which were associated with this disease. SEMG1 mRNA and protein expression were upregulated in asthenozoospermia patients compared with that in normozoospermia group [18]. CABYR and ROPN1 mRNA were significantly downregulated in asthenozoospermia patients' samples and a positive correlation was confirmed between the expression of the two genes, indicating that the co-expression of CABYR and ROPN1 was a prerequisite for normal sperm motility and flagellar function [19]. In addition, miRNAs also have dysregulated expressions in testicular biopsy samples and sperms of infertile men. The expression of miR-34b in the control group was significantly higher than that in the asthenozoospermia group, demonstrating that miR-34b might serve as a novel biomarker of male subfertility [20]. The downregulated expression of miR-525 and upregulated expression of miR-151a were also associated with asthenozoospermia and male infertility [18, 21].
In this study, we explored the differential expressed miRNAs (DEMs) and differentially expressed genes (DEGs) in asthenozoospermia patients through reanalyzing the data from published studies and a GEO dataset. Then, we conducted pathway and gene ontology analysis of DEGs using DAVID. The protein-protein interaction (PPI) network was constructed by STRING and DEMs' target genes were predicted by TargetScan. Our results proposed a novel insight into the related biological characteristics and molecular pathways of the DEMs and DEGs in asthenozoospermia. A miRNA-mRNA network further exhibited the potential mechanisms linked to asthenozoospermia.
Methods
Literature search of DEMs
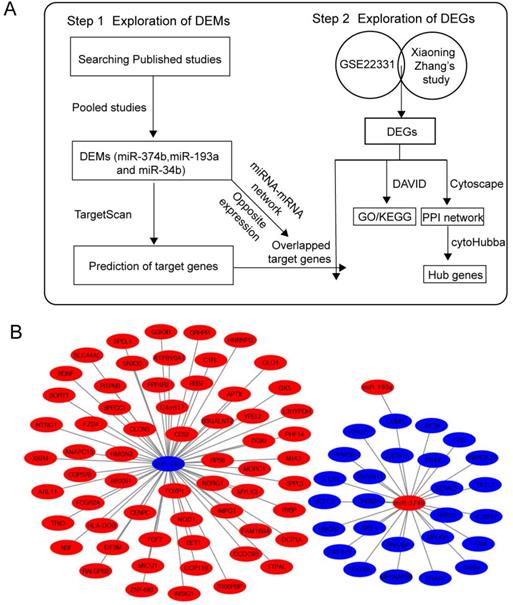
Firstly, we searched GEO database for published miRNA profiling data of ashenozoospermia, however, there was no available result. Then, we conducted a systematic search in PubMed and Web of Science and found 4 studies, which contained miRNA profiling results in ashenozoospermia [22-25]. All of samples used in 4 studies were semen samples and all of them used normal fertile individuals as controls. All studies identified the alteration trends of miRNAs (upregulated or downregulated). In order to investigate the involvement of crucial miRNAs in the modulation of ashenozoospermia, we pooled these 4 studies, and the overlapped miRNAs were presented with Venn diagram, using an online tool, Draw Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/). Next, we chose the miRNAs with consistent expression alteration trends in studies as DEMs in following analysis.
Data processing of DEGs
Xiaoning Zhang's study investigated the expression profiles of long noncoding RNA (lncRNA) and mRNA in mammalian sperm. mRNA between the normozoospermic and asthenozoospermic groups with q-value (FDR) ≤ 0.001 and fold-change (FC) ≥ 2 (|log2FC| > 1) was considered significantly changed, and we reanalyzed the changed mRNA in our following study [26]. Next, we searched GEO database for the available mRNA expression profile data using keywords “ashenozoospermia” and “Homo sapiens”, and the GSE22331 dataset was downloaded. GSE22331 had 2 groups from normozoospermic and asthenozoospermic men. Each group was pooled by 30 sperm samples to obtain enough total RNA. Then, we screened significantly changed mRNA between normozoospermia and asthenozoospermia groups based on FC ≥ 2. Since there was only 1 pooled sample in each group, the FDR could not be calculated. Next, we obtained the intersected elements of significantly changed mRNA between GSE22331 dataset and Xiaoning Zhang's study which were used as DEGs in following analysis.
KEGG and GO enrichment analysis of DEGs
We utilized DAVID (https://david.ncifcrf.gov/), an online biological information database, to visualize the DEGs enrichment of biological processes (BP), cellular components (CC), molecular function (MF) and biological pathways (P value < 0.05). The results of gene ontology and pathway analysis of DEGs were showed with histograms.
Sources leading to asthenozoospermia.
