Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(8):824-833. doi:10.7150/ijms.8358 This issue Cite
Research Paper
Frequent Co-Expression of miRNA-5p and -3p Species and Cross-Targeting in Induced Pluripotent Stem Cells
1. Department of Animal Science & Graduate Institute of Biotechnology, Chinese Culture University, Taipei, Taiwan;
2. Centre for Stem Cell Research, Universiti Tunku Abdul Rahman, Faculty of Medicine and Health Sciences, Kajang, Selangor, Malaysia;
3. Department of Preclinical Sciences, Universiti Tunku Abdul Rahman, Faculty of Medicine and Health Sciences, Kajang, Selangor, Malaysia;
4. Singapore BioImaging Consortium, Singapore;
5. Duke-NUS Graduate Medical School, Singapore;
6. Dean's Office, Universiti Tunku Abdul Rahman, Faculty of Medicine and Health Sciences, Kajang, Selangor, Malaysia;
7. Tissue Engineering Group, National Orthopaedic Centre of Excellence for Research and Learning, Department of Orthopaedic Surgery, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia.
Received 2013-12-14; Accepted 2014-5-14; Published 2014-6-5
Abstract
Background: A miRNA precursor generally gives rise to one major miRNA species derived from the 5' arm, and are called miRNA-5p. However, more recent studies have shown co-expression of miRNA-5p and -3p, albeit in different concentrations, in cancer cells targeting different sets of transcripts. Co-expression and regulation of the -5p and -3p miRNA species in stem cells, particularly in the reprogramming process, have not been studied.
Methods: In this work, we investigated co-expression and regulation of miRNA-5p and -3p species in human induced pluripotent stem cells (iPSCs), mesenchymal stem cells (MSCs) and embryonic stem cells (ESC) using a nanoliter-scale real-time PCR microarray platform that included 1,036 miRNAs.
Results: In comparing iPSC and ESC, only 32 miRNAs were found to be differentially expressed, in agreement of the ESC-like nature of iPSC. In the analysis of reprogramming process in iPSCs, 261 miRNAs were found to be differentially expressed compared with the parental MSC and pre-adipose tissue, indicating significant miRNA alternations in the reprogramming process. In iPSC reprogrammed from MSC, there were 88 miRNAs (33.7%), or 44 co-expressed 5p/3p pairs, clearly indicating frequent co-expression of both miRNA species on reprogramming. Of these, 40 pairs were either co-up- or co-downregulated indicating concerted 5p/3p regulation. The 5p/3p species of only 4 pairs were regulated in reverse directions. Furthermore, some 5p/3p species of the same miRNAs were found to target the same transcript and the same miRNA may cross-target different transcripts of proteins of the G1/S transition of the cell cycle; 5p/3p co-targeting was confirmed in stem-loop RT-PCR.
Conclusion: The observed cross- and co-regulation by paired miRNA species suggests a fail-proof scheme of miRNA regulation in iPSC, which may be important to iPSC pluripotency.
Keywords: Induced pluripotent stem cells, reprogramming, miRNA-5p/3p species, cell cycle control
Introduction
Human embryonic stem cells (ESC) are a unique cell type which derived from human blastocysts. ESC cell has an ability to undergo indefinite self-renewal and exhibited a full developmental potential to form various lineages [1]. These types of cells have been known as pluripotent cells and are utmost important in cell-based therapies and regenerative medicine. However, the issue of immune rejection stemming from incompatibility between the patient and donor cells has called for alternative approaches in generating pluripotent stem cells. In recent years, Takahashi and Yamanaka (2006) [2] demonstrated that retrovirus-mediated transfection with a combination of four transcription factors, Oct4, Sox2, Klf4 and c-Myc, was able to 'reprogram' mouse fibroblast and human somatic cells to undifferentiated pluripotent stem cells. Besides high similarity to ESC, these induced pluripotent stem cells (iPSCs) also offer invaluable therapeutic implications to human in term of in vitro disease modeling, pharmaceutical screening and cellular replacement therapies. Immune rejection issue can be easily overcome since iPSCs are derived from the same patient.
microRNAs (miRNAs) play an important role in gene regulation during pluripotency, self-renewal and differentiation of ESCs and iPSCs. miRNAs can be divided into two subgroups: pluripotent miRNAs and pro-differentiation miRNAs. Pluripotent miRNAs have been found to be involved in maintaining self-renewal and pluripotency of ESC. This class of miRNAs, including miR-137, miR-184, miR-200, miR-290, miR-302, and miR-9, was exclusively expressed in the pluripotent state and rapidly decreased upon differentiation stimuli [3]. Previous studies revealed that Dicer and Dgcr8-deficient ESC markedly delayed cell cycle progression [4,5]. By contrast, pro-differentiation miRNAs, such as let-7, miR-296, miR-134 and miR-470, have been found to regulate the differentiation processes in pluripotent cells [6,7]. These miRNAs were found to be upregulated during differentiation in ESC and inhibited the expression of pluripotency factors, including Nanog, Lin28, Sox2 and Klf4 [7,8].
In the miRNA biogenesis pathway, long primary transcript (pri-miRNA) is transcribed and then processed into a structure of 60 to 110 nt hairpin precursor miRNA (pre-miRNA) by cellular RNase enzyme III, Drosha, and double stranded RNA-binding domain protein, DGCR8 [9]. This pre-miRNA is then cleaved by another RNase III enzyme, Dicer, to generate ~22 nt miRNA: miRNA* duplex [10]. One strand of the duplex, complementary to the target, has been known as a functional guide strand (miRNA), whereas the other strand, which usually will be degraded, has been considered as a passenger strand (miRNA*) [11]. However, recent studies indicated that some miRNA* sequences were abundantly expressed as mature functional miRNAs [12-14]. In some cases, two mature miRNAs excised from the 5'- and 3'- arms of the same stem-loop pre-miRNA have been reported to be functional and target on different mRNAs [15,16]. To avoid confusion, human miRNA/miRNA* nomenclature has been retired. Instead, the miRNA-5p and -3p nomenclature is now being applied widely according to 5'- or 3'-arms derivation of the miRNA species.
miRNA 5p/3p pairs are co-expressed differently from tissue to tissue indicating tissue-dependent regulatory roles for the 5p/3p miRNA species [12]; co-existing miRNA pairs have also been reported in different cancer cells [16-20]. Besides cancer, the co-expressed let-7 and the mir-126 families have been demonstrated to play different roles in regulating ESC self-renewal, pluripotency, and differentiation [21,22].
