3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2024; 21(6):1064-1071. doi:10.7150/ijms.90341 This issue Cite
Research Paper
Association of IL-18 gene polymorphisms with clinical aspects of hyperlipidemia in middle-aged and early people in the community
1. School of Life Science, National Taiwan Normal University, Taipei, Taiwan.
2. Translational medicine center, Shin-Kong Wu Ho-Su Memorial Hospital, Taipei City, Taiwan.
3. Department of Medical Research, China Medical University Hospital, China Medical University, Taichung, Taiwan.
4. Department of Neurology, Shin Kong Wu Ho-Su Memorial Hospital, Taipei 111045, Taiwan.
5. School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan.
6. Department of Neurology, Shin Kong Wu Ho-Su Memorial Hospital, Taipei, Taiwan.
7. Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taipei 100025, Taiwan.
8. School of Medicine, Chang Gung University, Taoyuan 33302, Taiwan.
9. Department of Internal Medicine, Chang Gung Memorial Hospital, Keelung 204201, Taiwan.
10. Department of Respiratory Therapy, Fu Jen Catholic University, New Taipei City, Taiwan.
11. Department of Public Health, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
12. School of Public Health, College of Public Health, Taipei Medical University, Taipei, Taiwan.
Received 2023-9-20; Accepted 2024-3-16; Published 2024-4-22
Abstract
Hyperlipidemia is notorious for causing coronary artery disease (CAD). IL-18 is a proinflammtory cytokine that contributes to the pathogenesis of CAD. Previous reports have revealed that genetic polymorphism of IL-18 is associated with its expression level as well as the susceptibility to CAD. In the present study, we aim to investigate the relationship between IL-18 single nucleotide polymorphisms (SNPs) and hyperlipidemia in the Han Chinese population in Taiwan. A total of 580 participants older than 30 were recruited from the community. We collected the demographics, self-reported disease histories, and lifestyles. We also assessed the levels of lipid profiles including total cholesterol (CHOL), triglyceride, low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol. Two SNPs, rs3882891C/A (intron 5) and rs1946518A/C (promoter -607) of IL-18 were elucidated by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) methods. Our results revealed that rs3882891 AA was associated with lower risk of hypercholesterolemia, higher CHOL and LDL-C in subjects (p=0.003, p=0.000 and p=0.005 separately), and rs1946518 CC was associated with hypercholesterolemia, higher CHOL and LDL-C as well (p=0.021, p=0.003 and p=0.001 separately)
Furthermore, both SNPs were associated with IL-18 expression level, which was examined by Genotype-Tissue Expression (GTEx) Portal (p=0.042 and 0.016 separately). Finally, the haplotype of IL-18 was subsequently arranged in the order of rs3882891 and rs1946518. The result revealed that the AC haplotype of 2 IL-18 SNPs was also associated with lower risk of hypercholesterolemia, lower levels of CHOL and LDL-C (p=0.01, p=0.001 and 0.003). The current study is the first to report the association between IL-18 SNPs and hyperlipidemia in the Chinese Han population.
Introduction
Dyslipidemia is notorious for elevated levels of blood cholesterol (CHOL), low-density lipoprotein cholesterol (LDL-C), or triglyceride (TG), while the decreased level of high-density lipoprotein cholesterol (HDL-C) [1]. Dyslipidemia is one of the major risk factors related to coronary artery disease (CAD) [2], regarding to activate inflammatory responses caused by oxidation of LDL-C and very low density lipoprotein cholesterol (VLDL-C) [3, 4]. Genetic factor such as single nucleotide polymorphism (SNP) is reported to have critical roles in dyslipidemia [5].
The pathogenic mechanisms of CAD include endothelium dysfunction, lipid peroxidation, and inflammation [6]. With regard to the pivotal role of inflammation in hyperlipidemia-related CAD [7, 8], investigation of the association between SNPs of inflammatory genes and hyperlipidemia is urgent.
The interleukin-18 (IL-18) was first characterized as an interferon-γ-inducing cytokine, with pleiotropic proinflammatory activities which maintain immune responses [9]. Previous study shows that deletion of IL-18 develops dyslipidemia in mice [10]. Moreover, elevated expression of IL-18 is associated with hypertension, type 2 diabetes, metabolic syndrome, as well as the formation of atherosclerotic plaques [11-14], the development and pathogenesis of CAD [15, 16].