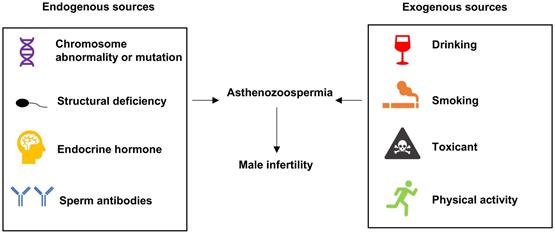
Explore the DEMs from 4 publications. (A) The Venn diagram showed the overlapped genes of 4 publications. (B) miR-374b, miR-193a and miR-34b had consistent expression alteration trends in studies and they were identified as DEMs in following analysis. DEMs, differentially expressed miRNAs.
Protein-protein interaction (PPI) network and module analysis
We utilized an online common software, STRING database (https://string-db.org), to build PPI network of DEGs. Then Cytoscape (www.cytoscape.org), a public source bioinformatics software platform, was applied to visualize and analyze the molecular interaction networks. The plugin, Molecular Complex Detection (MCODE), was applied to identify the most dense and significant module in PPI network based on degree cut-off = 2, node score cut-off = 0.2, max depth = 100 and k-score = 2. Another plugin, cytoHubba, was applied for screening out hub genes based on degrees. We chose the top 25 genes as hub genes and ClueGO and CluePedia plugins were performed to visualize biologic processes and pathways of hub genes.
Construction of miRNA-mRNA network
The putative target genes of DEMs were predicted by an online database, TargetScan (http://www.targetscan.org/mamm_31/), which was commonly used for miRNA target prediction. We obtained the intersection of predicted target genes and DEGs. Besides, the intersected genes of each miRNA-mRNA network should oppositely express with DEMs. The regulatory network of miRNA-mRNA was constructed by Cytoscape.
Hub gene expression in GEPIA
GEPIA (Gene Expression Profiling Interactive Analysis) was an online website which analyzed RNA sequencing expression data of tumors and normal samples from the TCGA and the GTEx projects (http://gepia.cancer-pku.cn). In our study, the expressions of hub genes in testicular cancer patients and controls were obtained from GEPIA.
Results
DEMs in asthenozoospermia
We have searched 146 differentially expression miRNAs from 4 previous studies (SI Table S1-4). Information about studies was shown in Table 1. By intersecting these 146 miRNAs of 4 studies (Figure 2A), 12 overlapped miRNAs that might play critical roles in ashenozoospermia were selected. Among these 12 miRNAs, miR-374b, miR-193a and miR-34b were finally identified as DEMs because of their consistent expression trends in above studies (Figure 2B).
DEGs in asthenozoospermia
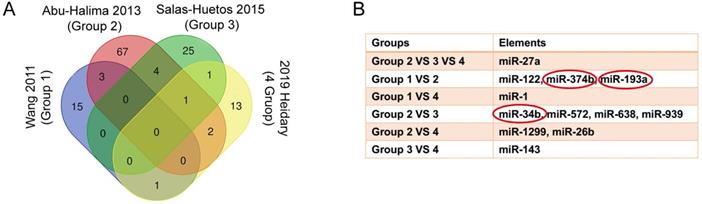
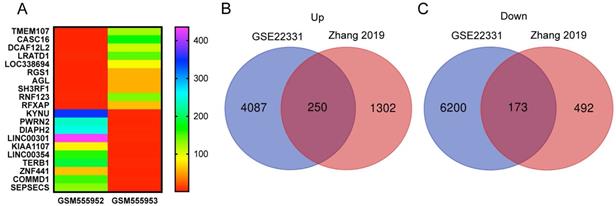
We reanalyzed a publicly available dataset GSE22331, comparing controls to asthenozoospermia, and identified 4337 upregulated and 6373 downregulated genes with the criteria of FC ≥ 2 (SI Table S5-6). The heat map showed the top 10 upregulated and the top 10 downregulated genes in GSE22331 (Figure 3A). Next, we reused the differentially expressed mRNA profile from Xiaoning Zhang's study in 2019 (data was downloaded from Supplementary data Table SIII). Changed upregulation and downregulation genes between asthenozoospermia group and the normal control group with FDR ≤ 0.001 and FC ≥ 2 were considered significant (SI Table S7-8). Then, Venn diagram software was performed to identify DEGs between GSE22331 dataset and Xiaoning Zhang's study. A total of 423 DEGs were detected, including 250 upregulated genes and 173 downregulated genes (Figure 3B-C).
