Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(18):2895-2904. doi:10.7150/ijms.51213 This issue Cite
Research Paper
Integrative Analysis of HNF1B mRNA in Human Cancers Based on Data Mining
1. Hepatobiliary and Pancreatic Interventional Treatment Center, Division of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, Zhejiang Province, China
2. Zhejiang Provincial Research Center for Diagnosis and Treatment of Hepatobiliary Diseases, Hangzhou 310003, Zhejiang Province, China
3. Zhejiang Clinical Research Center of Hepatobiliary and Pancreatic Diseases, Hangzhou 310003, Zhejiang Province, China
4. Department of Pharmacy, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
5. Bone Marrow Transplantation Center, Department of Hematology, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
Received 2020-7-28; Accepted 2020-10-7; Published 2020-10-18
Abstract
Cancer incidence is rapidly growing, and cancer is the leading cause of death worldwide in the 21st century. Hepatocyte nuclear factor 1B (HNF1B) is a transcription factor that involves the growth and development of multiple organs. The aim of this study was to explore the significance of HNF1B in human cancer by an integrative analysis of online databases. The UALCAN database, cBio cancer genomics portal, Cancer Regulome tools, Kaplan-Meier plotter and Tumor IMmune Estimation Resource (TIMER) website were used to perform the corresponding analysis. The results showed that HNF1B is dysregulated in various cancers and associated with the differential overall survival of cancer patients. HNF1B showed many mutation forms and high mutation levels in different cancer types. In addition, we found that HNF1B interacted with different genes in multiple aspects. Moreover, HNF1B expression is associated with many immune cell infiltration levels and influences the prognostic prediction of immune cells in some kinds of cancers. In conclusion, HNF1B plays a significant role in cancer and may be a potential target for cancer immunotherapy.
Keywords: HNF1B, cancer, immunotherapy, biomarker
Introduction
Cancer incidence and mortality are rapidly rising worldwide[1]. Moreover, cancer is the leading cause of death and an important barrier to extending life expectancy in the 21st century. Despite the achievements in our understanding of the pathogenesis of cancer and the development of multidisciplinary therapies, the clinical outcomes for patients remain poor[2-4]. Therefore, there is an urgent need to discover potential biomarkers of cancer to predict the overall prognosis and improve the therapeutic management of patients.
Hepatocyte nuclear factor 1B (HNF1B) is located at chromosome 17q12, and it involves the growth and development of multiple organs, including liver, pancreas, kidney, etc[5-7]. During kidney development, HNF1B mainly regulates the growth and development of renal tubules[8]. HNF1B plays an indispensable role in all stages of pancreas development, including pancreatic endocrine cells and duct development[9]. Genes encoding HNF1B transcription factors are prone to various types of mutations, causing the occurrence and progression of various diseases, including diabetes, renal insufficiency, and various malignant tumors[10-12].
In the present study, we evaluated expression and mutations of HNF1B in different types of cancer from The Cancer Genome Atlas (TCGA) database. The correlation between HNF1B expression and overall survival of cancer patients was further validated based on the Kaplan-Meier plotter. Moreover, we explored the association of HNF1B with immune cell infiltration levels and their prognostic values in cancer patients. The results of this study reveal the crucial role of HNF1B in cancer and an underlying mechanism between HNF1B and tumor-immune interactions.
Material & Methods
UALCAN database
The UALCAN database was employed to analyze the expression of HNF1B in tumors and normal tissues. This database includes RNA-seq and clinical information from the TCGA for 31 types of cancer[13].
Mutation analysis
The cBio cancer genomics portal was used to explore mutations and copy-number alterations of HNF1B in the TCGA pan-cancer studies. The cBio Cancer Genomics Portal provides rapid, intuitive, and high-quality access to the interactive analysis of multidimensional cancer genomics datasets[14].
Genome-wide association analysis
The Cancer Regulome tool was applied to explore the correlation between the expression levels of HNF1B and other genes in tumors. Spearman's correlation analysis based on the TCGA database was used to determine the correlation between two genes[15].
Kaplan-Meier plotter
The online Kaplan-Meier plotter database was used to explore the prognostic value of genes in cancer[16]. Kaplan-Meier survival plots were constructed to show the correlation between HNF1B expression and the overall survival of cancer patients. The statistical significance of the correlation was analyzed by the log-rank test.
Immune-associated analysis
The Tumor IMmune Estimation Resource (TIMER) database was applied to estimate the correlations between HNF1B expression and immune cell infiltration levels[17]. The Estimating the Proportion of Immune and Cancer cells (EPIC) algorithm was also adopted in this study[18]. Moreover, we explored the outcome of cancer patients by comprehensively considering immune cell infiltration and HNF1B expression levels.