The genetic association of IL-18 and its effect on the progression of CAD has been investigated previously. Several studies have discussed the role of genetic polymorphisms on the IL-18 expression level, which contributes to the prognosis of CAD [17-20]. Furthermore, rs1946518 polymorphism which located on IL-18 promoter region is associated with expression regulation [20]. Previous report showed that rs3882891 polymorphism is associated with an increased risk of acute myocardial infarction [21]. However, the association between these two IL-18 genetic polymorphism and dyslipidemia, which is the major cause of CAD, is rarely defined. Therefore, we evaluated two IL-18 SNPs (rs3882891C/A; NC_000011.10:112144037:C:A and rs1946518A/C; NC_000011.10:112164734:A:C) as candidate biomarkers for susceptibility to dyslipidemia. Our finding here reveals an obvious association between alternative alleles of rs3882891 and rs1946518 polymorphisms, and lower risk of hypercholesterolemia, lower circulating CHOL and LDL-C levels.
Materials and Methods
Subjects
The current study was a community-based prospective cohort study aimed to investigate individuals' the cardiovascular disease and cerebrovascular risk factors. The method of enrolled participant collection and the exclusion criteria were provided elsewhere [22]. All participants lived in Wen Shan and Shi Lin districts near Shin Kong Wu Ho-Su Memorial Hospital and Wan Fang Hospital. Only Chinese Han were included in this study. Exclusion criteria were listed: age ≤ 30, incomplete questionnaire, prior history of cancer or chronic kidney disease, refusal to blood collection or duplex ultrasound measured. Finally, 580 participants were included. All subjects signed a consent form to the original study; no names were published in the results. The study protocol received approval from the institutional ethics committee, and the investigations were conducted according to the Declaration of Helsinki for experiments involving humans. (IRB number: 96E086).
Data collection
Well-trained nurses interviewed all subjects using a structured questionnaire. Subjects' blood was collected after at least 8 hours of fasting. The samples were centrifuged for 15 minutes at 3000 g, separated into plasma, serum, and buffy coat tubes, and stored at 80°C. Technologists assessed CHOL, TG, LDL-C, and HDL-C levels in a standardized laboratory. Hypercholesterolemia is CHOL>250mg/dL or LDL-C>160mg/dL. Hypertriglyceridemia is defined as the level of TG>200mg/dL. Higher CHOL, TG, and LDL-C are defined separately as >200mg/dL, >150mg/dL, and >130mg/dL. Lower HDL-C is defined as <40mg/dL.
Genotyping
The genomic DNA was purified from the buffy coat by using the salting-out method as described in the previous study [23]. Genomic DNA was extracted from the buffy coat using the EasyPure® Genomic DNA Kit (Cat.No.GB100; Bioman Scientific Co., LTD, Taiwan), following the manufacturer's guidelines. The DNA was dissolved in TE buffer (10 mM Tris, pH 7.8, 1 mM EDTA) and preserved at -20°C until further analysis was performed. Genotyping of SNPs was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) methods.
The information of primers, restriction enzymes and PCR conditions for detecting IL-18 SNPs rs1946518A/C and rs3882891C/A was provided in the Supplementary information. Sequence validation and duplicate genotyping from 5% to 10% of blinded samples were done. In order to confirm accuracy, 60 samples were chosen at random and subjected to blind duplicate genotyping. The bases of an additional 30 samples were sequenced and compared to the results of genotyping.
Bioinformatics analysis
We conducted an analysis of genotype-phenotype correlation using data from the Genotype-Tissue Expression (GTEx) Portal (https://www.gtexportal.org/home/) [24], with a significance threshold set at p < 0.05. The following analysis investigated the relationship between rs1946518A/C and rs3882891C/A and IL-18 expression performance in whole blood utilizing information extracted from the GTEx database.
Statistical analysis
For continuous covariates, mean±standard deviation (SD) was showed, and number(percentage) was showed for categorical data. Student's t-test was used to compare continuous variables between two groups. The genotype distribution of each SNP was analyzed for Hardy-Weinberg equilibrium and confirmed by χ2 analysis. χ2 tests were performed to compare the categorical variables between the two groups. Adjusted odds ratios (AORs) with 95% confidence intervals (CIs) were conducted by multiple logistic regression analyses that controlled for age and gender. Haplotype frequencies were analyzed using Haploview, and statistical analysis of the haplotype structure was performed, as summarized in the manuscript [25]. All analyses were performed by using SPSS statistical software (version 25). Statistical significance was set at p < 0.05.