Despite reports on the involvement of specific miRNAs in ESC and iPSC, genome-wide studies focusing on the participation of miRNA-5p/3p pairs in the cell cycle process are still lacking. This study aimed to systematically investigate co-expression and regulation of 5p/3p paired miRNA species in iPSC self-renewal maintenance.
Materials and Methods
Cell lines and RNA preparation
The adipose stem cell (ASC) was obtained from Invitrogen (Carlsbad, CA, USA). The human white pre-adipocyte (HWP) and the human adipose-derived MSC (MSC-AT) were obtained from PromoCell (Heidelberg, Germany). Derivation of characterization of the induced pluripotent stem cell (iPSC) lines, HWP-derived iPCS (HWP-iPSC), ASC-derived iPSC (ASC-iPSC) and MSC-iPSC are described by Sugii et al. [23,24] (Table 1).
miRNA Profiling
Total RNA (1 µg) extracted from each cell line were analyzed using a nanoscale miRNA real-time qRT-PCR array (SmartChip Human MicroRNA Panel v3; WaferGen Biosystems, Fremont, CA, USA) containing 1036 miRNA-specific reactions in quadruplicate for a total of 5184 reactions per sample. miRNA was first ligated to a pre-adenylated linker (3' adapter) by RNA ligase 2 at 22 °C for 60 min. The ligated RNA was subjected to one-step on-chip real-time qRT-PCR reaction at 52 °C for 5 min, 95 °C for 10 min, 95 °C for 1 min and 52 °C for 1 min, followed by 39 cycles of PCR amplification at 95 °C for 1 min and 60 °C for 1 min to synthesize cDNA and to amplify target on the SmartChip Cycler (Wafergen Biosystems). In this analysis, an additional 7 endogenous and 4 exogenous controls were included for data quality control.
Selection Criteria for Differentially Expressed miRNAs in Pairwise Comparison
The microarray data for iPSC were compared with the data for parental cells. For calculations of expression levels, the All-Mean normalization method was employed, where mean Ct of all expressed genes were used [25,26]. To compute the expression level of expressed miRNAs, the Cts of each sample were compared to its average Ct (All-Mean) to obtain the ΔCt values. ΔΔCt was then calculated by the two ΔCt values between the iPSC and its parental cell type. The log2 (fold change) was log2 {Fold change (2^-[delta][delta]Ct)}. The selection criteria for differentially expression of miRNA was the log2 (fold change) ≥1.5 or ≤-1.5 with p<0.05.
MicroRNA Quantitative Real-Time RT-PCR
Real-time qRT-PCR was performed using the NCode SYBR GreenER miRNA qRT-PCR kit (Invitrogen, Carlsbad, CA, USA) following the supplier's instructions in a Rotor-Gene Q real-time PCR cycler (Qiagen). Following miRNA poly(A) tailing, first-strand cDNA was synthesized using the Superscript III RT/RNaseOUT enzyme mix provided in the kit, followed by real-time RT-PCR using SYBR select master mix (Applied Biosystems, USA) in Rotor-Gene Q. Amplification was carried out for 40 cycles at 95 °C for 15 s and primer annealing at 60 °C for 1 min. Experiments were performed in triplicates and were normalized to the data of the small nuclear RNA (snRNA) U6. Primers used for miRNA quantification were as follows: forward primer (miR-9-F) 5'-TCTTTGGTTATCTAGCTGTATGA-3' and universal primer (provided in NCode SYBR GreenER miRNA qRT-PCR kit) as reverse primer. The U6 oligonucleotide 5'-CACCACGTTTATACGCCGGTG-3' was used as the normalization control. Relative miRNA levels were calculated using the comparative ΔΔCт method.
miRNA Stem-Loop RT-PCR
Primers for detection of mature miRNAs were designed according to Chen et al. (2005) [27]. cDNAs were synthesized according to the manufacturer's manual (Invitrogen). Briefly, the annealing program for the stem-loop primers (Additional file 1: Supplementary Table 1) was 5 min at 65 °C. Stem-loop products were then added to an RT reaction using Superscript III reverse transcriptase (Invitrogen) containing 4 μL first-strand buffer, 2 μL 0.1 M DTT, 0.1 μL RNaseOUT, and 0.25 μL (50 units) SuperScript III reverse transcriptase. The reaction was performed with the following incubation conditions: 16 °C for 30 min, followed by 60 cycles of 30 °C for 30 s, 42 °C for 30 s, 50 °C for 1 s. The enzyme was inactivated by incubation at 85 °C for 5 min. The cDNA was used at a dilution of 1:10 in water in subsequent PCR reactions. The PCR products were then detected by electrophoresis on 4% agarose gels, and cDNA loading controls were normalized with U6.
Bioinformatics
Putative miRNA target transcripts were obtained from TargetScan, miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) and DIANA-microT v5.0 (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) [28]. Only target transcripts predicted by at least two different algorithms were selected.