Gene ontology and pathway enrichment analysis
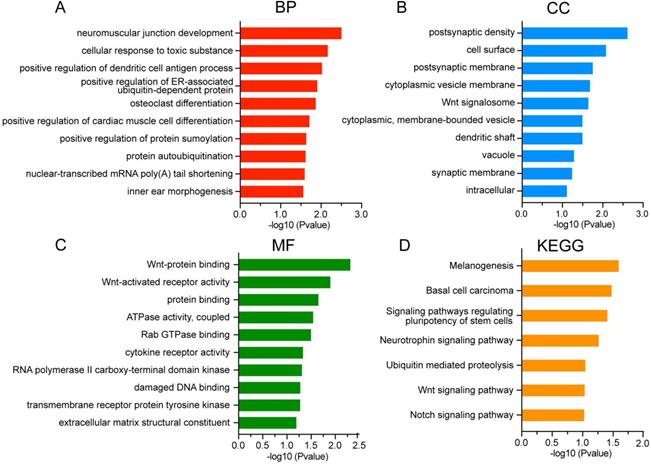
To further characterize the biological classifications of DEGs, GO terms and KEGG pathways enrichment analysis were performed by DAVID. GO analysis revealed that changes in BP of DEGs were mainly enriched in neuromuscular junction development, cellular response to toxic substance, and positive regulation of dendritic cell antigen process (Figure 4A). Changes in CC of DEGs were significantly enriched in postsynaptic density, cell surface and postsynaptic membrane (Figure 4B). Changes in MF of DEGs were significantly enriched in Wnt-protein binding, Wnt-activated receptor activity and protein binding (Figure 4C). The DEGs were significantly enriched in 7 KEGG pathways including signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway and Notch signaling pathway (Figure 4D).
Explore the DEGs between GSE22331 and Xiaoning Zhang's study. (A) Heatmap visualization of the top 10 significantly upregulated and the top 10 significantly downregulated genes in GSE22331. (B-C) DEGs were selected between GSE22331 and Xiaoning Zhang's study. The 2 datasets showed the overlaps of 250 upregulated DEGs and 173 downregulated DEGs. DEGs: differentially expressed genes.
GO and KEGG analysis of DEGs in asthenozoospermia. (A-C) GO analysis of DEGs. (D) KEGG analysis of DEGs. DEGs, differentially expressed genes; BP, biological processes; CC, cellular components; MF, molecular functions.
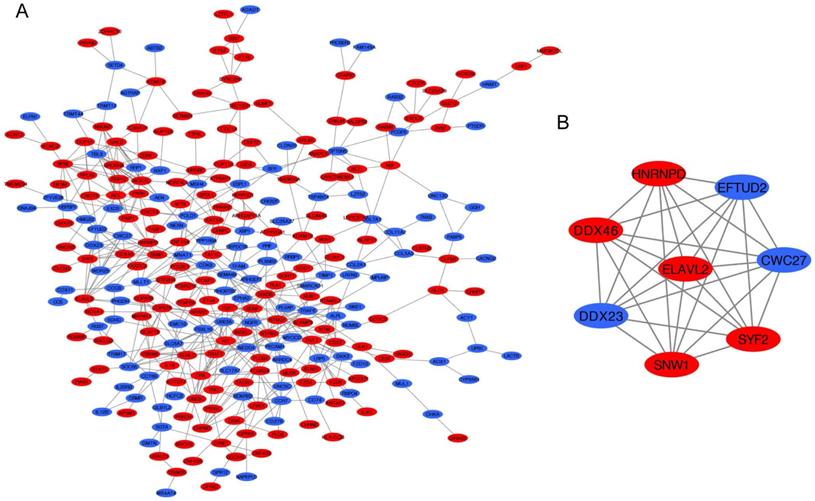
PPI network analysis. (A) Construction of the PPI network. (B) The most significant module was selected from PPI network using the MCODE. Upregulated DEGs were marked in red nodes, and downregulated DEGs were marked in blue nodes. PPI, protein-protein interaction; DEGs: differentially expressed genes.
Information of 4 studies
| Study | Samples | Inclusion criteria of miRNAs |
|---|---|---|
| Wang 2011 | A = a pool of 58 samples; N = a pool of 100 samples | Copy numbers > 50 and FC > 2 |
| Abu-Halima 2013 | A = 9; N = 9 | FC > 2 and P value < 0.05 |
| Salas-Huetos 2015 | A = 10; N = 10 | Significant difference of the mean normCt value between two groups (P value < 0.05) |
| Heidary 2019 | A = 10; N = 10 | FC > 2 and P value < 0.05 |
A: asthenozoospermia; N: normozoospermia; FC: fold change; normCt: normalized threshold cycle.
PPI network of DEGs and module analysis
The PPI network of DEGs was constructed by STRING database including 276 nodes and 550 edges (Figure 5A). Furthermore, the most significant module was established using MCODE which comprised 5 upregulated genes and 3 downregulated genes (Figure 5B).