Results
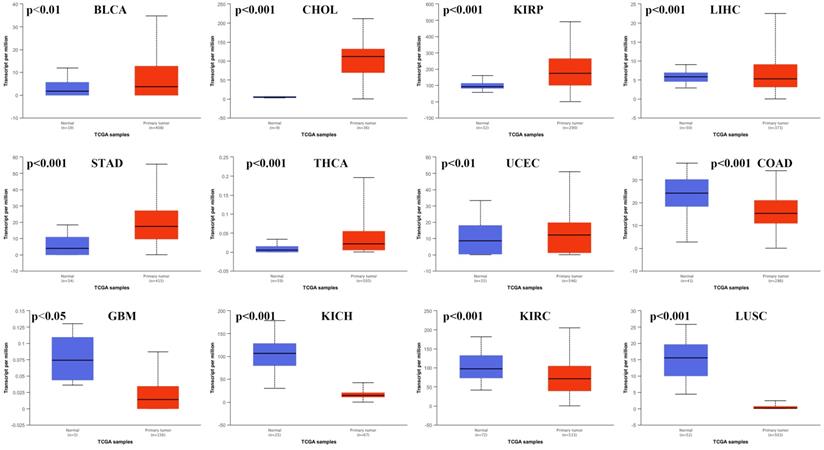
Abnormal expression of HNF1B in cancer
We used the UALCAN database to analyze the expression of HNF1B in different types of cancer. HNF1B levels in cancer tissues and in normal tissues were compared based on data from the TCGA. The results showed that HNF1B expression in various cancers was obviously different from that in normal tissues (Figure 1). The expression of HNF1B was upregulated in bladder urothelial carcinoma (BLCA), cholangiocarcinoma (CHOL), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC), while the expression of HNF1B was downregulated in colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), and lung squamous cell carcinoma (LUSC). However, no significant differences were revealed in breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), or rectum adenocarcinoma (READ) (Supplementary Figure 1).
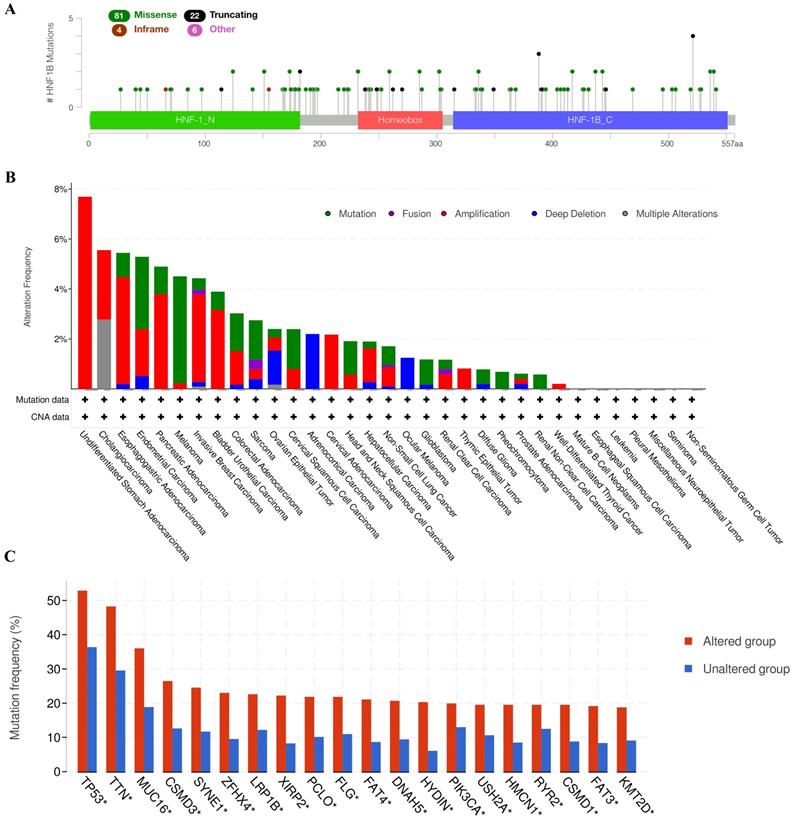
HNF1B mutation in cancer
We applied the cBio cancer genomics portal to explore the mutations of HNF1B in different cancers. As shown in Figure 2A, the mutation types of HNF1B included missense mutations, truncating mutations, in-frame mutations and other mutations. Moreover, HNF1B showed high mutation levels in Esophagogastric Adenocarcinoma, Endometrial Carcinoma, Pancreatic Adenocarcinoma, Melanoma, Invasive Breast Carcinoma, Bladder Urothelial Carcinoma, Colorectal Adenocarcinoma, Sarcoma, Ovarian Epithelial Tumor, Cervical Squamous Cell Carcinoma, Head and Neck Squamous Cell Carcinoma, Glioblastoma, Diffuse Glioma, Pheochromocytoma and Renal Non-Clear Cell Carcinoma. Additionally, cancer patients with HNF1B mutations are more susceptible to many other gene mutations, including TP53, TTN, MUC16, CSMD3, SYNE1, ZFHX4, LRP1B, XIRP2, PCLO, FLG, FAT4, DNAH5, HYDIN, PIK3CA, USH2A, HMCN1, RYR2, CSMD1, FAT3 and KMT2D (Figure 2C).