Results and Discussion
Differential Expression of miRNAs in Different Stem Cell Types
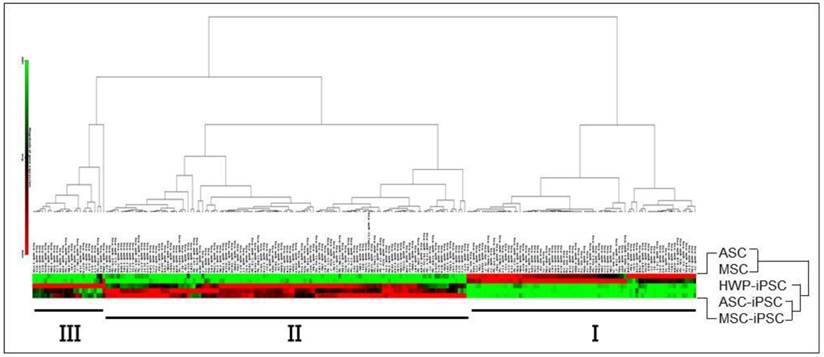
To identify differentially expressed miRNAs in iPS cells relative to the parental cells from which they are derived via reprogramming, a HWP cell line, two human adipose-derived mesenchymal stem cell lines, designated as ASC and hMSC-AT, and the three induced pluripotent stem iPS cell lines derived from them were used (Table 1). Two well-characterized ESC lines, H6 and H9 were also included for comparison. Total RNAs prepared from these eight cell lines were subjected to quantitative miRNA profiling using a nanolitre-scale real-time RT-PCR microarray platform that included 1,036 miRNA species. On obtaining the microarray data, hierarchical clustering analysis of the miRNA profiles was performed between the two MSC and the three iPSC (Fig. 1). Since the HWP cells are unipotent and not multipotent cells [29], HWP was omitted from the two multipotent MSC cell lines in the hierarchical clustering analysis. The clustergram showed that the miRNAs were clustered into two major (I & II) and one minor (III) clusters. Cluster I included miRNAs that were expressed in high levels in the MSC lines whereas cluster II included miRNAs highly expressed in iPSC. In each of these two clusters, there were also subclusters generated based on different miRNA levels observed. The data clearly showed that different miRNA signatures in the multipotent MSC relative to the pluripotent iPSC. Furthermore, miRNAs in cluster III were in high expression levels specifically in the HWP-derived iPSC in various lower expression levels in the other two MSC-derived iPSC lines, indicating iPSC derived from various sources are not entirely identical.
Hierarchical clustering analysis of miRNA profiles of MSC and iPSC. Level of gene expression is shown in the colour code shown at the bottom, ranging from minimal expression levels in green, average or weak miRNA expression in black and maximal levels in red. The analysis was performed using miScript miRNA PCR Array Data Analysis Web Portal.
Stem cell lines used in this work
| Cell type | Cell line | Abbreviation |
|---|---|---|
| ESC | HuES6 | H6 |
| H9 | H9 | |
| MSC | Adipose Stem Cell | ASC |
| Adipose-derived MSC | hMSC-AT | |
| Preadipocytes | Human white preadipocytes | HWP |
| iPSC | ASC-derived | ASC-iPSC |
| hMSC-AT-derived | MSC-iPSC | |
| HWP-derived | HWP-iPSC |
Pairwise comparisons between iPSC and ESC, and iPSC and MSC were next performed by using the cut-off threshold of log2 (fold change) ≥1.5 or ≤-1.5, and also the criteria that valid data were available for all the cell lines under consideration with statistical significance (p<0.05). Furthermore, it was defined by the platform that a miRNA that had a threshold value of Ct ≥ 30 was considered undetectable in expression level. Hence, in the iPSC-MSC comparison, a miRNA was considered activated in iPSC when this miRNA was in the detectable range in iPSC, but was undetectable (Ct ≥ 30) in MSC. Likewise, a miRNA was considered shutdown in iPSC when the miRNA was detectable in MSC but was undetectable in iPSC. On the other hand, up- or down-regulated expression was used to describe increased or decreased detectable miRNA levels in pairwise comparison.
When the miRNA expression data of the two iPSC lines were first compared pairwise with their respective parental MSC lines, 441-445 (42.6-43.0%) miRNAs were found to have altered in expression levels as defined (Table 2). On reprogramming of the monopotent HWP, 494 miRNAs (47.7%) were differentially expressed (Table 2). If the data were considered collectively under the criteria defined above, 261 miRNAs (25.2%) were found to be differentially expressed in the three iPSC relative to the two MSC lines (Table 3 & Additional file 1: Supplementary Table 2) clearly indicating extensive changes in the miRNA profiles when MSC was reprogrammed to iPSC. On the other hand, when the miRNA expression data of the three iPSC lines were collectively compared with those of the two ESC lines, only 32 miRNAs (3.1%) were found to be differentially expressed (Table 3 & Additional file 1: Supplementary Table 3), consistent with ESC-like characteristics of iPSC.
miRNAs altered in expression levels in pairwise comparison between MSC/HWP and the derived iPS cells
| Stem cell line | No. differentially expressed miRNA (% of miRNA analyzed) | |
|---|---|---|
| iPSC | MSC/HWP | |
| hMSC-AT-iPSC vs | hMSC-AT | 441 (42.6%) |
| ASC-iPSC vs | ASC | 445 (43.0%) |
| HWP-iPSC vs | HWP | 494 (47.7%) |
Differentially expressed miRNAs in stem cells
| iPSC vs ESC1 | iPSC vs MSC2 | |
|---|---|---|
| Activated | 12 (37.5%) | 66 (25.3%) |
| Up-regulated | 13 (40.6%) | 111 (42.5%) |
| Subtotal: | 25 (78.1%) | 177 (67.8%) |
| Shut-down | 3 (9.4%) | 17 (6.5%) |
| Down-regulated | 4 (12.5%) | 67 (25.7%) |
| Subtotal: | 7 (21.9%) | 84 (32.2%) |
| Total | 32 | 261 |
1Data based on comparing 3 iPSC vs 2 ESC. 2Data based on 3 iPSC vs 2 MSC; In both columns, data were log2(fold change) ≥1.5 or ≤-1.5.
In further iPSC-ESC pairwise comparison, 25 (78.1%) of the 32 differentially expressed miRNAs were found to be activated/upregulated while 7 (21.9%) miRNAs were shutdown/downregulated (Table 3). In MSC-iPSC comparison, 177 (67.8%) were activated /upregulated and 83 (32.2%) miRNAs were shutdown/downregulated (Table 3). The data indicated that in pluripotency, two-fold more miRNAs are activated/upregulated than shutdown/downregulated. The data indicated that on reprogramming to pluripotency, many more target genes are shutdown or downregulated, which may be a significant event in rendering pluripotent stem cells the potential to differentiate into all cell types. The observation is consistent with the pluripotency of iPSC, and philosophically suggests that iPSC is life on hold, waiting for appropriate signals to release different sets of brakes to enter into differentiation into different tissues and organs.