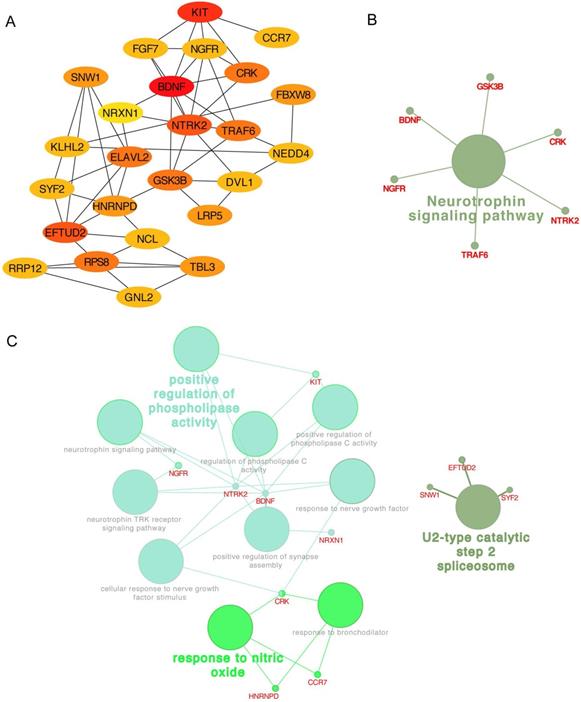
In addition, the top 25 genes filtered by connectivity degree in PPI network were identified as hub genes using cytoHubba (Figure 6A), and the most significant gene was BDNF with connectivity degree of 8, followed by NTRK2, HNRNPD, EFTUD2, GSK3B, NGFR, ELAVL2, RPS8, KIT, TRAF6, DVL1, FGF7, SNW1, SYF2, NCL, CRK, NEDD4, TBL3, RRP12, GNL2, NRXN1, KLHL2, LRP5, FBXW8, and CCR7. The pathway enrichment and biological process analysis of hub genes were shown in Figure 6B-C. Neurotrophin signaling pathway was in the center position associated with multiple hub genes.
Construction of miRNA-mRNA network
Then, we explored the target genes of 3 DEMs using TargetScan database and we obtained the intersection elements between predicted target genes and DGEs. Since the expression of target gene is opposite to its upstream miRNA, we selected the overlapped genes with opposite expressions to their upstream DEMs (Figure 7A). The network of the selected target genes and 3 DEMs was shown in Figure 7B using Cystoscape. In this network, miR-193a and miR-374b both negatively regulated OSMR.
Expression in testicular cancer
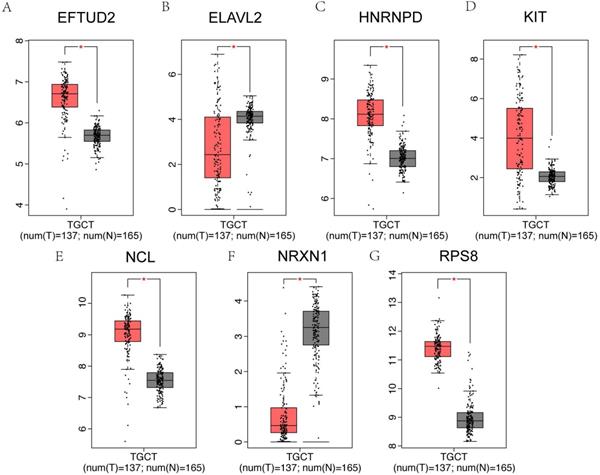
It has been reported that patients with testicular cancer might have subnormal sperm motility [27], and we further investigated the alterations of hub genes in testicular cancer patients and normal controls. Among 25 hub genes, EFTUD2, HNRNPD, KIT, NCL and RPS8 were significantly upregulated in testicular cancer patients, however, ELAVL2 and NRXN1 were significantly downregulated using GEPIA online database (Figure 8A-G).
Hub genes analysis. (A) The 25 most important hub genes selected by connectivity degree were screened out using the cytoHubba. The color of node from yellow to red represented the connectivity degree from low to high. (B) The KEGG analysis of hub genes was performed using the ClueGO and the CluePedia. Different color of node denoted different functional annotation of ontology. (C) The GO analysis of hub genes was performed using the ClueGO and the CluePedia. Different color of node denoted different functional annotation of ontology.