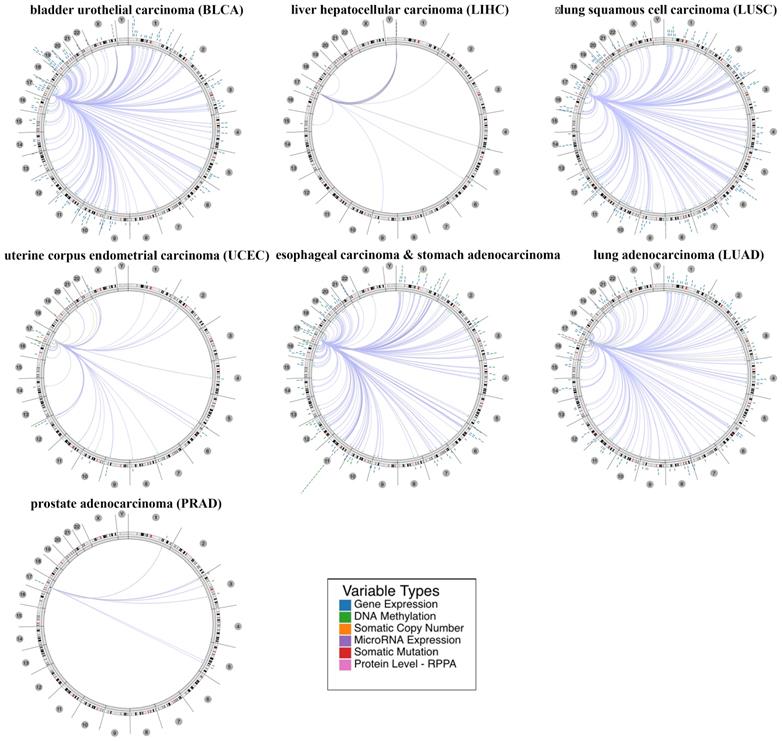
Genome-wide association of HNF1B in cancer
Furthermore, we explored the relevant human genome location and the association between certain genes and HNF1B in human cancer based on a regulome explorer. Circus plots were drawn to reveal the correlation between HNF1B and other genes according to the association among genes, DNA methylation, somatic copy-number alteration, somatic mutation and protein level. As shown in Figure 3, HNF1B was associated with other genes that could be detected in bladder urothelial carcinoma, liver hepatocellular carcinoma, lung squamous cell carcinoma, uterine corpus endometrial carcinoma, esophageal carcinoma and stomach adenocarcinoma, lung adenocarcinoma and prostate adenocarcinoma.
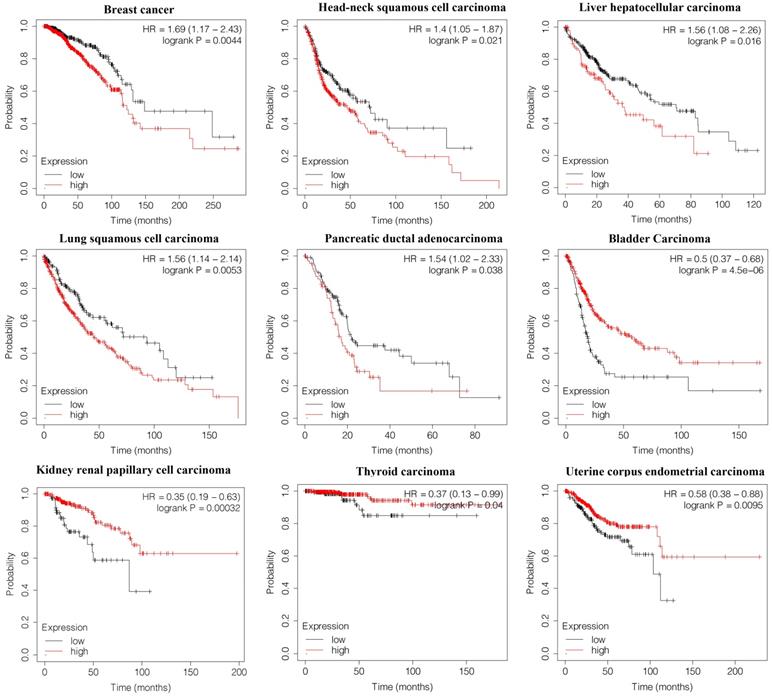
Survival analysis of HNF1B in cancer
To determine the potential value of HNF1B in predicting the overall survival of cancer patients, we constructed Kaplan-Meier survival curves. HNF1B was found to be negatively associated with overall survival in breast cancer (HR=1.69, p=0.0044), head-neck squamous cell carcinoma (HR=1.4, p=0.021), liver hepatocellular carcinoma (HR=1.56, p=0.016), lung squamous cell carcinoma (HR=1.56, p=0.0053), and pancreatic ductal adenocarcinoma (HR=1.54, p=0.038). However, higher levels of HNF1B indicated better overall survival in bladder carcinoma (HR=0.5, p=4.5e-06), kidney renal papillary cell carcinoma (HR=0.35, p=0.00032), (HR=0.37, p=0.04), and uterine corpus endometrial carcinoma (HR=0.58, p=0.0095) (Figure 4).