In silico validation of Differentially Expressed miRNA in iPSC on Reprogramming
The array of differentially expressed miRNAs when MSC/HWP was reprogrammed into iPSC was cross-checked with what was available in the literature (Table 4).
In silico validation of WaferGen data on miRNAs that are differential expressed in iPSC relative to MSC/HWP
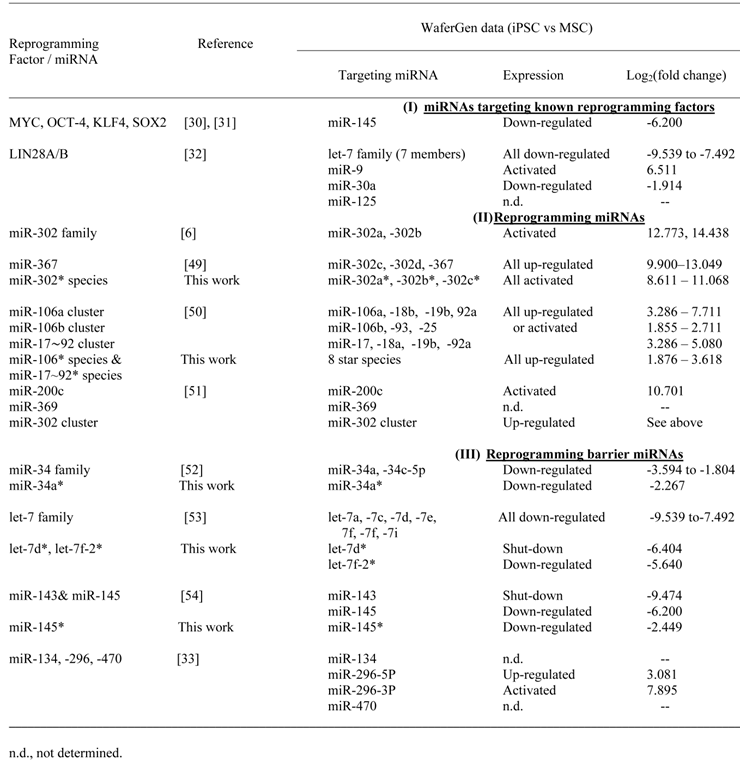
In the first group of miRNAs targeting known reprogramming factors, miR-145, which was shown to modulate the Yamanaka factors, Myc, Oct-4, Klf4 and Sox2 [30,31], would be predicted to be downregulated in iPSC on reprogramming. A downregulated log2 (fold change) of -6.2 was discerned on our analysis. Likewise, the let-7 family and miR-30A, which were shown to target LIN28A/B [32], all 7 let-7 family members and miR-30A were downregulated in our analysis. However, miR-9 was found to be upregulated by 6.511 fold, which appeared to be inconsistent with the predicted downregulation. To resolve the discrepancy, pairwise real-time RT-PCR was performed between the MSC/HWP and the derived iPSC lines (Table 5). In all the three pairs, upregulated levels in iPSC were consistently obtained with a statistically significant mean upregulated level of 3.14-fold, confirming the microarray data. It remains to be confirmed and investigated if miR-9 does modulate LIN28A/B to resolve the discrepancy. In the category of known reprogramming miRNAs (Table 4), all were found to be activated or upregulated in our microarray dataset as would be predicted, further supporting the validity of our results. There are also a group of miRNAs known to block reprogramming (Table 4, category III). With the exceptions of miR-134, -296 and -470, which were all suggested by Tay et al. (2008) [33] to target the coding sequences of Oct4 and Sox2 transcripts, all other known reprogramming barrier miRNAs were downregulated or shutdown on reprogramming of MSC/HWP to iPSC in this work.
Validation of miR-9 up-regulation in iPSC relative to MSC/MCS
| MSC/HWP vs iPSC | Log2(fold change) |
|---|---|
| ASC vs ASC-iPSC | 2.73 ± 0.34 |
| hMSC-AT vs AT-iPSC | 2.96 ± 0.34 |
| HWP vs HWP-iPSC | 3.72 ± 0.79 |
| Mean | 3.14 ± 0.52 (p<0.01) |
Data were obtained by real-time PCR analysis in three independent experiments.
Recent miRNA studies are beginning to document frequent co-expression of both the miRNA and the miRNA* strands derived from the 5'- and 3' arms of the pre-miRNA duplex [16,19,21,22]. When available in the microarray dataset in our analysis, the miRNA* species were identified and are included in this study (Additional file 1: Supplementary Table 2). It is interesting to note that the identified miRNA* species were co-up- or co-downregulated with their sister strand despite the fact that the two miRNA species carry different sequences and should be targeting different transcripts. Possible involvement of miRNA-miRNA* pairs is further investigated below. In summary, our microarray data were largely supported by published reports, supporting the robustness of the microarray platform used. Identification of co-expressed miRNA and miRNA* species may deserve further attention.
Co-expression of miRNA-5p/3p Pairs in Stem Cells
We next focused on co-expression of the miRNA and miRNA* pairs in stem cells. miRBase has recently retired the human miRNA/miRNA* nomenclature but advised the use of miRNA-5p and -3p nomenclature based on derivation from the 5'- or 3'-arm of the pre-miRNA precursor. In the subsequent sections in this paper, the 5p/3p nomenclature is used, and the original miR-miR* names are also listed alongside. Results of 5p/3p pairs included in the microarray data with each of the pair showing log2 (fold change) ≥1.5 or ≤-1.5 and p<0.05 were extracted from the dataset (Table 6).