Results
All study subjects were Chinese Han. The total 580 participants were enrolled in the current study. The average age of subjects was approximately 60 years old. About 56% of subjects were male and more than 80% of the subjects had dyslipidemia including hypercholesterolemia (CHOL>250mg/dL or LDL-C>160mg/dL) or and hypertriglyceridemia (TG>200mg/dL). After adjustment of age and gender, the AOR to the risk of lipid abnormalities in IL-18 genotypes are provided in Tables 2. All genotypic frequencies were in Hardy-Weinberang equilibrium (p > 0.05). In both control and case groups, the highest distribution frequencies for rs3882891 and rs1946518 were heterozygous for CA and AC, respectively. The recessive models of rs3882891 showed lower risks of hypercholesterolemia, high CHOL and high LDL-C in participants with higher significant variation (AOR=0.431, CI=0.226-0.823, p=0.011; AOR=0.411, CI=0.263-0.644, p=0.000; AOR=0.602, CI=0.397-0.913, p=0.017). Furthermore, the AA genotype of rs3882891 was associated with the lower risks of hypercholesterolemia, high CHOL and high LDL-C in participants (AOR=0.314, CI=0.148-0.667, p=0.003; AOR=0.337, CI=0.198-0.574, p=0.000; AOR=0.488, CI=0.295-0.807, p=0.005 separately) (Table 2).The recessive models of rs1946518 were significantly associated with lower CHOL level and LDL-C (AOR=0.512, CI=0.325-0.807, p=0.004; AOR=0.544, CI=0.350-0.847, p=0.007) and CC genotype of rs1946518 showed similar phenomenon, with lower risk to hypercholesterolemia (AOR=0.421, CI=0.202-0.877, p=0.021), high CHOL (AOR=0.435, CI=0.254-0.747, p=0.003) and high LDL-C levels (AOR=0.409, CI=0.242-0.692, p=0.001) (Table 2). In our investigation, the AA genotype of rs3882891 and CC genotype of rs1946518 shows lower risk of developing lipid abnormalities.
Demographical characteristics in 580 subjects in current study.
| General Characteristics and Serum Lipid Levels of the Subjects | |
|---|---|
| All subjects (N=580) | Mean±SD or N (%) |
| Age (yrs) | 60.83± 11.468 |
| Gender | |
| Male | 326 (56.2%) |
| Female | 254 (43.8%) |
| Hypertension | SBP>140, DBP>90 |
| No | 343 (59.1%) |
| Yes | 237 (40.9%) |
| Hypercholesterolemia | CHOL>250 or LDL-C>160 |
| No | 483 (89.5%) |
| Yes | 97 (10.5%) |
| Hypertriglyceridemia | TG>200 |
| No | 482 (83.1%) |
| Yes | 98 (16.9%) |
| CHOL_subgroup | >200 |
| No | 351 (60.5%) |
| Yes | 229 (39.5%) |
| TG_subgroup | >150 |
| No | 321 (55.3%) |
| Yes | 259 (44.7%) |
| LDL-C_subgroup | >130 |
| No | 340 (58.6%) |
| Yes | 240 (41.4%) |
| HDL-C_subgroup | <40 |
| No | 382 (65.9%) |
| Yes | 198 (34.1%) |
| SBP | 132.20 ± 22.559 |
| DBP | 82.31 ± 40.190 |
| CHOL | 183.81 ± 59.285 |
| TRIG | 152.09 ± 84.310 |
| LDL | 125.52 ± 32.724 |
| HDL | 47.59 ± 14.663 |
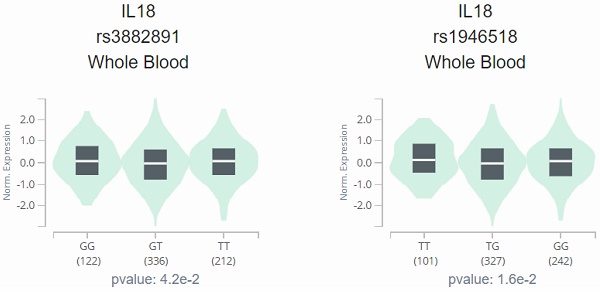
The analysis results of Genotype-Tissue Expression (GTEx) Portal based on IL-18 SNPs (rs1946518 and rs3882891) indicated the inverse relationship between both SNPs and IL-18 expression, and the individuals with the AA genotype of rs3882891 and CC genotype of rs1946518 showed lower expression of IL-18 than reference genotypes in a whole blood sample (p=0.042 and 0.016, Figure 1).