HNF1B expression is correlated with immune infiltration levels in cancer
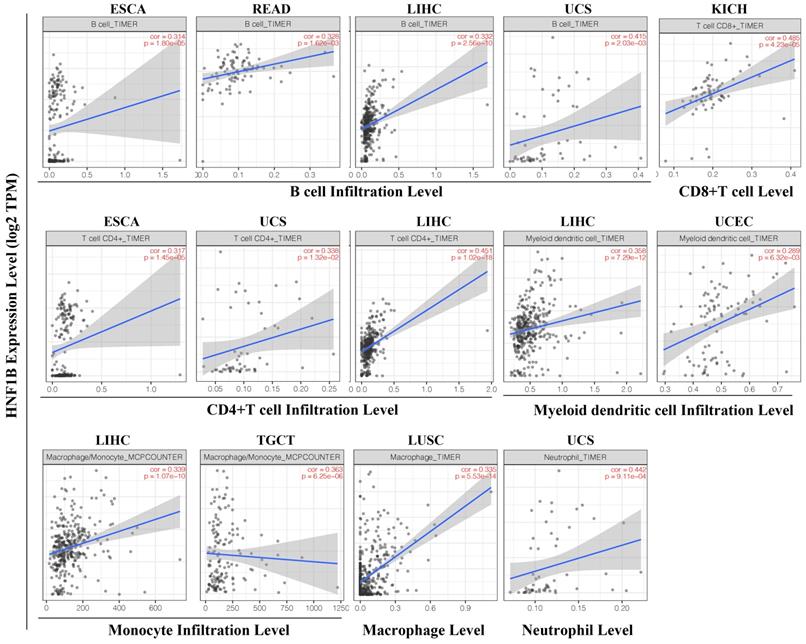
Based on the TCGA dataset, we evaluated the correlation of HNF1B with levels of immune cell infiltration in cancer from TIMER. As shown in Figure 5, HNF1B was significantly associated with B cells, CD8+ T cells, CD4+ T cells, myeloid dendritic cells (DCs), monocytes, macrophages and neutrophils in multiple tumors. A positive correlation between HNF1B expression and B-cell infiltration levels was found in esophageal carcinoma (ESCA, r = 0.314, p =1.80e-6), Rectum adenocarcinoma (READ, r=0.328, p=1.62e-03), liver hepatocellular carcinoma (LIHC, r= 0.332, p = 2.56e-10), Uterine Carcinosarcoma (UCS, r = 0.415, p =2.03e-03). A positive correlation between HNF1B expression and CD8+ T cell levels was found in Kidney Chromophobe (KICH, r=0.485, p=4.23e-05). Moreover, there was a strong positive correlation between HNF1B expression and CD4+ T cell levels in esophageal carcinoma (ESCA, r = 0.317, p =1.45e-5), Uterine Carcinosarcoma (UCS, r = 0.338, p =1.32e-02), liver hepatocellular carcinoma (LIHC, r= 0.451, p = 1.02e-18). In addition, the results also revealed a positive correlation between HNF1B expression and myeloid DC infiltration levels in liver hepatocellular carcinoma (LIHC, r= 0.358, p = 7.29e-12), Uterine Corpus Endometrial Carcinoma (UCEC, r=0.289, p=6.32e-03). The positive correlation between HNF1B expression and monocyte infiltration level was found in liver hepatocellular carcinoma (LIHC, r= 0.339, p = 1.07e-10) and Testicular Germ Cell Tumors (TGCT, r=0.363, p=6.25e-06). In Lung squamous cell carcinoma (LUSC), HNF1B was correlated with the macrophage infiltration (r=0.335, p=5.53e-14). In Uterine Carcinosarcoma (UCS), HNF1B was associated with the neutrophil infiltration (r=0.442, p=9.11e-04). Moreover, the EPIC algorithm analysis showed that HNF1B was significantly associated with NK cell and cancer-associated fibroblasts in cancer (Supplementary Figure 2).