Out of 32 miRNAs that were differentially expressed in iPSC relative to ESC, there were only three 5p/3p pairs (Table 6). Of these, the 5p/3p pairs of miR-199a and -181c were co-upregulated. However, the 5p/3p pair of miR-855-5p was shutdown in iPSC relative to ESC whilst miR-855-3p counterpart was activated in iPSC. Of the 261miRNAs that are differentially expressed in iPSC relative to MSC, 88 miRNAs (44 pairs, 33.7%) were co-expressed in 5p/3p pairs (Table 6). The alterations of the 44 5p/3p pairs observed in iPSC relative to MSC ranged from significant up-regulation by 10.366-fold in miR-200c-3p to down-regulation by -12.420-fold in miR-10a-5p (Table 6). On further examination, the 26 (59.1%) 5p/3p miRNA pairs were co-upregulated/co-activated and 14 (31.8%) pairs were co-downregulated/co-shutdown. Four pairs (9.1%), miR-139, -146b, -196b and -876, showed reverse directions of 5p/3p co-expression (Table 6). Taken together, the data showed frequent (33.7%) co-expression of 5p/3p miRNAs in iPSC on reprogramming, and that the majority (91.9%) of the co-expressed 5p/3p pairs was co-up- or co-down-regulated in the same direction strongly suggesting concerted regulation of miRNA sister pairs in the reprogramming process. Since miR-5p and -3p species have different sequences which are presumably targeting different target transcripts, the biological significance of 5p/3p co-expression deserves further attention.
miRNA families and expression levels of co-regulated 5p-3p pairs in iPSC relative to ESC and MSC
| No | miRNA family | Chromos'l site | miRNA-5p | Log2(FC) | miRNA-3p | Log2(FC) |
|---|---|---|---|---|---|---|
| iPSC vs ESC (n=3) A1. Co-upregulated/-activated pairs (n=2) | ||||||
| 1 | mir-199 | 19p13.2 | miR-199a-5p | 3.290±0.81 | miR-199a-3P | 2.913±0.93 |
| 2 | mir-181 | 19q13.13 | miR-181c | 2.082±0.38 | miR-181c* | 1.525±0.06 |
| A2. Pairs with reverse regulation (n=1) | ||||||
| 3 | mir-885 | 3p25.3 | miR-885-5p | -3.388±0.19 | miR-885-3p | 2.125±0.86 |
| iPSC vs MSC (n=44) B1. Co-upregulated/-activated pairs (n=26) | ||||||
| 1 | mir-8 | 12p13.31 | miR-141-5p | 2.110±0.35** | miR-141-3p | 6.567±0.35** |
| 2 | miR-200c-5p | 1.583±1.16 | miR-200c-3p | 10.366±0.69** | ||
| 3 | mir-17 | 13q31.3 | miR-18a-5p | 5.823±1.29** | miR-18a-3p | 4.133±1.04** |
| 4 | miR-20a-5p | 4.226±1.06** | miR-20a-3p | 4.096±1.30** | ||
| 5 | Xq26.2 | miR-18b-5p | 7.376±0.11** | miR-18b-3p | 2.453±1.05* | |
| 6 | miR-20b-5p | 7.561±4.23* | miR-20b-3p | 3.023±0.93** | ||
| 7 | miR-106a-5p | 4.640±1.08** | miR-106a-3p | 2.871±1.49* | ||
| 8 | mir-25 | 1q22 | miR-92b-5p | 2.920±0.46** | miR-92b-3p | 4.721±4.02 |
| 9 | 7q22.1 | miR-25-5p | 2.000±1.17* | miR-25-3p | 1.983±0.53** | |
| 10 | 13q31.3 | miR-92a-1-5p | 3.251±1.07** | miR-92a-3p | 2.923±0.57* | |
| 11 | mir-126 | 9q34.3 | miR-126-5p | 2.133±1.10** | miR-126-3p | 3.186±0.55** |
| 12 | mir-130 | 11q12.1 | miR-130a-5p | 1.610±0.15 | miR-130a-3p | 3.770±1.84 |
| 13 | mir-135 | 3p21.2 | miR-135a-5p | 9.107±0.77** | miR-135a-3p | 5.600±0.23 |
| 14 | mir-148 | 7p15.2 | miR-148a-5p | 4.960±0.58** | miR-148a-3p | 1.650±1.27* |
| 15 | mir-290 | 19q13.42 | miR-371a-5p | 6.256±1.14** | miR-371a-3p | 4.1±0.06* |
| 16 | mir-296 | 20q13.32 | miR-296-5p | 3.083±0.44** | miR-296-3p | 7.560±0.85** |
| 17 | mir-335 | 7q32.2 | miR-335-5p | 4.416±1.65* | miR-335-3p | 5.066±2.11** |
| 18 | mir-339 | 7p22.3 | miR-339-5p | 2.690±0.72* | miR-339-3p | 5.825±0.14** |
| 19 | mir-340 | 5q35.3 | miR-340-5p | 3.283±1.10** | miR-340-3p | 2.625±1.41** |
| 20 | mir-373 | 19q13.42 | miR-373-5p | 2.500±1.27* | miR-373-3p | 7.240±1.39** |
| 21 | mir-515 | miR-515-5p | 7.053±0.63** | miR-515-3p | 4.083±0.45** | |
| 22 | mir-515 | 19q13.42 | miR-518f-5p | 3.901±0.51** | miR-518-3p | 6.971±0.59** |
| 23 | miR-519e-5p | 3.320±0.44** | miR-519e-3p | 3.046±0.55** | ||
| 24 | mir-589 | 7p22.1 | miR-589-5p | 2.410±1.04** | miR-589-3p | 2.221±1.05* |
| 25 | mir-744 | 7p12 | miR-744-5p | 1.536±0.27 | miR-744-3p | 3.393±1.03** |
| 26 | mir-3180 | ? | miR-3180-5p | 4.471±3.73 | miR-3180-3p | 6.253±1.31** |
| B2. Co-downregulated/-shutdown pairs (n=14) | ||||||
| 1* | let-7 | 9q22.32 | let-7d-5p | -9.120±0.35** | let-7d-3p | -6.090±0.55** |
| 2* | mir-10 | 7q21.32 | miR-10a-5p | -12.420±0.40** | miR-10a-3p | -2.790±0.34 |
| 3 | mir-22 | 17p13.3 | miR-22-5p | -7.880±0.04** | miR-22-3p | -4.230±1.84 |
| 4 | mir-24 | 9q22.32 | miR-24-1-5p | -1.927±0.41 | miR-24-3p | -3.300±1.59 |