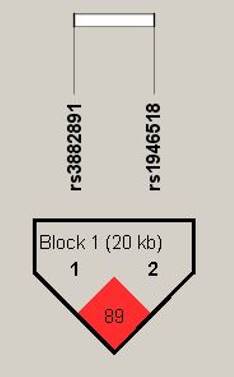
Haplotype analysis based on the genotype data in the order of rs3882891 and rs1946518, revealing these two SNPs were in high linkage disequilibrium (89%) in halpoblock (Figure 2). Both minor alleles of rs3882891 and rs1946518 which compose the AC haplotype has lower risk of developing lipid abnormalities, such as hypercholesterolemia (OR=0.439, CI=0.231-0.835, p=0.01), higher CHOL (OR=0.665, CI=0.521-0.849, p=0.001), and higher LDL-C (OR=0.696, CI=0.546-0.887, p = 0.003) (Table 3).
Odds ratio (OR), adjusted odds ratio (AOR), and related 95% confidence interval (CI) of hyperlipidemia associated with IL-18 genotypic frequencies.
| All subjects (N=580) n (%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Hypercholesterolemia (T-chol>250mg/dL, LDL-C>160mg/dL) | ||||||||
| Gene (SNP) | Genetic model | No (N=483) | Yes (N=97) | OR (95% CI) | p values | AOR (95% CI) | p values | |
| rs3882891 | Dominant | CC | 106 (22%) | 31 (31.6%) | 1 | 1 | ||
| CA+AA | 376 (78%) | 67 (68.4%) | 0.609 (0.378-0.982)* | 0.041 | 0.581 (0.357-0.946)* | 0.029 | ||
| Recessive | CC+CA | 363 (75.3%) | 86 (87.8%) | 1 | 1 | |||
| AA | 119 (24.7%) | 12 (12.2%) | 0.426 (0.225-0.806)* | 0.007 | 0.431 (0.226-0.823)* | 0.011 | ||
| Additive | CC | 106 (22%) | 31 (31.6%) | 1 | 1 | |||
| CA | 257 (53.3%) | 55 (56.1%) | 0.732 (0.446-1.200) | 0.215 | 0.703 (0.425-1.162) | 0.169 | ||
| AA | 119 (24.7%) | 12 (12.2%) | 0.345 (0.169-0.706)* | 0.003 | 0.314 (0.148-0.667)* | 0.003 | ||
| rs1946518 | Dominant | AA | 108 (22.4%) | 29 (29.6%) | 1 | 1 | ||
| AC+CC | 374 (77.6%) | 69 (70.4%) | 0.687 (0.424-1.114) | 0.127 | 0.626 (0.381-1.028) | 0.064 | ||
| Recessive | AA+AC | 379 (78.6%) | 85 (86.7%) | 1 | 1 | |||
| CC | 103 (21.4%) | 13 (13.3%) | 0.563 (0.302-1.049) | 0.067 | 0.564 (0.300-1.061) | 0.076 | ||
| Additive | AA | 108 (22.4%) | 29 (29.6%) | 1 | 1 | |||
| AC | 271 (56.2%) | 56 (57.1%) | 0.770 (0.466-1.270) | 0.304 | 0.710 (0.426-1.184) | 0.189 | ||
| CC | 103 (21.4%) | 13 (13.3%) | 0.470 (0.232-0.954)* | 0.034 | 0.421 (0.202-0.877)* | 0.021 | ||
| Hypertriglyceridemia (TG>200mg/dL) | ||||||||
| Gene (SNP) | Genetic model | No (N=465) | Yes (N=115) | OR (95% CI) | p values | AOR (95% CI) | p values | |
| rs3882891 | Dominant | CC | 107 (23%) | 30 (26.1%) | 1 | 1 | ||
| CA+AA | 358 (77%) | 85 (73.9%) | 0.847 (0.530-1.353) | 0.487 | 0.832 (0.519-1.35) | 0.446 | ||
| Recessive | CC+CA | 356 (76.6%) | 93 (80.9%) | 1 | 1 | |||
| AA | 109 (23.4%) | 22 (19.1%) | 0.773 (0.463-1.289) | 0.322 | 0.796 (0.475-1.333) | 0.385 | ||
| Additive | CC | 109 (22.6%) | 28 (28.9%) | 1 | 1 | |||
| CA | 261 (54%) | 51 (52.6%) | 0.902 (0.553-1.473) | 0.681 | 0.877 (0.535-1.438) | 0.603 | ||
| AA | 113 (23.4%) | 18 (18.6%) | 0.72 (0.391-1.327) | 0.291 | 0.727 (0.393-1.346) | 0.31 | ||
| rs1946518 | Dominant | AA | 107 (23%) | 30 (26.1%) | 1 | 1 | ||