Discussion
Male infertility is a complicated multifactorial pathological condition affecting around 7% male population [28]. Particularly, sperm motility is a crucial factor for fertilization, and over 80% of male infertility is induced by sperm motility impairment [8]. Sperm functional or structural defects, deleterious effect of seminal plasma or a combination of these may impair sperm motility [29]. Asthenozoospermia, also known as asthenospermia, is a common condition characterized by low sperm motility [30]. Interestingly, recent studies have reported that sperm RNAs, including a wide variety of mRNA, miRNA and lncRNA were emerging as critical roles regulating asthenozoospermia [31]. Evidence has revealed certain associations between asthenozoospermia and specific variations of sperm transcriptome profile, and also identified some RNA molecules were differentially expressed in asthenozoospermia patients. Our study aimed to investigate significant dysregulated miRNAs and genes in asthenozoospermia and explore the potential regulatory network in asthenozoospermia pathological condition.
First, we identified 3 overlapped DEMs including miR-374b, miR-193a and miR-34b by screening out previous studies. miR-374b seemed to be one of the best normalizing miRNA candidates associated with spermatogenesis and embryogenesis, and it also exhibited a particular behavior and a stable expression in human fertile individuals [32]. In addition, decreased miR-374b expression could act as a first indication of increased sperm DNA fragmentation index, indicating that miR-374b would potentially become a diagnostic biomarker of idiopathic infertile males [33]. miR-193a was upregulated in sterile triploid fish which might take part in sperm activity and testicular development by targeting functional genes [34]. The expression of miR-34b in asthenozoospermia group was significantly lower than control group [20]. Besides, the frequency of methylation of the promoter region of miR-34b in infertile men was higher than fertile men, and the highest frequency of methylation was observed in asthenoteratospermia patients [35]. These 3 DEMs were proven to be associated spermatogenesis and they were promising for severing as novel biomarkers that could enhance the diagnosis of male infertility. We could further explore the epigenetic regulatory mechanisms of miR-374b, miR-193a and miR-34b in asthenozoospermia.
We next identified 423 DEGs between GSE22331 dataset and Xiaoning Zhang's study. Among these DGEs, 250 genes were upregulated, and 173 genes were downregulated in asthenozoospermia. We observed that DGEs were enriched in development-related pathways, such as signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway and Notch signaling pathway. Studies have reported the germ cell lineage originated in the early stage of development and underwent a series of complicate developmental processes that culminate in the generation of the fully matured gametes, the oocytes and the spermatozoa. Human gametogenesis might be reconstituted from pluripotent stem cells, which would facilitate our understanding of germ cell development and fertilization [36]. Another study has indicated that post-transcriptional Wnt signaling could affect spermatozoa through GSK3 through inhibiting protein phosphatase 1 to initiate sperm motility [37]. The identification of DEGs and DEMs in asthenozoospermia could provide an insight into the molecular mechanisms driving spermatogenesis and open the possibility of new therapeutic targets to repair impaired sperm physiological function and quality.
miRNA-mRNA network. (A) Flow chart of network construction process. (B) Network of DEMs and target DEGs. DEMs, differentially expressed miRNAs; DEGs: differentially expressed genes.
Hub genes expression in testicular cancer. (A-G) The 7 genes of 25 hub genes exhibited significantly different expressions in testicular cancer. EFTUD2, HNRNPD, KIT, NCL and RPS8 were upregulated, ELAVL2 and NRXN1 were downregulated in testicular cancer patients.
Further investigation of PPI network, BDNF, NTRK2, HNRNPD, EFTUD2, GSK3B, NGFR, ELAVL2, RPS8, KIT, TRAF6, DVL1, FGF7, SNW1, SYF2, NCL, CRK, NEDD4, TBL3, RRP12, GNL2, NRXN1, KLHL2, LRP5, FBXW8, and CCR7 were identified as hub genes. The most significant gene was BDNF, a member of the nerve growth factor family. BDNF was detected in the head, neck, and tail of human spermatozoa [38], and exogenous BDNF at 0.133 nM could significantly influence viability and motility of human sperm [39]. Interestingly, another neurotrophic factors, NGF, has been reported to rescue Sertoli cell viability [40]. NGFR, a low affinity receptor which can bind to NGF, exerted a role in the processes of rat Leydig cell postnatal differentiation and in the regulation of functional activities [41]. The expression of miR-4485 was significantly downregulated in the asthenozoospermia patients compared to controls, and KIT, which acted as its target gene, was related to male infertility by bioinformatic analysis [42]. A recent study has uncovered a decrease in abundancy of CCR7 receptor in the high-fertile boars [43]. In particular, these hub genes were enriched in the neurotrophin signaling pathway, indicating that they were critical for differentiation of nerve cells as well as playing critical roles in the development of reproductive system and the maintenance of spermatozoa normal function. Many aspects of fertility depended on intact neurologic function. Therefore, these similar results provided a comprehensive overview that nerve-related genes might also impact male fertilization, and their relationships required to be investigated further. We also constructed the miRNA-mRNA network, and all of these central interactions suggested us new clues to explore underlying mechanisms between miRNA and its target mRNA in asthenozoospermia.