| 5 | mir-29 | 7q32.3 | miR-29a-5p | -1.807±2.01 | miR-29a-3p | -5.980±0.73 |
| 6 | 7q32.3 | miR-29b-1-5p | -5.503±1.55** | miR-29b-3p | -3.963±1.70 | |
| 7 | mir-31 | 9p21.3 | miR-31-5p | -4.536±0.70* | miR-31-3p | -4.447±0.71 |
| 8 | mir-145 | 5q32 | miR-145-5p | -4.807±2.67 | miR-145-3p | -2.097±1.46 |
| 9 | mir-193 | 17q11.2 | miR-193a-5p | -4.6307±0.66** | miR-193a-3p | -2.026±2.66 |
| 10 | mir-199 | 19p13.2 | miR-199a-5p | -7.677±4.11* | miR-199a-3P | -8.274±2.58 |
| 11 | mir-214 | 1q24.3 | miR-214-5p | -2.869±0.22* | miR-214-3p | -3.834±1.23 |
| 12 | mir-218 | 5q34 | miR-218-5p | -3.257±0.52 | miR-218-2-3p | -1.540±0.68 |
| 13 | mir-221 | Xp11.3 | miR-221-5p | -3.277±0.40** | miR-221-3p | -3.080±0.82* |
| 14 | mir-322 | Xq26.3 | miR-424-5p | -3.650±0.18** | miR-424-3p | -3.934±0.98* |
| B3. Pairs with reverse regulation (n=4) | ||||||
| 1 | mir-139 | 11q13.4 | miR-139-5p | -9.810±0.61** | miR-139-3p | 1.943±0.87* |
| 2 | mir-146 | 10q24.32 | miR-146b-5p | -4.367±2.07 | miR-146b-3p | 1.500±0.31 |
| 3 | mir-196 | 7p15.2 | miR-196b-5p | -5.587±0.86** | miR-196b-3p | 1.500±0.66** |
| 4 | mir-876 | 9p21.1 | miR-876-5p | -3.797±0.72* | miR-876-3p | 1.510±1.30** |
*p<0.05; **p<0.01
Frequent co-expression of the miRNA-5p/3p pairs in many cell types has only recently begun to be reported [20,34-37]. In this study, out of the 261 miRNAs that were found to be differentially expressed in iPSC on reprogramming from MSC, 88 miRNAs (33.7%) were 5p/3p pairs fitting our stringent selection criteria (Table 6) indicating frequent co-expression of miR-5p/3p pairs. Furthermore, out of the 44 co-expressed pairs, 26 and 14 pairs were either co-upregulated or co-down-regulated, respectively, making a total of 40 pairs (90.1%) co-regulated in the same direction (Table 6), consistent with coordinated selection of the 5'- and 3'-arm of the pre-miRNA precursors. Interestingly, four pairs showed reversed directions of expression in iPSC on reprogramming.
Selection of either or both the 5p or 3p miRNA species has been reported to be dependent on temporal, spatial and physiological and pathological conditions [12,14,38,39]. Specific arm selection is thought to be thermodynamically regulated [40,41]. Changes in strand selection in cancer cells and in different tissues and developmental stages are probably associated with the presence of abundant spatial- and temporal-specific signals [12,14,38,42,43]. In cancer cells, minute changes in the steady-state levels of the double-stranded RNA binding protein (dsRBP) and other dsRBP-associating enzymes may have effects on 5p/3p strand selection of concurrent expression [44]. In summary, co-regulation of the 5p/3p miRNA species in normal and pluripotent tissues and cells is most likely to be complex, subjecting to subtle physiological changes pre-miRNA processing enzymes and signals.
Cross-regulation of Targets of Related Biological Functions by miRNA-5p/3p Species
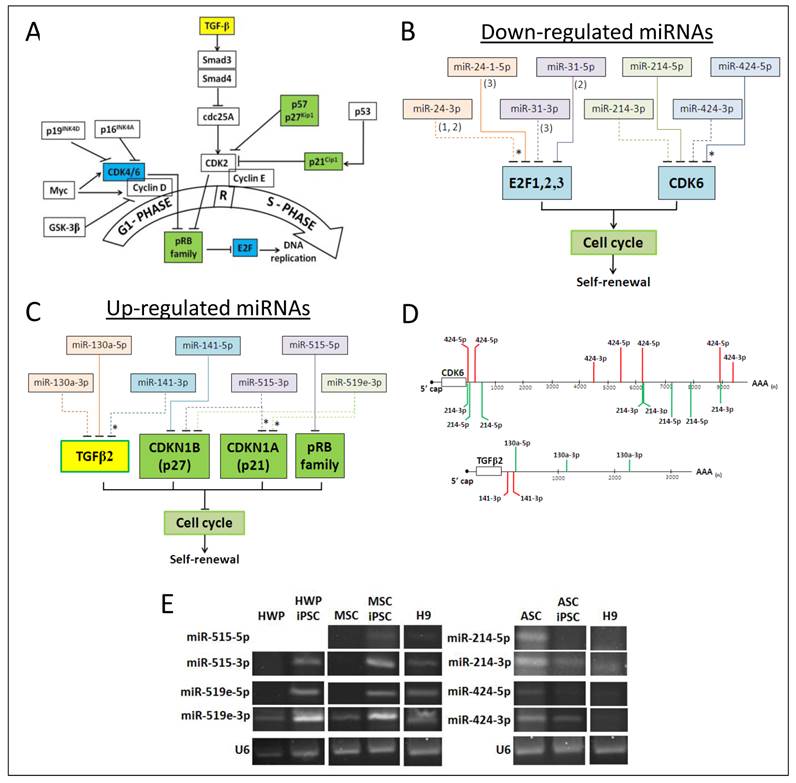
The 5p and 3p miRNA species of the same precursor have partial reverse complementary sequences and should generally be targeting different transcripts. The fact that about one-third of the miRNAs that are differentially expressed in iPSC on reprogramming are paired 5p/3p species with different sequences (Table 6), it may be predicted that different sets of transcripts are regulated by the 5p/3p pair, extending the biological significance of the paired expression. Indeed, when the predicted targets of the paired miRNAs are mined from the various databases, a wide array of targets are found, albeit with many targets falling into similar functional groups of assorted signalling pathways and cell cycle control (Additional file 1: Supplementary Table 4). To further explore cross targeting, targeted cell cycle proteins were further examined since one of the main features of iPSC is continued self-renewal involving active entry from the G1 phase into S phase of the cell cycle (see Fig. 2A). Progression through the G1/S phase check point of the cell cycle is essential to maintain the undifferentiated state of the cells; differentiation occurs when the G1 phase is stalled [45]. ESC has a short cell cycle mode due to the absence of the G1/S restriction which enables cells to rapidly move through the G1 phase and enter the S phase to ensure continuous self-renewal, a defining feature of stem cells [46,47].