| AC+CC | 358 (77%) | 85 (73.9%) | 0.847 (0.530-1.353) | 0.487 | 0.809 (0.504-1.300) | 0.382 | ||
| Recessive | AA+AC | 366 (78.7%) | 98 (85.2%) | 1 | 1 | |||
| CC | 99 (21.3%) | 17 (14.8%) | 0.641 (0.366-1.124) | 0.118 | 0.651 (0.370-1.145) | 0.136 | ||
| Additive | AA | 107 (23%) | 30 (26.1%) | 1 | 1 | |||
| AC | 259 (55.7%) | 68 (59.1%) | 0.936 (0.577-1.521) | 0.791 | 0.889 (0.544-1.451) | 0.637 | ||
| CC | 99 (21.3%) | 17 (14.8%) | 0.612 (0.318-1.179) | 0.14 | 0.599 (0.31-1.159) | 0.128 | ||
| Higher CHOL (>200 mg/dL) | ||||||||
| Gene (SNP) | Genetic model | No (N=351) | Yes (N=229) | OR (95% CI) | p values | AOR (95% CI) | p values | |
| rs3882891 | Dominant | CC | 71 (20.2%) | 66 (28.8%) | 1 | 1 | ||
| CA+AA | 280 (79.8%) | 163 (71.2%) | 0.626 (0.425-0.922)* | 0.017 | 0.605 (0.407-0.898)* | 0.013 | ||
| Recessive | CC+CA | 252 (71.8%) | 197 (86.0%) | 1 | 1 | |||
| AA | 99 (28.2%) | 32 (14.0%) | 0.413 (0.266-0.642)* | 0.000 | 0.411 (0.263-0.644)* | 0.000 | ||
| Additive | CC | 71 (20.2%) | 66 (28.8%) | 1 | 1 | |||
| CA | 181 (51.6%) | 131 (57.2%) | 0.779 (0.520-1.166) | 0.224 | 0.751 (0.497-1.134) | 0.174 | ||
| AA | 99 (28.2%) | 32 (14%) | 0.348 (0.207-0.585)* | 0.000 | 0.337 (0.198-0.574)* | 0.000 | ||
| rs1946518 | Dominant | AA | 75 (21.4%) | 62 (27.1%) | 1 | 1 | ||
| AC+CC | 276 (78.6%) | 167 (72.9%) | 0.732 (0.497-1.078) | 0.114 | 0.685 (0.461-1.019) | 0.062 | ||
| Recessive | AA+AC | 267 (76.1%) | 197 (86.0%) | 1 | 1 | |||
| CC | 84 (23.9%) | 32 (14.0%) | 0.516 (0.330-0.807)* | 0.003 | 0.512 (0.325-0.807)* | 0.004 | ||
| Additive | AA | 75 (21.4%) | 62 (27.1%) | 1 | 1 | |||
| AC | 192 (54.7%) | 135 (59%) | 0.851 (0.569-1.271) | 0.43 | 0.794 (0.526-1.198) | 0.272 | ||
| CC | 84 (23.9%) | 32 (14%) | 0.461 (0.272-0.781)* | 0.004 | 0.435 (0.254-0.747)* | 0.003 | ||
| Higher TG (>150 mg/dL) | ||||||||
| Gene (SNP) | Genetic model | No (N=321) | Yes (N=259) | OR (95% CI) | p values | AOR (95% CI) | p values | |
| rs3882891 | Dominant | CC | 73 (22.7%) | 64 (24.7%) | 1 | 1 | ||
| CA+AA | 248 (77.3%) | 195 (75.3%) | 0.897 (0.611-1.317) | 0.579 | 0.897 (0.610-1.318) | 0.579 | ||
| Recessive | CC+CA | 247 (76.9%) | 202 (78.0%) | 1 | 1 | |||
| AA | 74 (23.1%) | 57 (22.0%) | 0.942 (0.636-1.394) | 0.765 | 0.944 (0.637-1.399) | 0.775 | ||
| Additive | CC | 73 (22.7%) | 64 (24.7%) | 1 | 1 | |||
| CA | 174 (54.2%) | 138 (53.3%) | 0.905 (0.6041-1.354) | 0.626 | 0.904 (0.603-1.354) | 0.623 | ||
| AA | 74 (23.1%) | 57 (22%) | 0.879 (0.543-1.422) | 0.598 | 0.880 (0.543-1.426) | 0.605 | ||
| rs1946518 | Dominant | AA | 75 (23.4%) | 62 (23.9%) | 1 | 1 | ||
| AC+CC | 246 (76.6%) | 197 (76.1%) | 0.969 (0.659-1.424) | 0.872 | 0.971 (0.659-1.428) | 0.879 | ||
| Recessive | AA+AC | 258 (80.4%) | 206 (79.5%) | 1 | 1 | |||
| CC | 63 (19.6%) | 53 (20.5%) | 1.054 (0.700-1.585) | 0.802 | 1.056 (0.701-1.590) | 0.795 | ||
| Additive | AA | 75 (23.4%) | 62 (23.9%) | 1 | 1 | |||
