3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(1):169-175. doi:10.7150/ijms.49906 This issue Cite
Research Paper
The expression and clinical significance of TPM4 in hepatocellular carcinoma
1. Department of Clinical Laboratory Center, The Second Hospital of Lanzhou University, Lanzhou 730000, P.R. China.
2. The First School of Clinical Medicine, Southern Medical University, Guangzhou, China, 510515.
3. Center of Reproductive Medicine, Department of Obstetrics and Gynecology, First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China, 510080.
*These authors contributed equally to this article.
Received 2020-6-24; Accepted 2020-10-7; Published 2021-1-1
Abstract
Hepatocellular carcinoma (HCC) is known as the fifth most common cancer in the world for its poor prognosis. New diagnostic markers and treatments are urgent to discover. To evaluate the protein expression of Tropomyosin4 (TPM4) and investigate its prognostic value in HCC, we collected 110 patients with different degrees of HCC and 10 patients with normal hepatic tissues and performed immunohistochemistry. Western bot was used to evaluate the expression of TPM4 in three HCC cell lines (HepG2, Huh7, SMMC-7721) and normal liver cell line LO2, as well as 7 HCC tissues and 7 normal hepatic tissues. The results of TPM4 staining revealed that TPM4 expression in HCC was higher than that in normal hepatic tissues, which was positive in 51.8% (n=57) and negative in 48.2% (n=53) while in normal hepatic tissues positive staining was in 10% (n=1) and negative staining was in 90% (n=9) (P=0.011). And the expression of TPM4 was related to pT status, grade and stage (P<0.001, P=0.015 and P<0.001, respectively). Western blot results indicated that TPM4 was high expressed in HCC cell line and HCC tissues. In conclusion, we believe that TPM4 can be applied as a diagnostic and prognostic marker to assist the management of HCC.
Keywords: hepatocellular carcinoma, TPM4, biological marker, clinicopathological parameters, diagnosis
Introduction
Liver cancer can be classified as primary and secondary. According to the statistics, liver cancer is the dominating cause of cancer mortality in males of Southern China [1]. It occupies the fifth position in tumor and ranks fourth among cancer subtypes, and accounts for more than 50% of the world's total in China [1,2]. Primary liver cancer is considered as one of the most aggressive cancers. Its occurrence is a multifactorial and complex progress with poor prognosis. Moreover, many patients are insensitive to the relevant drugs during the treatment. The majority of primary liver cancer is hepatocellular carcinoma (HCC), which is always diagnosed in the advanced stage [3]. From 2007 to 2016, the incidence of liver cancer increased rapidly at the rate of 2% to 3% [4]. The dominating inducement of HCC is the chronic infection based on hepatitis B virus (HBV) and hepatitis C virus (HCV), and the latter was rarely detected [4,5]. No matter what kind of liver cancer it is, operation will be the most appropriate surgical treatment in the early stage and radiofrequency ablation will be used in the last stage. However, chemical treatments are not recommended for its low efficiency [2]. At present, some biomarkers can be applied to analyze the prognosis of HCC, for instance, the significant advantage of albumin-bilirubin grade in HCC and other liver diseases have been widely acknowledged [6]. In spite of this, some biological markers which are able to assist diagnosis need to be discovered. With the advance of modern medicine, the situation could be improved.
Tropomyosin4 (TPM4) belongs to the tropomyosin family, member of which are actin-binding proteins. It helps regulate actomyosin interaction in muscle by extending α-helical coiled-coil homodimer [7]. TPM4 is expressed in blood platelet and has an effect on the volume and number of platelets. It has been confirmed that TPM4 is the independent risk factor of stroke or heart diseases [8], while some specific tumors in human bodies may express abnormal levels of TPM4. Therefore, TPM4 is likely to be used as a marker for several cancers. By changing actin cytoskeleton, TPM4 enhances the migration of tumor cells in lung cancer [9]. High expression of TPM4 in ovarian tumor tissue was first reported in 2004 [10]. Detecting the expression of proteins TPM4, ANXA6, HSP27, PRDX2 and NCF2 may contribute to the pathological and cytological diagnosis and prognosis assessment in squamous cervical cancer [11]. However, there are few cases of diagnosing HCC by testing the content of TPM4. Therefore, it's highly desirable to explore the connection between the expression of TPM4 and HCC, including its TNM classification, so that it may provide reference information for the future medicine.