Testicular cancer is one of the most common cancers in men with major semen parameter disturbances [44, 45]. We further tested whether these putative hub genes in asthenozoospermia could extensively affect testicular cancer. We compared the mRNA expression of 25 hub genes between testicular cancer patients and normal controls by GEPIA dataset. As expected, EFTUD2, ELAVL2, HNRNPD, KIT, NCL, NRXN1 and RPS8 were significantly varied in testicular cancer patients.
In our study, the promising miRNAs and mRNA in asthenozoospermia were filtered. They might be dysregulated and exhibited diagnostic values in asthenozoospermia. Additionally, we constructed the miRNA-mRNA network which revealed the underlying mechanism in asthenozoospermia. The development of therapies through direct targeting this network might contribute to the effective treatment of asthenozoospermia. Overall, our findings provided several clues to future diagnosis and clinical treatment of asthenozoospermia.
There were still certain limitations in our current study. There was no available data by searching the GEO database for published microarray results of miRNAs in asthenozoospermia. Therefore, we reused microarray assays data of miRNAs in asthenozoospermia from 4 published studies by applying a systematic search of PubMed and Web of Science. However, these search results might be incomprehensive, and the inclusion criteria of GEMs were different. Additionally, DGEs in GSE22331 only based on FC ≥ 2 without FDR, since there was only one pooled sample in each group. Hence, the repeatability of the results might not be reliable. The findings of the miRNA-mRNA network did not reveal a high degree of several DEMs, because of our strict including criteria of miRNA and mRNA and limited miRNA target prediction database.
Conclusion
In summary, we were the first to pool published studies and a GEO dataset to identify potential DEMs and DEGs that were involved in asthenozoospermia. Importantly, we also clearly demonstrated the relationship between DEMs and DEGs and suggested regulatory pathways in asthenozoospermia including signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway and Notch signaling pathway. We provided key information to profoundly understand the pathological process of asthenozoospermia and presented theoretical basis to discover novel therapeutic interventions. Further studies in vivo and in vitro are required to confirm these intriguing mechanistic possibilities.
Abbreviations
miRNA: microRNA; lncRNA: long noncoding RNA; DEMs: differentially expressed miRNAs; DEGs: differentially expressed genes; BP: biological processes; CC: cellular components; MF: molecular functions; PPI: protein-protein interaction.
Supplementary Material
Supplementary tables.
Acknowledgements
We would like to acknowledge the excellent technical assistance of Yejinpeng Wang.
Funding
This study was funded by Zhongnan Hospital of Wuhan University, Excellent doctor Fund Project (grant number: ZNYB2020003). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
Literature research results are available from the authors upon reasonable request.
Author contributions
LM.L. and S.C. conceived and designed the study, LM.L. performed the analysis procedures, LM.L. and S.C. analyzed the results, LM.L. and S.C. contributed analysis tools, LM.L. contributed to the writing of the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dutta UR, Suttur MS, Venugopal VS, Posanapally LP, Gopalasetty S, Talwar S, Anand S, Billapati S, Jesudasan RA, Dalal A. Cytogenetic and molecular study of 370 infertile men in South India highlighting the importance of copy number variations by multiplex ligation-dependent probe amplification. Andrologia 2020:e13761.