| AC | 183 (57%) | 144 (55.6%) | 0.952 (0.637-1.422) | 0.81 | 0.953 (0.637-1.426) | 0.815 | ||
| CC | 63 (19.6%) | 53 (20.5%) | 1.018 (0.619-1.672) | 0.945 | 1.021 (0.621-1.679) | 0.935 | ||
| Higher LDL-C (>130mg/dL) | ||||||||
| Gene (SNP) | Genetic model | No (N=340) | Yes (N=240) | OR (95% CI) | p values | AOR (95% CI) | p values | |
| rs3882891 | Dominant | CC | 70 (20.6%) | 67 (27.9%) | 1 | 1 | ||
| CA+AA | 270 (79.4%) | 173 (72.1%) | 0.669 (0.455-0.984)* | 0.041 | 0.656 (0.444-0.970)* | 0.034 | ||
| Recessive | CC+CA | 251 (73.8%) | 198 (82.5%) | 1 | 1 | |||
| AA | 89 (26.2%) | 42 (17.5%) | 0.598 (0.396-0.903)* | 0.014 | 0.602 (0.397-0.913)* | 0.017 | ||
| Additive | CC | 70 (20.6%) | 67 (27.9%) | 1 | 1 | |||
| CA | 181 (53.2%) | 131 (54.6%) | 0.756 (0.505-1.132) | 0.174 | 0.738 (0.491-1.111) | 0.146 | ||
| AA | 89 (26.2%) | 42 (17.5%) | 0.493 (0.3-0.81)* | 0.005 | 0.488 (0.295-0.807)* | 0.005 | ||
| rs1946518 | Dominant | AA | 68 (20%) | 69 (28.7%) | 1 | 1 | ||
| AC+CC | 272 (80%) | 171 (71.3%) | 0.620 (0.421-0.911)* | 0.015 | 0.589 (0.398-0.872)* | 0.008 | ||
| Recessive | AA+AC | 259 (76.2%) | 205 (85.4%) | 1 | 1 | |||
| CC | 81 (23.8%) | 35 (14.6%) | 0.546 (0.353-0.845)* | 0.006 | 0.544 (0.350-0.847)* | 0.007 | ||
| Additive | AA | 68 (20%) | 69 (28.7%) | 1 | 1 | |||
| AC | 191 (56.2%) | 136 (56.7%) | 0.702 (0.47-1.048) | 0.083 | 0.666 (0.443-1) | 0.05 | ||
| CC | 81 (23.8%) | 35 (14.6%) | 0.426 (0.253-0.716)* | 0.001 | 0.409 (0.242-0.692)* | 0.001 | ||
| Lower HDL-C (<40 mg/dL) | ||||||||
| Gene (SNP) | Genetic model | No (N=382) | Yes (N=198) | OR (95% CI) | p values | AOR (95% CI) | p values | |
| rs3882891 | Dominant | CC | 87 (22.8%) | 50 (25.3%) | 1 | 1 | ||
| CA+AA | 295 (77.2%) | 148 (74.7%) | 0.873 (0.585-1.302) | 0.505 | 0.868 (0.570-1.320) | 0.508 | ||
| Recessive | CC+CA | 288 (75.4%) | 161 (81.3%) | 1 | 1 | |||
| AA | 94 (24.6%) | 37 (18.7%) | 0.704 (0.460-1.079) | 0.106 | 0.693 (0.444-1.080) | 0.105 | ||
| Additive | CC | 87 (22.8%) | 50 (25.3%) | 1 | 1 | |||
| CA | 201 (52.6%) | 111 (56.1%) | 0.961 (0.633-1.459) | 0.852 | 0.96 (0.619-1.49) | 0.857 | ||
| AA | 94 (24.6%) | 37 (18.7%) | 0.685 (0.409-1.147) | 0.149 | 0.674 (0.393-1.154) | 0.15 | ||
| rs1946518 | Dominant | AA | 85 (22.3%) | 52 (26.3%) | 1 | 1 | ||
| AC+CC | 297 (77.7%) | 146 (73.7%) | 0.804 (0.540-1.196) | 0.281 | 0.805 (0.530-1.223) | 0.309 | ||
| Recessive | AA+AC | 301 (78.8%) | 163 (82.3%) | 1 | 1 | |||
| CC | 81 (21.2%) | 35 (17.7%) | 0.798 (0.514-1.239) | 0.314 | 0.793 (0.502-1.254) | 0.322 | ||
| Additive | AA | 85 (22.3%) | 52 (26.3%) | 1 | 1 | |||
| AC | 216 (56.5%) | 111 (56.1%) | 0.840 (0.555-1.271) | 0.409 | 0.844 (0.546-1.304) | 0.444 | ||
| CC | 81 (21.2%) | 35 (17.7%) | 0.706 (0.418-1.195) | 0.194 | 0.705 (0.407-1.22) | 0.211 | ||
Odds ratio (OR) and 95% confidence interval (CI) of hyperlipidemia associated with IL-18 rs3882891/rs1946518 haplotype frequencies.