Materials and Methods
Cell lines and cultures
Human hepatic-derived hepatoma G2 cell (HepG2) was purchased from American Type Culture Collection (ATCC HB-8065, Rockville, MD, USA) and it was cultivated in Dulbecco's Modified Eagle Medium (DMEM, Gibco, USA), supplemented with 10% fetal bovine serum (FBS, Gibco, Life Technologies, Melbourne, Australia). Human HCC cell line Huh7 was saved in laboratory and maintained in DMEM added 10% FBS, 10 mM HEPES, 100 U/ml of penicillin, and 100 μg/ml streptomycin. Human HCC cell line SMMC-7721 was preserved in laboratory and cultured in RMPI 1640 (1640, Gibco, USA) which contained 10% FBS. Human normal cells LO2 was obtained from laboratory conservation, maintained in DMEM, 10% FBS. All cells were maintained in a humidified incubator at 37°C with 5% CO2.
Tissue samples
We have gathered the information of patients with hepatoma from Second affiliated hospital of Lanzhou university between 2015 and 2019. There were 120 pathological sections of hepatic in total, among which there were 110 samples from patients with hepatoma without treatment and the remaining 10 samples were from normal tissues. Other information included age, sex, pathology diagnosis, histology grade, clinical stage and pTNM. The mean age was 49.1 and the rest details were demonstrated in Table 2. The study obeyed the protocol of the Ethic Committee of Second affiliated hospital of Lanzhou university and written informed consents were also obtained.
Western Blot
In the presence of the protease inhibitors, total protein was isolated from HCC cells by Radio-Immunoprecipitation Assay (RIPA) buffer (Beyo-time, China), which contained 1mM PMSF (Beyo-time, China). The protein concentration was then measured using BCA Protein Assay Kit (Beyo-time, China) and normalized using the standard BSA curve. These samples were dissolved in loading buffer and unstructured for 10 mins by boiling. In the end, equivalent amounts of proteins were separated by 10% SDS-PAGE gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, USA) on a standard wet-transfer equipment (Trans-blot SD, Bio-Rad, USA). The membranes were then blocked in Tris-Buffer Saline Tween 20 (1×TBST) containing 5% non-fat milk at room temperature. After 2 hours, membranes would be incubated with TPM4 (rabbit polyclonal, 1:1000, Proteintech, China) primary antibody and GAPDH primary antibody (Mouse monoclonal, 1:2000, Proteintech, China) at 4°C for a night. After three times washing with 1×TBST for 10 mins, we again incubated the membranes with secondary antibodies labeled with horseradish peroxidase-(HRP) (Go-at anti-Rabbit IgG, 1:5000, MultiSciences, China; Goat anti-Mouse IgG, 1:5000, MultiSciences, China) at room temperature for 1.5 hours. Afterwards, the membranes were then washed with 1×TBST for 3 times, followed by the enhanced chemiluminescence reagents (BeyoECL Plus Kit, Beyotime, China), data scanning and protein bands were recorded by Tanon-5200 (Tanon, Shanghai, China). The optical density of protein bands was quantitatively determined.
Tissue microarrays (TMA) construction and immunohistochemistry
The construction of TMA covered 110 pathological sections of HCC and 10 normal samples according to the standard method [12]. Continuous 5 μm portion of pathological section was removed in TMA block mass for immunohistochemical analysis, followed by staining basing on the standard procedures. Subsequently, the slices were dewaxed in xylene and rehydrated in fractional ethanol. Activity of endogenous catalase would be inhibited by 0.3% H2O2. After antigen recovery, the citrate buffer was heated with sodium for 20 mins in a self-cleavage tank for 20 mins. Sections were then incubated and 1×phosphate-buffered saline (PBS) which contained 5% normal goat serum for blocking was used for 30 mins. Sections were cultured at 4°C with primary antibody from TPM4 (rabbit polyclonal, Proteintech, China) for a night, following being washed for 3 times by 1×PBS and cultivated with HRP conjugated-secondary antibodies at room temperature. 1×PBS would be applied for additional wash after 30 mins. Under the use of hematoxylin and dehydrated before mounting, these sections were stained. Immunostaining was performed by DAB Horseradish Peroxidase Color Development Kit (Beyotime, China). Phosphate buffered saline took the place of anti-TPM4 antibody to act as a negative control.