HNF1B mRNA expression was abnormal in various cancers based on UALCAN database analysis.
A. Mutation diagram of HNF1B in different cancer types across protein domains. B. HNF1B mutation and copy-number alterations in the TCGA pan-cancer studies. C. Genes with the top 20 highest frequency in any group (HNF1B-altered group and HNF1B-unaltered group). TP53-tumor protein p53; TTN-titin; MUC16-mucin 16; CSMD3-CUB and Sushi multiple domains 3; SYNE1-spectrin repeat containing nuclear envelope protein 1; ZFHX4 zinc finger homeobox 4; LRP1B-LDL receptor related protein 1B; XIRP2-xin actin binding repeat containing 2; PCLO-piccolo presynaptic cytomatrix protein; FLG-filaggrin; FAT4-FAT atypical cadherin 4; DNAH5-dynein axonemal heavy chain 5; HYDIN-HYDIN axonemal central pair apparatus protein; PIK3CA-phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; USH2A-usherin; HMCN1-hemicentin 1; RYR2-ryanodine receptor 2; CSMD1-CUB and Sushi multiple domains 1; FAT3-FAT atypical cadherin 3; KMT2D-lysine methyltransferase 2D.
Comprehensive prognostic analysis of HNF1B and immune infiltration levels in tumors
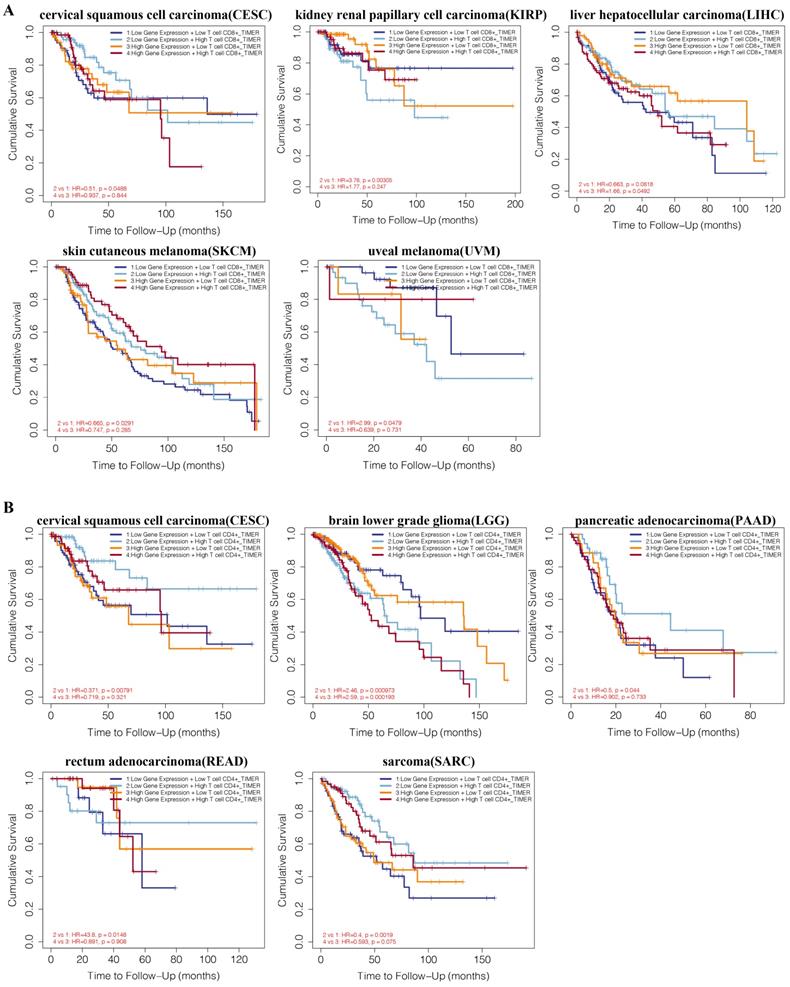
We performed a prognosis analysis based on the HNF1B expression levels and immune cells in different tumors via TIMER. As shown in Figure 6A, CD8+ T cell levels were negatively associated with overall survival in the low HNF1B expression group of kidney renal papillary cell carcinoma (KIRP, HR=3.76, P=0.00305) and uveal melanoma (UVM, HR=2.99, P=0.0479). CD8+ T cells were found to be positively associated with overall survival in the low HNF1B expression group of cervical squamous cell carcinoma (CESC, HR=0.51, p=0.0488) and skin cutaneous melanoma (SKCM, HR=0.665, p=0.0291). In the low HNF1B expression group of liver hepatocellular carcinoma (LIHC), CD8+ T cells were revealed to be positively associated with overall survival. Additionally, the combination of HNF1B expression and CD4+ T cell infiltration levels showed prognostic value in cervical squamous cell carcinoma (CESC), brain lower grade glioma (LGG), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), sarcoma (SARC) (Figure 6B). In bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), kidney renal clear cell carcinoma (KIRC), brain lower grade glioma (LGG) and mesothelioma (MESO), the expression level of HNF1B also influenced the prognostic value of B-cell infiltration levels (Supplementary Figure 3).