| All subjects (N=1160) n (%) | ||||
|---|---|---|---|---|
| Hypercholesterolemia (T-chol>250mg/dL, LDL-C>160mg/dL) | ||||
| Haplotype | No | Yes | OR (95% CI) | p values |
| rs3882891, rs1946518 | ||||
| CA 2 | 344 (43.7%) | 81 (82.7%) | 1 | |
| AC 3 | 116 (24.1%) | 12 (12.2%) | 0.439 (0.231-0.835)* | 0.01 |
| CC 1 | 19 (3.9%) | 5 (5.1%) | 0.895 (0.324-2.468) | 0.83 |
| AA 4 | 3 (0.6%) | 0 (0%) | 0.809 (0.773-0.848) | 0.401 |
| Higher CHOL (>200 mg/dL) | ||||
| Haplotype | No | Yes | OR (95% CI) | p values |
| rs3882891, rs1946518 | ||||
| CA 2 | 307 (43.7%) | 251 (54.8%) | 1 | |
| AC 3 | 344 (49.0%) | 187 (40.8%) | 0.665 (0.521-0.849)* | 0.001 |
| CC 1 | 16 (2.3%) | 12 (2.6%) | 1.090 (0.506-2.347) | 0.825 |
| AA 4 | 35 (5%) | 8 (1.7%) | 0.280 (0.127-0.614)* | 0.001 |
| Higher LDL-C (>130mg/dL) | ||||
| Haplotype | No | Yes | OR (95% CI) | p values |
| rs3882891, rs1946518 | ||||
| CA 2 | 302 (44.4%) | 256 (53.3%) | 1 | |
| AC 3 | 334 (49.1%) | 197 (41.0%) | 0.696 (0.546-0.887)* | 0.003 |
| CC 1 | 19 (2.8%) | 9 (1.9%) | 1.790 (0.796-4.024) | 0.154 |
| AA 4 | 25 (3.7%) | 18 (3.8%) | 0.849 (0.453-1.592) | 0.61 |
Discussion
The main findings of this work were the obvious associations between genotypes AA and CC of two IL-18 SNPs (rs3882891 and rs1946518) and lower risk of hypercholesterolemia, lower circulating CHOL and LDL-C levels, which may contribute to the progression of CAD. The data also found that subjects who carry genotypic variants show lower circulating IL-18 levels than wild type.
Genetic variation of IL-18 has been proven to associate with or even affect the circulating level of IL-18 in previous reports. The previous study evaluated 5 SNPs located in promoter and 5'UTR regions of IL-18, and the data showed that in response to stimulation, haplotypes 1 (G-C-G-T-C) and 3 (T-A-G-T-C) could enhance promoter transcriptional activity than haplotype 2 (T-A-C-G-T) [20]. Another study investigated IL-18 expression after lipopolysaccharide (LPS) exposure, and the results indicated that the subjects who carried the -137 GG genotype produced more IL-18 than the heterozygous genotype [26]. The IL-18-5848 T>C was associated with IL-18 levels in coronary artery bypass graft (CABG) patients, not only in the baseline but also 6 hours postoperative [27]. Here, we found that both IL-18 SNPs (rs3882891 and rs1946518) were associated with IL-18 expression by utilizing the online Genotype-Tissue Expression (GTEx) Portal. However, the previous study has found no significant association of IL-18 rs1946518 and IL-18 serum level in CAD patients [28]. This could contribute to ethnic differences which lead to a different distribution of the allele frequency.