Since miRNAs are reported to play a central role in contributing to this unique abbreviated cell cycle structure [5,48], 5p/3p paired miRNAs that were differentially expressed in iPSC on reprogramming were extracted from the dataset and the putative target transcripts of the cell cycle control were identified (Fig. 2). Four miRNA pairs, namely miR-24 and miR-31 are predicted or validated to regulate the E2F factors, miR-214 and miR-424 target CDK6 (Fig. 2B). Importantly, both the 5p and 3p miRNA species are predicted to be targeting at the same transcript despite different sequences. In CDK6, seven miR-424-5p/3p and eight miR-214-5p/3p putative targeting sites are mapped on the ~10 kb 3'-untranslated region (3'UTR) of the CDK6 mRNA (Fig. 2D, upper panel), clearly indicating cross and co-regulation of CDK by both the 5p and 3p species of two independent miRNAs. Down-regulated expression of both 5p and 3p species of miR-424 and -214 was confirmed in stem-loop RT-PCR (Fig. 2E, right panel). Downregulation of miR-24, -31, -214 and -424 in iPSC on reprogramming would have resulted in the upregulation of the E2F factors leading to entry into the S phase in active replication, consistent with the self-renewal characteristic of iPSC. Likewise, TGFβ2, p27 (CDKN1B), p21 (CDKN1A) and the pRB proteins are predicted or validated to be cross-regulated by the 5p and/or 3p species of two or more miRNAs (Fig. 2C). Three miR-130a-5p/3p and two miR-141-3p sites are mapped in the shorter 3'UTR of the TGFβ2 mRNA (Fig. 2D, bottom panel). Upregulation of both the 5p and 3p species of miR-515 and -519e were also confirmed in stem-loop RT-PCR (Fig. 2E, left panel).
miRNA cross-regulation of targets of cell cycle control proteins suggests a fail-proof mode of miRNA regulation in iPSC to ensure that when any one of the regulatory miRNAs is disabled by mutations or by transcriptional or post-transcriptional suppression, other miRNA species in the regulatory circuit are still available to continue to exert the crucial biological function. Co-participation of the 5p/3p species adds further advantages to the fail-proof execution of miRNA regulation. In cancer cells, experimentally validated 5p/3p pairs that often suppress transcripts in related pathogenesis pathways have been reported [20,36,37].
Cell cycle control by co-expressed miRNA pairs iPSC. (A) Simplified view of the G1 to S phase of the cell cycle passing through the restricted (R) point. Factors that are in colored boxes are those that are further investigated. (B) Targeting of the cell cycle factors, E2F1, 2 and 3 and CDK6 by co-expressed miRNA pairs that were down-regulated in iPSC. In E2F targeting, (1), (2) and (3) indicate E2F1, E2F2 and E2F3, respectively. Solid and dashed lines indicate targeting by the 5p or 3p species, respectively. Asterisks indicated validated miRNA targeting. (C) Cell cycle promotion by co-expressed miRNA pairs that were up-regulated in iPSC. See also legend to (C) above. (D) Predicted miRNA targeting sites in the 3'UTR sequences of CDK6 and TGFβ2 as determined by DIANA-microT v5.0. Only 7-and 8-mericalignments between the miRNA seed sequence and the target transcript are depicted. (E) Co-expression of miRNA pairs as determined by stem-loop RT-PCR. PCR products were analyzed in 4% agarose gels. The U6 snRNA was used as an internal control. HWP, human white pre-adipocyte; HWP-iPSC, HWP-derived induced pluripotent stem cell; ASC, adipose stem cell, ASC-iPSC: ASC-derived iPSC; MSC, mesenchymal stem cell from adipose tissue; MSC-iPSC, MSC-derived iPSC.
Conclusions
In this work, frequent co-expression and concerted regulation of miRNA-5p/3p pairs is demonstrated in iPSC. Some 5p and 3p species of the same miRNAs were found to target the same transcript and the same miRNA may cross-target different transcripts of proteins of the G1/S phase transition of the cell cycle in an apparent fail-proof scheme of miRNA regulation. Our data suggest the importance in further elucidation of possible clinical significance of co-existing miRNA-5p/3p pairs in stem cells to explain pluripotency.
Abbreviations
miR, microRNA; iPSCs, induced pluripotent stem cells; MSCs, mesenchymal stem cells; ESC, embryonic stem cells; hES, Human embryonic stem; HWP, human white pre-adipocyte; HWP-iPSC, HWP-derived induced pluripotent stem cell; ASC, adipose stem cell; ASC-iPSC, ASC-derived iPSC; snRNA, small nuclear RNA.
Supplementary Material
Additional File 1Supplementary Table 1-Table 4.