Discussion
Hepatocyte nuclear factor (HNF) is a type of transcription factor that is distributed in the liver, pancreas, intestine, kidney and other organs, regulating the specific expression of a series of related genes[19]. HNF1, HNF3, HNF4 and HNF6 are its main subtypes, and these factors work together to coordinate and maintain the body's steady state. HNF1B is a member of the HNF family, participates in regulating the metabolism of lipids, carbohydrates and proteins, and plays an important role in regulating hepatocyte differentiation and liver development[20]. Recent studies have shown that HNF1B has a significant correlation with the occurrence and development of various tumors, such as prostate cancer and pancreatic cancer [21,22]. In this study, we found that the expression of HNF1B was upregulated at different levels in BLCA, CHOL, KIRP, LIHC, STAD, THCA, and UCEC and downregulated in COAD, GBM, KICH, KIRC, and LUSC. Based on these results, HNF1B may be a good potential diagnostic biomarker for different cancers.
The correlation between HNF1B and other genes from the TCGA database was determined using the Cancer Regulome tool. Only genes with a correlation of Abs = 0.5 and p <0.01 are shown in the circus plots (200 max results).
Correlation between HNF1B expression and the overall survival of cancer patients (Kaplan-Meier analysis).
Consistent with the above results, the significance of HNF1B in hepatocellular carcinoma has been examined. Shim et al revealed that the expression of HNF1B predicted disease recurrence and hepatocellular carcinoma-specific death after liver transplantation in patients with hepatocellular carcinoma[23]. Another clinicopathological study showed that the HNF1B expression in hepatocellular carcinoma may be related with the change of phenotype on recurrence[24]. The authors also found that HNF1B expression was associated with poorer disease outcome, being an independent risk factor for both disease-free survival and overall survival. Moreover, it has been reported that the single nucleotide polymorphism (SNP) of HNF1B can affect the susceptibility of endometrial tumors. Spurdle et al. conducted gene sequencing studies on endometrial cancer patients and control groups and found that HNF1B gene SNP (rs4430796, G→A) can reduce the incidence of endometrial cancer[25]. Meta-analysis of large samples also confirmed this result[26]. At the same time, the HNF1B gene can independently affect the prognosis of endometrial tumors[27].
The HNF1B gene is located on chromosome 17q12 and contains 9 exons, which have 3 different functional domains[28]. Most HNF1B mutations are clustered in the first 4 exons of the gene. HNF1B has frequent spontaneous neonatal mutations, and the spontaneous deletion rate is up to 50%. A total of 106 HNF1B gene mutations, including gene deletion (34%), missense mutation (31%), frameshift deletion or insertion mutation (15%), nonsense mutation (11%) and splicing point mutation (8%) have been reported[29,30]. According to these results, our study found that HNF1B mutations occurred widely in human cancers and that the most common type is missense mutations. Additionally, patients with HNF1B mutations are more prone to mutations in other genes, such as TP53, TTN and MUC16. Survival analysis revealed that HNF1B expression indicated distinct overall survival rates of cancer patients. The distinct prognostic value of HNF1B may be caused by the different mutations and interactions between HNF1B and other genes in different types of cancers.
Immune cells that infiltrate tumors play crucial roles in tumor development[31]. Evading immune destruction is one of the hallmarks of cancer[32]. The role of the immune system in cancer oncogenesis is increasingly being investigated, and immunotherapy has become a powerful clinical strategy for treating cancer[33,34]. Our study showed that HNF1B expression is associated with diverse immune cell infiltration levels in different cancers. The results demonstrated a strong positive correlation between HNF1B expression and the infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and DCs in cancer. Additionally, we divided patients into a low HNF1B expression group and a high HNF1B expression group, and CD8+ T cell, CD4+ T cell and B-cell levels were distinctly associated with overall survival of cancer patients in the different groups. The association of HNF1B and immune cells has rarely been studied[35] and needs to be further investigated. HNF1B expression may be a considerable factor for patients who are going to adopt immunotherapy.