IHC Evaluation
There were two authors who accounted for the immunostaining results of TPM4 on a semi-quantitative scale of staining intensity and percentage. They did not possess the prior knowledge about the characteristics of the patients. Cytoplasmic staining was measured as the positive immunostaining. Percentage fractions were 5 increments (0, 5%, 10%, ..., 100%). Only when the differences were harmonized did the two authors stop re-estimating.
Selection of cut-off score
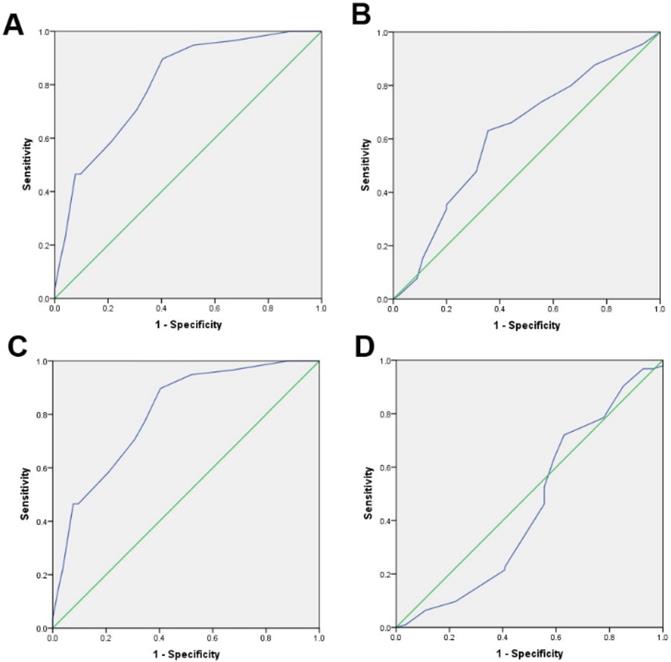
The optimal cut-off score was obtained through Receiver operating characteristic (ROC) curve analysis, and the 0, 1-criterion was created by referring to the incremental expression of TPM4 [13]. We divided the clinicopathological parameters analyzed by ROC into two groups: histological grade (G0-G2, G3), pT stage (T0-T2, T3-T4), clinical stage (I-II, III-IV). Subsequently, the ROC curves were drawn based on the specificity and sensitivity of each TPM4 score. The point closest to the given point [0.0, 1.0] would be considered as the cut-off score. Therefore, the tumor would be regarded as TPM4 “negative” if the last score had been below the “cut-off score”. Otherwise, the tumor would be regarded as TPM4 “positive”.
Statistical analysis
SPSS 20.0 software (SPSS, Chicago, IL, USA) was used to complete statistical estimations. Data was expressed as mean ± standard deviation with no less than three independent experiments included. Optimal cut-off point of TPM4 positive was obtained by ROC being introduced. Applied T-test was used to search for expression distinction between two groups. The chi-square served as a role to evaluate the connection between TPM4 expression and the clinicopathological parameters of hepatoma. If P values were less than 0.05, a distinction would be made as statistically significant.
Results
TPM4 expression in HCC cells
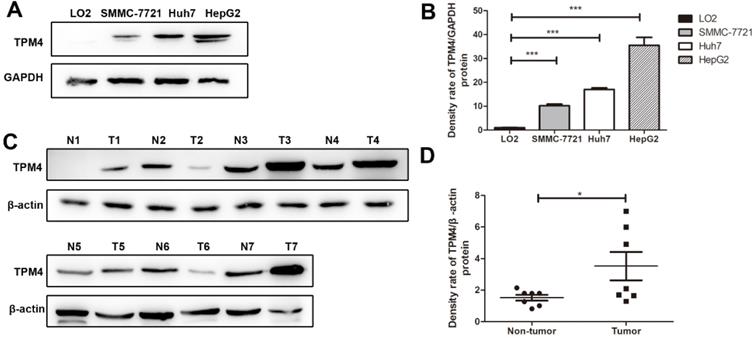
Through western blot, we successfully examined the expression of TPM4 in three human HCC cell lines, HepG2, Huh7, SMMC-7721 and human normal immortalized liver cell line, LO2. The results showed that TPM4 expression in HCC cell lines were distinctly higher than that in human normal liver cell line (Figure 1).