2. Luo G, Hou M, Wang B, Liu Z, Liu W, Han T, Zhang D, Zhou X, Jia W, Tan Y. et al. Tsga10 is essential for arrangement of mitochondrial sheath and male fertility in mice. Andrology. 2020
3. Manfrevola F, Chioccarelli T, Cobellis G, Fasano S, Ferraro B, Sellitto C, Marella G, Pierantoni R, Chianese R. CircRNA Role and circRNA-Dependent Network (ceRNET) in Asthenozoospermia. Front Endocrinol (Lausanne). 2020;11:395
4. Petousis S, Prapas Y, Papatheodorou A, Margioula-Siarkou C, Papatzikas G, Panagiotidis Y, Karkanaki A, Ravanos K, Prapas N. Fluorescence in situ hybridisation sperm examination is significantly impaired in all categories of male infertility. Andrologia. 2018 50
5. Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, Bidart M, Yassine S, Pierre V, Delaroche J, Hennebicq S. et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95-104
6. Coutton C, Vargas AS, Amiri-Yekta A, Kherraf ZE, Ben Mustapha SF, Le Tanno P, Wambergue-Legrand C, Karaouzene T, Martinez G, Crouzy S. et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 2018;9:686
7. Dong FN, Amiri-Yekta A, Martinez G, Saut A, Tek J, Stouvenel L, Lores P, Karaouzene T, Thierry-Mieg N, Satre V. et al. Absence of CFAP69 Causes Male Infertility due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am J Hum Genet. 2018;102:636-648
8. Shen Y, Zhang F, Li F, Jiang X, Yang Y, Li X, Li W, Wang X, Cheng J, Liu M. et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat Commun. 2019;10:433
9. Homonnai ZT, Peled M, Paz GF. Changes in semen quality and fertility in response to endocrine treatment of subfertile men. Gynecol Obstet Invest. 1978;9:244-255
10. Tourmente M, Villar-Moya P, Rial E, Roldan ER. Differences in ATP Generation Via Glycolysis and Oxidative Phosphorylation and Relationships with Sperm Motility in Mouse Species. J Biol Chem. 2015;290:20613-20626
11. Lores P, Dacheux D, Kherraf ZE, Nsota Mbango JF, Coutton C, Stouvenel L, Ialy-Radio C, Amiri-Yekta A, Whitfield M, Schmitt A. et al. Mutations in TTC29, Encoding an Evolutionarily Conserved Axonemal Protein, Result in Asthenozoospermia and Male Infertility. Am J Hum Genet. 2019;105:1148-1167
12. Shibahara H, Shiraishi Y, Suzuki M. Diagnosis and treatment of immunologically infertile males with antisperm antibodies. Reprod Med Biol. 2005;4:133-141
13. Eslamian G, Amirjannati N, Rashidkhani B, Sadeghi MR, Baghestani AR, Hekmatdoost A. Adherence to the Western Pattern Is Potentially an Unfavorable Indicator of Asthenozoospermia Risk: A Case-Control Study. J Am Coll Nutr. 2016;35:50-58
14. Lopez Teijon M, Garcia F, Serra O, Moragas M, Rabanal A, Olivares R, Alvarez JG. Semen quality in a population of volunteers from the province of Barcelona. Reprod Biomed Online. 2007;15:434-444
15. Gaur DS, Talekar MS, Pathak VP. Alcohol intake and cigarette smoking: impact of two major lifestyle factors on male fertility. Indian J Pathol Microbiol. 2010;53:35-40
16. Littarru GP, Tiano L. Clinical aspects of coenzyme Q10: an update. Nutrition. 2010;26:250-254
17. Daoud S, Sellami A, Bouassida M, Kebaili S, Ammar Keskes L, Rebai T, Chakroun Feki N. Routine assessment of occupational exposure and its relation to semen quality in infertile men: a cross-sectional study. Turk J Med Sci. 2017;47:902-907
18. Zhou QZ, Guo XB, Zhang WS, Zhou JH, Yang C, Bian J, Chen MK, Guo WB, Wang P, Qi T. et al. Expressions of miR-525-3p and its target gene SEMG1 in the spermatozoa of patients with asthenozoospermia. Andrology. 2019;7:220-227
19. Pelloni M, Paoli D, Majoli M, Pallotti F, Carlini T, Lenzi A, Lombardo F. Molecular study of human sperm RNA: Ropporin and CABYR in asthenozoospermia. J Endocrinol Invest. 2018;41:781-787
20. Eikmans M, J DHA, Blijleven L, Meuleman T, van Beelen E, van der Hoorn MP, Claas FHJ. Optimization of microRNA Acquirement from Seminal Plasma and Identification of Diminished Seminal microRNA-34b as Indicator of Low Semen Concentration. Int J Mol Sci. 2020 21
21. Zhou R, Wang R, Qin Y, Ji J, Xu M, Wu W, Chen M, Wu D, Song L, Shen H. et al. Mitochondria-related miR-151a-5p reduces cellular ATP production by targeting CYTB in asthenozoospermia. Sci Rep. 2015;5:17743
22. Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, Li L, Wang J, Li X, Shao Y. et al. Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem. 2011;57:1722-1731
23. Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, Backes C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril. 2013;99:1249-1255 e1216
24. Salas-Huetos A, Blanco J, Vidal F, Godo A, Grossmann M, Pons MC, S FF, Garrido N, Anton E. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil Steril. 2015;104:591-601
25. Heidary Z, Zaki-Dizaji M, Saliminejad K, Khorram Khorshid HR. MicroRNA profiling in spermatozoa of men with unexplained asthenozoospermia. Andrologia. 2019;51:e13284
26. Zhang X, Zhang P, Song D, Xiong S, Zhang H, Fu J, Gao F, Chen H, Zeng X. Expression profiles and characteristics of human lncRNA in normal and asthenozoospermia spermdagger. Biol Reprod. 2019;100:982-993
27. Hansen PV, Trykker H, Andersen J, Helkjaer PE. Germ cell function and hormonal status in patients with testicular cancer. Cancer. 1989;64:956-961
28. Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15:369-384
29. Curi SM, Ariagno JI, Chenlo PH, Mendeluk GR, Pugliese MN, Sardi Segovia LM, Repetto HE, Blanco AM. Asthenozoospermia: analysis of a large population. Arch Androl. 2003;49:343-349
30. Askari M, Kordi-Tamandani DM, Almadani N, McElreavey K, Totonchi M. Identification of a homozygous GFPT2 variant in a family with asthenozoospermia. Gene. 2019;699:16-23
31. Corral-Vazquez C, Salas-Huetos A, Blanco J, Vidal F, Sarrate Z, Anton E. Sperm microRNA pairs: new perspectives in the search for male fertility biomarkers. Fertil Steril. 2019;112:831-841
32. Salas-Huetos A, Blanco J, Vidal F, Mercader JM, Garrido N, Anton E. New insights into the expression profile and function of micro-ribonucleic acid in human spermatozoa. Fertil Steril. 2014;102:213-222 e214
33. Li L, Li H, Tian Y, Hu M, Le F, Wang L, Liu X, Jin F. Differential microRNAs expression in seminal plasma of normospermic patients with different sperm DNA fragmentation indexes. Reprod Toxicol. 2020;94:8-12
34. Tao M, Zhou Y, Li S, Zhong H, Hu H, Yuan L, Luo M, Chen J, Ren L, Luo J. et al. MicroRNA Alternations in the Testes Related to the Sterility of Triploid Fish. Mar Biotechnol (NY). 2018;20:739-749
35. Momeni A, Najafipour R, Hamta A, Jahani S, Moghbelinejad S. Expression and Methylation Pattern of hsa-miR-34 Family in Sperm Samples of Infertile Men. Reprod Sci. 2020;27:301-308
36. Saitou M, Miyauchi H. Gametogenesis from Pluripotent Stem Cells. Cell Stem Cell. 2016;18:721-735
37. Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. Post-transcriptional Wnt Signaling Governs Epididymal Sperm Maturation. Cell. 2015;163:1225-1236
38. Zheng L, Li C, Sun Y, Liu Z, Zhou X. Expression of brain-derived neurotrophic factor in mature spermatozoa from fertile and infertile men. Clin Chim Acta. 2011;412:44-47
39. Safari H, Khanlarkhani N, Sobhani A, Najafi A, Amidi F. Effect of brain-derived neurotrophic factor (BDNF) on sperm quality of normozoospermic men. Hum Fertil (Camb). 2018;21:248-254
40. Muller D, Davidoff MS, Bargheer O, Paust HJ, Pusch W, Koeva Y, Jezek D, Holstein AF, Middendorff R. The expression of neurotrophins and their receptors in the prenatal and adult human testis: evidence for functions in Leydig cells. Histochem Cell Biol. 2006;126:199-211
41. Koeva YA. Immunolocalization of neurotrophic factors and their receptors in the Leydig cells of rat during postnatal development. Folia Med (Plovdiv). 2002;44:27-31
42. Heidary Z, Zaki-Dizaji M, Saliminejad K, Edalatkhah H, Khorram Khorshid HR. MiR-4485-3p expression reduced in spermatozoa of men with idiopathic asthenozoospermia. Andrologia. 2020;52:e13539
43. Alvarez-Rodriguez M, Martinez C, Wright D, Barranco I, Roca J, Rodriguez-Martinez H. The Transcriptome of Pig Spermatozoa, and Its Role in Fertility. Int J Mol Sci. 2020 21
44. Panner Selvam MK, Agarwal A, Pushparaj PN. Altered Molecular Pathways in the Proteome of Cryopreserved Sperm in Testicular Cancer Patients before Treatment. Int J Mol Sci. 2019 20
45. Wang CY, Hong YH, Syu JS, Tsai YC, Liu XY, Chen TY, Su YM, Kuo PL, Lin YM, Teng YN. LRWD1 Regulates Microtubule Nucleation and Proper Cell Cycle Progression in the Human Testicular Embryonic Carcinoma Cells. J Cell Biochem. 2018;119:314-326
Author contact
![]() Corresponding author: Dr. Song Chen, E-mail: song.chenedu.cn, Tel: +86-27-6781-3103.
Corresponding author: Dr. Song Chen, E-mail: song.chenedu.cn, Tel: +86-27-6781-3103.

 Global reach, higher impact
Global reach, higher impact