In conclusion, our study found that HNF1B mRNA is dysregulated in various cancers and that the differential expression of HNF1B indicates different prognoses. HNF1B mutations are widely observed in tumors and interact with different genes in different cancer types, which may be the cause of the distinct prognostic values in cancers. Moreover, HNF1B is associated with the level and prognosis prediction of immune cells in some kinds of cancers.
HNF1B expression is correlated with the immune infiltration levels in cancer (TIMER algorithm). ESCA: esophageal carcinoma, READ: rectum adenocarcinoma, LIHC: liver hepatocellular carcinoma, UCS: uterine carcinosarcoma, KICH: kidney chromophobe, UCEC: uterine corpus endometrial carcinoma, TGCTs: testicular germ cell tumors, LUSC: lung squamous cell carcinoma.
A. Comprehensive prognostic value of HNF1B expression and CD8+ T cell infiltration levels based on TIMER. B. Comprehensive prognostic value of HNF1B expression and CD4+ T cell infiltration levels based on TIMER.
Abbreviation
HNF1B: Hepatocyte nuclear factor 1B; TIMER: Tumor IMmune Estimation Resource; TCGA: The Cancer Genome Atlas; EPIC: Estimating the Proportion of Immune and Cancer cells; BLCA: bladder urothelial carcinoma; CHOL: cholangiocarcinoma; KIRP: kidney renal papillary cell carcinoma; LIHC: liver hepatocellular carcinoma; STAD: stomach adenocarcinoma; THCA: thyroid carcinoma; UCEC: uterine corpus endometrial carcinoma; COAD: colon adenocarcinoma; GBM: glioblastoma multiforme; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; LUSC: lung squamous cell carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma; ESCA: esophageal carcinoma; HNSC: head and neck squamous cell carcinoma; LUAD: lung adenocarcinoma; PAAD: pancreatic adenocarcinoma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; UCS: Uterine Carcinosarcoma; UVM: uveal melanoma; SKCM: skin cutaneous melanoma; DCs: dendritic cells; LGG: brain lower grade glioma; SARC: sarcoma; MESO: mesothelioma.
Supplementary Material
Supplementary figures.
Acknowledgements
The authors thank the TIMER, UALCAN, cBio cancer genomics portal and Kaplan-Meier plotter for the availability of the data.
Funding
The study was supported by the Medical and Health Science and Technology Project of Zhejiang Province (Project No.2019KY070, No.2019KY380); Zhejiang Provincial Natural Science Foundation of China (No. LYQ20H310005).
Author Contributions
Chunhui Nie conceived and designed the study; Bei Wang obtained the datasets; Baoquan Wang performed data analysis; Ning Lv wrote the paper. Enfan Zhang revised the manuscript. All authors reviewed and approved the final manuscript.
Data Availability Statement
The data that support the findings of this study are available in UALCAN database and TIMER database.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
2. Tompkins KD, Thorburn A. Regulation of Apoptosis by Autophagy to Enhance Cancer Therapy. Yale J Biol Med. 2019;92:707-718
3. Ma W, Mao Q, Xia W, Dong G, Yu C, Jiang F. Gut Microbiota Shapes the Efficiency of Cancer Therapy. Front Microbiol. 2019;10:1050
4. Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nature reviews Cancer. 2017;17:20-37
5. Ferrè S, Igarashi P. New insights into the role of HNF-1β in kidney (patho)physiology. Pediatr Nephrol. 2019;34:1325-1335
6. Roelandt P, Antoniou A, Libbrecht L, Van Steenbergen W, Laleman W, Verslype C. et al. HNF1B deficiency causes ciliary defects in human cholangiocytes. Hepatology. 2012;56:1178-1181
7. Haldorsen IS, Vesterhus M, Raeder H, Jensen DK, Søvik O, Molven A. et al. Lack of pancreatic body and tail in HNF1B mutation carriers. Diabet Med. 2008;25:782-787
8. Massa F, Garbay S, Bouvier R, Sugitani Y, Noda T, Gubler MC. et al. Hepatocyte nuclear factor 1β controls nephron tubular development. Development. 2013;140:886-896
9. De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, Haumaitre C. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development. 2015;142:871-882
10. Pillai R, Paglialunga S, Hoang M, Cousteils K, Prentice KJ, Bombardier E. et al. Deletion of ARNT/HIF1β in pancreatic beta cells does not impair glucose homeostasis in mice, but is associated with defective glucose sensing ex vivo. Diabetologia. 2015;58:2832-2842
11. Heidet L, Decramer S, Pawtowski A, Morinière V, Bandin F, Knebelmann B. et al. Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin J Am Soc Nephrol. 2010;5:1079-1090