Acknowledgements
This work was supported by University of Malaya HIR-MoE Grant Initiative (Reference number -UM.C/625/1/HIR/MOHE/CHAN/03, account number - A000003 - 50001) to KBC, SKC and TK. CJH was supported by a grant (NSC-101-2313-B-034-003) from the National Science Council, Taiwan.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309(5965):255-6
2. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663-76
3. Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5(2):351-8
4. Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40(12):1478-83
5. Wang Y, Melton C, Li YP. et al. miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell reports. 2013;4(1):99-109
6. Lin SL, Chang DC, Chang-Lin S. et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14(10):2115-24
7. Anokye-Danso F, Trivedi CM, Juhr D. et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376-88
8. Liao B, Bao X, Liu L. et al. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011;286(19):17359-64
9. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205
10. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663-70
11. O'Toole AS, Miller S, Haines N, Zink MC, Serra MJ. Comprehensive thermodynamic analysis of 3' double-nucleotide overhangs neighboring Watson-Crick terminal base pairs. Nucleic Acids Res. 2006;34(11):3338-44
12. Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35(17):5944-53
13. Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat Struct Mol Biol. 2008;15(4):354-63
14. Jagadeeswaran G, Zheng Y, Sumathipala N. et al. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC genomics. 2010;11:52
15. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140-4
16. Almeida MI, Nicoloso MS, Zeng L. et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142(4):886-96 e9
17. Jazdzewski K, Liyanarachchi S, Swierniak M. et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA. 2009;106(5):1502-5
18. Jiang L, Huang Q, Zhang S. et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC cancer. 2010;10:318
19. Martin EC, Elliott S, Rhodes LV. et al. Preferential star strand biogenesis of pre-miR-24-2 targets PKC-alpha and suppresses cell survival in MCF-7 breast cancer cells. Mol Carcinog. 2012 [Epub ahead of print]
20. Shan SW, Fang L, Shatseva T. et al. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J Cell Sci. 2013;126(Pt 6):1517-30
21. Koh W, Sheng CT, Tan B. et al. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC genomics. 2010;11(Suppl 1):S6
22. Huang X, Gschweng E, Van Handel B, Cheng D, Mikkola HK, Witte ON. Regulated expression of microRNAs-126/126* inhibits erythropoiesis from human embryonic stem cells. Blood. 2011;117(7):2157-65
23. Sugii S, Kida Y, Kawamura T. et al. Human and mouse adipose-derived cells support feeder independent induction of pluripotent stem cells. Proc Natl Acad Sci USA. 2010;107(8):3558-63
24. Sugii S, Kida Y, Berggren WT, Evans RM. Feeder-dependent and feeder-independent iPS cell derivation from human and mouse adipose stem cells. Nat Protoc. 2011;6(3):346-58
25. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358-69
26. Mestdagh P, van Vlierberghe P, de Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64
27. Chen C, Ridzon D A, Broomer A J. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179
28. Paraskevopoulou MD, Georgakilas G, Kostoulas N. et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169-73
29. Ong WK, Sugii S. Adipose-derived stem cells: fatty potentials for therapy. Int J Biochem Cell Biol. 2013;45(6):1083-6
30. Sachdeva M, Zhu S, Wu F. et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106(9):3207-12
31. Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647-58
32. Zhong X, Li N, Liang S, Huang Q, Coukos G, Zhang L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem. 2010;285(53):41961-71
33. Tay YM, Tam WL, Ang YS. et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26(1):17-29
34. Mah SM, Buske C, Humphries RK, Kuchenbauer F. miRNA*: a passenger stranded in RNA-induced silencing complex? Crit Rev Eukaryot Gene Expr. 2010;20(2):141-8
35. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347-55
36. Zhang T, Luo Y, Wang T, Yang JY. MicroRNA-297b-5p/3p target Mllt3/Af9 to suppress lymphoma cell proliferation, migration and invasion in vitro and tumor growth in nude mice. Leuk Lymphoma. 2012;53(10):2033-40
37. Zhang Y, Yang P, Sun T. et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nature cell biology. 2013;15(3):284-94
38. Yang JS, Phillips MD, Betel D. et al. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17(2):312-26
39. Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5(6):e11387
40. Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209-16
41. Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199-208
42. Griffiths-Jones S, Hui JH, Marco A, Ronshaugen M. MicroRNA evolution by arm switching. EMBO reports. 2011;12(2):172-7
43. Marco A, Macpherson JI, Ronshaugen M, Griffiths-Jones S. MicroRNAs from the same precursor have different targeting properties. Silence. 2012;3(1):8
44. Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013;19(5):639-48
45. Miller JP, Yeh N, Vidal A, Koff A. Interweaving the cell cycle machinery with cell differentiation. Cell Cycle. 2007;6(23):2932-8
46. Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69(10):4093-6
47. Ghule PN, Medina R, Lengner CJ. et al. Reprogramming the pluripotent cell cycle: restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J Cell Physiol. 2011;226(5):1149-56
48. Stadler B, Ivanovska I, Mehta K. et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19(7):935-50
49. Kuo CH, Deng JH, Deng Q, Ying SY. A novel role of miR-302/367 in reprogramming. Biochem Biophys Res Commun. 2012;417(1):11-6
50. Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30(5):823-34
51. Miyoshi N, Ishii H, Nagano H. et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8(6):633-8
52. Choi YJ, Lin CP, Ho JJ. et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13(11):1353-60
53. Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97-100
54. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529-33
Author contact
![]() Corresponding author: Chiu-Jung Huang, PhD, Professor, Department of Animal Science, School of Agriculture, Chinese Culture University, 55, Hwa-Kang Road, Yang Ming Shan, Taipei 111, Taiwan. Tel: +886 2 28610511x31231; Fax: +886 2 28613100; E-mail: hqr2pccu.edu.tw (C.-J. Huang)
Corresponding author: Chiu-Jung Huang, PhD, Professor, Department of Animal Science, School of Agriculture, Chinese Culture University, 55, Hwa-Kang Road, Yang Ming Shan, Taipei 111, Taiwan. Tel: +886 2 28610511x31231; Fax: +886 2 28613100; E-mail: hqr2pccu.edu.tw (C.-J. Huang)

 Global reach, higher impact
Global reach, higher impact