Western blot results of TPM4 expression in liver cell lines and tissues. A. Western blot indicated significant up-regulation in hepatocellular carcinoma cells (SMMC-7721, Huh7, HepG2) compared to in the human normal immortalized cell lines (LO2). GAPDH was used as control. C. Western blot results of TPM4 in 7 hepatocellular carcinoma tissues(T1-T7) and 7 normal hepatocellular tissues(N1-N7) B.D. Western blot results were measured as optical density value and expressed graphically. TPM4 protein expressions were significantly higher in hepatocellular carcinoma cells and tissues (*P<0.05, **P<0.01, ***P<0.001).
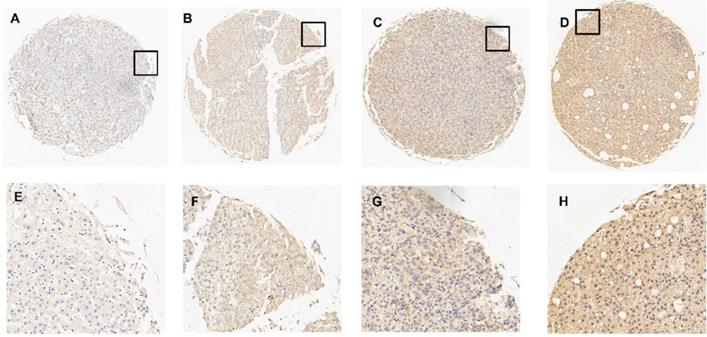
IHC results of the expression of TPM4 in normal hepatic tissues and hepatocellular carcinoma tissues. A. 5% positive expression of TPM4 was detected in hepatic normal tissue (case 117) (10×). B. 40% positive expression of TPM4 was detected in a hepatocellular carcinoma tissue (case 80) (10×). C. 80% positive expression of TPM4 was showed in a hepatocellular carcinoma tissue (case 45) (10×). D. 95% positive expression of TPM4 was showed in hepatocellular carcinoma (case 61) (10×). E-H. Demonstrate the higher magnification (40×) from the area of black box in (A-D), respectively.
TPM4 expression in tissue samples of HCC
Compared with normal hepatic tissues, we evaluated the content of TPM4 in HCC tissues by applying immunohistochemistry (IHC). Immunoreactivity could be detected in the cell membrane of tumor cells. Figure 2 has shown 4 typical samples of different levels of TPM4 IHC staining. The optimal cut-off score was determined by the analysis from Receiver operating Characteristic curve (ROC). Figure 3 showed that the clinical stage and pT status were both chosen for the shortest distance from point (0.0, 1.0). Therefore, 67.5% would be considered as the optimal cut-off score, above which was supposed to be positive and otherwise was supposed to be negative. According to the cut-off score, 51.8% (n=57) remained to be positive and the rest 48.2% (n=53) remained to be negative after TPM4 staining. In normal hepatic tissues with TPM4 staining, 10% (n=1) of normal cases suggested to be positive while 90% (n=9) suggested to be negative. Using Chi-square test for further analysis, we tried to explore the connection between the expression of TPM4 and HCC and normal hepatic tissue which showed that TPM4 expression in HCC was distinctly higher than that in human normal hepatocytes (χ2=6.419 P=0.011) (Table 1).
Correlation between TPM4 and clinicpathological parameters
In addition, we separately analyzed the connection between TPM4 expression and clinicopathological parameters of HCC cell lines. Table 2 indicated that TPM4 positive was related to pT status, histological grade and clinical stage (all P < 0.05). While, there was not enough evidence to prove that sex and age were correlated to TPM4 positive (all P > 0.05).