12. Bártů M, Dundr P, Němejcová K, Tichá I, Hojný H, Hájková N. The Role of HNF1B in Tumorigenesis of Solid Tumours: a Review of Current Knowledge. Folia Biol (Praha). 2018;64:71-83
13. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658
14. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404
15. Li QT, Huang ZZ, Chen YB, Yao HY, Ke ZH, He XX. et al. Integrative Analysis of Siglec-15 mRNA in Human Cancers Based on Data Mining. Journal of Cancer. 2020;11:2453-2464
16. Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX. et al. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Frontiers in immunology. 2019;10:6
17. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer research. 2017;77:e108-e110
18. Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017 6
19. Lau HH, Ng NHJ, Loo LSW, Jasmen JB, Teo AKK. The molecular functions of hepatocyte nuclear factors - In and beyond the liver. J Hepatol. 2018;68:1033-1048
20. El-Khairi R, Vallier L. The role of hepatocyte nuclear factor 1β in disease and development. Diabetes Obes Metab. 2016;18(Suppl 1):23-32
21. Quilichini E, Fabre M, Dirami T, Stedman A, De Vas M, Ozguc O. et al. Pancreatic Ductal Deletion of Hnf1b Disrupts Exocrine Homeostasis, Leads to Pancreatitis, and Facilitates Tumorigenesis. Cell Mol Gastroenterol Hepatol. 2019;8:487-511
22. Dan C, Zhang H, Zeng W, Huang L, Gong X, Li H. et al. HNF1B expression regulates ECI2 gene expression, potentially serving a role in prostate cancer progression. Oncol Lett. 2019;17:1094-1100
23. Shim JH, Lee HC, Han S, Kang HJ, Yu E, Lee SG. Hepatocyte nuclear factor 1β is a novel prognostic marker independent of the Milan criteria in transplantable hepatocellular carcinoma: a retrospective analysis based on tissue microarrays. Liver Transpl. 2013;19:336-345
24. Yu DD, Jing YY, Guo SW, Ye F, Lu W, Li Q. et al. Overexpression Of Hepatocyte Nuclear Factor-1beta Predicting Poor Prognosis Is Associated With Biliary Phenotype In Patients With Hepatocellular Carcinoma. Scientific reports. 2015;5:13319
25. Spurdle AB, Thompson DJ, Ahmed S, Ferguson K, Healey CS, O'Mara T. et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat Genet. 2011;43:451-454
26. Setiawan VW, Haessler J, Schumacher F, Cote ML, Deelman E, Fesinmeyer MD. et al. HNF1B and endometrial cancer risk: results from the PAGE study. PLoS One. 2012;7:e30390
27. Mandato VD, Farnetti E, Torricelli F, Abrate M, Casali B, Ciarlini G. et al. HNF1B polymorphism influences the prognosis of endometrial cancer patients: a cohort study. BMC Cancer. 2015;15:229
28. Clissold RL, Ashfield B, Burrage J, Hannon E, Bingham C, Mill J. et al. Genome-wide methylomic analysis in individuals with HNF1B intragenic mutation and 17q12 microdeletion. Clin Epigenetics. 2018;10:97
29. Sneha P, Doss CG. Elucidating the Mutational Landscape in Hepatocyte Nuclear Factor 1β (HNF1B) by Computational Approach. Adv Protein Chem Struct Biol. 2017;107:283-306
30. Alvelos MI, Rodrigues M, Lobo L, Medeira A, Sousa AB, Simão C. et al. A novel mutation of the HNF1B gene associated with hypoplastic glomerulocystic kidney disease and neonatal renal failure: a case report and mutation update. Medicine (Baltimore). 2015;94:e469
31. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883-899
32. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. cell. 2011;144:646-674 %@ 0092-8674
33. Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18:175-196
34. Satge D. A Tumor Profile in Primary Immune Deficiencies Challenges the Cancer Immune Surveillance Concept. Frontiers in immunology. 2018;9:1149
35. Oda K, Hamanishi J, Matsuo K, Hasegawa K. Genomics to immunotherapy of ovarian clear cell carcinoma: Unique opportunities for management. Gynecol Oncol. 2018;151:381-389
Author contact
![]() Corresponding author: Enfan Zhang, Bone Marrow Transplantation Center, Department of Hematology, The First Affifiliated Hospital, School of Medicine, Zhejiang University, No. 79, Qingchun Road, Hangzhou, Zhejiang Province, China, Tel: +86-571-87236706, Fax: +86-571-87236706, E-mail: efzhangedu.cn
Corresponding author: Enfan Zhang, Bone Marrow Transplantation Center, Department of Hematology, The First Affifiliated Hospital, School of Medicine, Zhejiang University, No. 79, Qingchun Road, Hangzhou, Zhejiang Province, China, Tel: +86-571-87236706, Fax: +86-571-87236706, E-mail: efzhangedu.cn

 Global reach, higher impact
Global reach, higher impact