Relationship of TPM4 expression between hepatic tissues and hepatocellular carcinoma tissues
| TPM4 staining | All cases (%) | Negative expression (%) | Positive expression (%) | P value |
|---|---|---|---|---|
| Hepatocellular carcinoma tissues | 110 | 53(48.2%) | 57(51.8%) | 0.011 |
| Normal hepatic tissues | 10 | 9(90.0%) | 1(10.0%) |
Relationship of TPM4 expression and clinicopathological features in hepatocellular carcinoma
| Variables | TPM4 staining | |||
|---|---|---|---|---|
| Negative (%) | Positive (%) | Total | P valueb | |
| Sex | ||||
| Male | 44 | 42 | 86 | 0.236 |
| Female | 9 | 15 | 24 | |
| Age (year) | ||||
| ≤49.1a | 23 | 27 | 50 | 0.676 |
| >49.1 | 30 | 30 | 60 | |
| pT Status | ||||
| T1 | 1 (100.0%) | 0 (0.0%) | 1 | 0.000 |
| T2 | 35 (68.6%) | 16 (31.4%) | 51 | |
| T3 | 17 (30.9%) | 38 (69.1%) | 55 | |
| T4 | 0 (0.0%) | 3 (100.0%) | 3 | |
| Grade | ||||
| 1 | 0 (0.0%) | 2 (100.0%) | 2 | 0.015 |
| 2 | 24 (38.1%) | 39 (61.9%) | 63 | |
| 3 | 21 (60.0%) | 14 (40.0%) | 35 | |
| 4 | 8 (80.0%) | 2 (20.0%) | 10 | |
| Stage | ||||
| I | 1 (100.0%) | 0 (0.0%) | 1 | 0.000 |
| II | 35 (68.6%) | 16 (31.4%) | 51 | |
| III | 16 (29.1%) | 39 (70.9%) | 55 | |
| IV | 1 (33.3%) | 2 (66.7%) | 3 | |
a: Mean age;
b: P values are from Chi-square test.
Receiver Operating Characteristic (ROC) was used to select the optimal cut-off score for positive expression of TPM4: A. pT stage. B. Histological grade. C. Clinical stage. D. Sex. The cut-off score is 67.5%.
Discussion
HCC is acknowledged as a serious kind of cancer, which is recognized around the world. Currently, major breakthroughs have been made not only in the treatment of HCC but also in the prediction of tumor recurrence after clinical treatment [14]. TPM4 is involved in the regulation of actin binding proteins, and TPM4 was reported that it related the progress in several cancers. It was reported that TPM4 was highly detected in the serum of ovarian patients. And TPM4 may act as an oncogene and a potential early diagnosis marker in breast cancer, lung cancer and esophageal squamous cell carcinoma [15]. However, the relationship of TPM4 and HCC is unknown.
To reveal the expression pattern of TPM4 in cells from HCC, our experiment was conducted on three HCC cells lines (HepG2, Huh7, SMMC-7721) and one normal hepatic immortalized human urothelial cell line (LO2) by applying western blot. The results of the experiments indicated that the content of TPM4 in three HCC cell lines were up-regulated apparently compared with the normal hepatic immortalized human urothelial cell line (P < 0.05). Cell lines are models in vitro and can explain the expression of TPM4 in some degree. We hypothesized that TPM4 was highly expressed in HCC. Futhermore, 7 pairs of HCC tissues and normal hepatic tissues were used to verify the hypothesis. The results indicated that TPM4 was high expressed in HCC (P < 0.05).
However, the role of TPM4 in the diagnosis of HCC is still unclear. To evaluate the clinical application value of TPM4, there were 110 hepatoma tissues and 10 normal hepatic tissues collected and they were constructed TMA stained with TPM4. Subsequently, we obtained the optimal cut-off score by adopting ROC curve. Basing on 4 clinicopathological parameters including histological grade, clinical stage, pT stage and sex, ROC curves were then created. “67.5%” was defined as the optimal cut-off score, staining above which would be regarded as “TPM4” positive. Statistic result represented that expression of TPM4 in HCC tissues were higher than that in normal hepatic tissues. More detailed analysis, higher expression of TPM4 was correlated with histological grade, clinical stage, pT stage but had nothing to do with sex and age. In the study, we found that the expression of TPM4 will increase with the aggravation of histological grade, pT and clinical stage while the content is low in the normal hepatic cells.
Many studies focused on identifying the candidate biomarkers for HCC, such as genetic mutations, gene expression and non-coding RNAs. Alpha-fetoprotein (AFP) is a widely used marker for detecting and monitoring HCC, however, AFP serum levels stay normal in nearly 30% of advanced HCC patients [16]. Other markers have been reported, but most performed an unsatisfactory sensitivity or specificity. There are experiments which can prove that the progression of HCC can be inhibited by raising the expression of three main genes which are cysteine sulfinic acid decarboxylase (CSAD), glutamic-oxaloacetic transaminase 2 (GOT2) and suppressor of cytokine signaling 2 (SOCS2) through METTL14 [17,18,19]. Ribosomal protein S11 (RPS11) overexpression is affiliated with variety of malignancies and tumor recurrence, which can be used as a potential biomarker for predicting the prognosis of patients who have received radical resection of HCC [20]. Besides, studies suggested that p38γ is able to interact with CDKs acting as a type of CDK-like-kinase and thus regulating the cell cycle. And high expression of p38γ can be detected in human HCC and might be considered as a therapeutic target [21]. JNK pathway plays an initial role in maintaining hepatic homeostasis. Numerous diseases are correlated with its dysregulation, including steatosis, fibrosis, cirrhosis and hepatoma [22, 23, 24]. The abnormality or deficiency of JNK will inhibit the proliferation of HCC cells [25, 26, 27]. However, the factors related to the pathogenesis have not been completely identified because of its complicated mechanism, and the roles of TPM4 in HCC need to be further studied.
Finally, a conclusion can be drawn from the experiment that TPM4 is highly expressed in hepatoma cells compared with normal hepatic tissues. It is also affiliated with clinicopathological parameters of HCC, from which we can infer TPM4 can serve as a specific biological marker to diagnose HCC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82060531), Internationally Technological Cooperation Project of Gansu Province (18YF1WA117), Longyuan Youth Creative and Innovative talents Project of Gansu Province (2020RCXM149), Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital (CY2018-MS11), Science and Technology Development Guidance Project of Lanzhou city (2019-ZD-68).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lei H Y, Chang C P. Lectin of Concanavalin A as an anti-hepatoma therapeutic agent[J]. J Biomed Sci. 2009;16:10
2. Wang Q, Zhang P, Li Z. et al. Evaluation of Polymer Nanoformulations in Hepatoma Therapy by Established Rodent Models[J]. Theranostics. 2019;9(5):1426-1452
3. Neuhaus P, Jonas S, Bechstein W O. Hepatoma of the liver-resection or transplantation?[J]. Langenbecks Arch Surg. 2000;385(3):171-178
4. Siegel R L, Miller K D, Jemal A. Cancer statistics, 2020[J]. CA Cancer J Clin. 2020;70(1):7-30
5. Zhang W, Fu T, Guo Z. et al. Serum miR-375 Levels Are Closely Related to Disease Progression from HBV Infection to HBV-Related Hepatocellular Carcinoma[J]. Biomed Res Int. 2020;2020:5819385
6. Feng D, Wang M, Hu J. et al. Prognostic value of the albumin-bilirubin grade in patients with hepatocellular carcinoma and other liver diseases[J]. Ann Transl Med. 2020;8(8):553
7. James J K, Nanda V. Comparative dynamics of tropomyosin in vertebrates and invertebrates[J]. Proteins. 2020;88(2):265-273
8. Kim Y J, Cho A R, Sul H J. et al. The effects of crizotinib in a transgenic Drosophila model expressing the human TPM4-ALK fusion gene or TPM4[J]. Biol Open. 2019;8(7):bio044362
9. Zhao X, Jiang M, Wang Z. TPM4 promotes cell migration by modulating F-actin formation in lung cancer[J]. Onco Targets Ther. 2019;12:4055-4063
10. Dube D K, Dube S, Abbott L. et al. Cloning, Sequencing, and the Expression of the Elusive Sarcomeric TPM4alpha Isoform in Humans[J]. Mol Biol Int. 2016;2016:3105478
11. Lomnytska M I, Becker S, Bodin I. et al. Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during uterine cervix carcinogenesis: diagnostic and prognostic value[J]. Br J Cancer. 2011;104(1):110-119
12. Zhou H, Shi R, Wei M. et al. The expression and clinical significance of HERC4 in breast cancer[J]. Cancer Cell Int. 2013;13(1):113
13. Feng Y, Zhang Q, Fu M. et al. Low expression of PinX1 is associated with malignant behavior in basal-like breast cancer. Oncol Rep. 2017;38(1):109-119
14. Bandopadhyay M, Bharadwaj M. Exosomal miRNAs in hepatitis B virus related liver disease: a new hope for biomarker[J]. Gut Pathog. 2020;12:23
15. Tang HY, Beer LA, Tanyi JL. et al. Speicher DW. Protein isoform-specific validation defines multiple chloride intracellular channel and tropomyosin isoforms as serological biomarkers of ovarian cancer. J Proteomics. 2013;89:165-178
16. Li Z, Li F, Peng Y. et al. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma[J]. Cancer Med. 2020;00:1-13
17. Zhang D M, Luo Y, Yishake D. et al. Prediagnostic dietary intakes of vitamin A and beta-carotene are associated with hepatocellular-carcinoma survival[J]. Food Funct. 2020;11(1):759-767
18. Di Palo A, Siniscalchi C, Mosca N. et al. Proto-oncogene Zbtb7a represses miR-125a-5p transcription in hepatocellular carcinoma cells[J]. Mol Biol Rep. 2020;47(6):4875-4878
19. Cheng X Y, Chen Y S, Zi J G. et al. Dietary habits and Risk of Hepatocellular Carcinoma among Hepatitis B Surface Antigen Carriers: A Prospective Cohort Study in China[J]. J Dig Dis. 2020;21(7):406-415
20. Zhou C, Sun J, Zheng Z. et al. High RPS11 level in hepatocellular carcinoma associates with poor prognosis after curative resection[J]. Ann Transl Med. 2020;8(7):466
21. Tomas-Loba A, Manieri E, Gonzalez-Teran B. et al. p38γ is essential for cell cycle progression and liver tumorigenesis[J]. Nature. 2019;568(7753):557-560
22. Nakagawa H, Maeda S. Molecular mechanisms of liver injury and hepatocarcinogenesis: focusing on the role of stress-activated MAPK[J]. Patholog Res Int. 2012;2012:172894
23. Papa S, Bubici C, Zazzeroni F. et al. Mechanisms of liver disease: cross-talk between the NF-kappaB and JNK pathways[J]. Biol Chem. 2009;390(10):965-976
24. Schwabe R F. Cell death in the liver-all roads lead to JNK[J]. Gastroenterology. 2006;131(1):314-316
25. Tournier C, Hess P, Yang D D. et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway[J]. Science. 2000;288(5467):870-874
26. Sugioka Y, Watanabe T, Inagaki Y. et al. c-Jun NH2-terminal kinase pathway is involved in constitutive matrix metalloproteinase-1 expression in a hepatocellular carcinoma-derived cell line[J]. Int J Cancer. 2004;109(6):867-874
27. Khan S, Zaki H. Crosstalk between NLRP12 and JNK during Hepatocellular Carcinoma[J]. Int J Mol Sci. 2020;21(2):496
Author contact
![]() Corresponding author: Linjing Li, Department of Clinical Laboratory Center, The Second Hospital of Lanzhou University, 82 Cuiying Gate Road, Lanzhou, Gansu, 730000, P.R. China. E-mail: lilinjedu.cn.
Corresponding author: Linjing Li, Department of Clinical Laboratory Center, The Second Hospital of Lanzhou University, 82 Cuiying Gate Road, Lanzhou, Gansu, 730000, P.R. China. E-mail: lilinjedu.cn.

 Global reach, higher impact
Global reach, higher impact