IL-18 displays a significant eQTL association with rs3882891 (A) and rs1946518 (B) genotypes in lung tissues (GTEx data set).
Displaying the pairwise linkage disequilibrium (LD) patterns of IL-18. A schematic illustration of IL-18 is provided, highlighting the positions of SNP polymorphisms and depicting the D' values for pairwise linkage disequilibrium. The D' values for each SNP are visually- represented using a grayscale, with white denoting low D' and darker shades indicating high D'.
Although the previous report has provided evidence that IL-18 rs1946518 polymorphism was associated with susceptibility to CAD in East Asians [29], there is no relevant report that investigates the association between IL-18 rs1946518 and dyslipidemia, one of the major causes involved in the pathogenesis of CAD. In accordance with previous findings [29], our results indicate that the genotypic variants of IL-18 rs1946518 is associated with lower circulating CHOL and LDL-C levels. Furthermore, previous study shows that rs1946518 polymorphism, located within the IL-18 promoter region, is implicated in the regulation of IL-18 expression [20]. This finding proposes a mechanism by which rs1946518 polymorphism is related to gene regulation of IL-18, which in turn affects lipid levels in circulation and thus contributing to the progression of CAD. However, the previous study reveals that IL-18 knockout (IL-18 -/-) mice develop hypercholesterolemia, hyper-high-density-lipoprotein cholesterolemia, and hypertriglyceridemia while aging [10]. With regard to the pleiotropic property of IL-18 in biological function, complete knockout of IL-18 gene expression may elicit some adverse effects. However, the molecular mechanism involved in lipid metabolism by IL-18 expression should be examined in the future.
The subjects who carry the AA genotypic variant of rs3882891 show a lower prevalence of circulating CHOL and LDL-C levels. Furthermore, the rs3882891 CC genetic variant shows lower expression of IL-18. A previous study indicated a correlation between the rs3882891 polymorphism and an elevated risk of acute myocardial infarction [21]. As high levels of IL-18 have been correlated with an elevated risk of developing cardiovascular diseases, including acute myocardial infarction, and are considered a therapeutic target [30], the impact of the rs3882891 polymorphism on IL-18 expression, as well as its association with dyslipidemia, need to be elucidated in future studies.
Several limitations in this study should be addressed. Although subjects were enrolled from communities in Taiwan, only individuals of Chinese Han were included in this study and the sample size was small exclusion criteria for participant enrollment. Moreover, the present work provided observation results, and the molecular mechanism should be delineated to confirm our findings here.
Supplementary Material
Supplementary information and figures.
Acknowledgements
Institutional Review Board StatementThis study was conducted in accordance with the guidelines laid down in the Declaration of Helsinki. The study protocol was approved by the Institutional review board of the Shin Kong Wu Ho-Su Memorial Hospital (IRB number: 96E086). Eligible participants provided written informed consent.
Funding
This work was supported by grants from Ministry of Science and Technology (MOST107-2314-B-038-072-MY3, MOST110-2314-B-038-056-MY3). Shin-Kong Wu Ho-Su Memorial Hospital and Fu Jen Catholic University (109-SKH-FJU-04, 110-SKH-FJU-04, 111-SKH-FJU-03).
Consent to participate
All of the participants' written informed consent was received before they participated in the study.
Availability of data and materials
The data presented in this study are available from the corresponding author upon request. The data are not publicly available due to privacy restrictions.
Author contributions
Study conception and design: P. C. Chen, C. H. Bai; data collection: W. H. Chen, L. M. Lien; analysis and interpretation of results: P. C. Chen, C. H. Bai; draft manuscript preparation: P.C. Chen; supervision and project administration: C. H. Bai; resources and funding acquisition: C. C. Chou, C. C. Chao.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Michos ED, McEvoy JW, Blumenthal RS. Lipid Management for the Prevention of Atherosclerotic Cardiovascular Disease. The New England journal of medicine. 2019;381:1557-67
2. Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ. et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e596-e646
3. Cushing SD, Berliner JA, Valente AJ, Territo MC, Navab M, Parhami F. et al. Minimally modified low density lipoprotein induces monocyte chemotactic protein 1 in human endothelial cells and smooth muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:5134-8
4. Rajavashisth TB, Andalibi A, Territo MC, Berliner JA, Navab M, Fogelman AM. et al. Induction of endothelial cell expression of granulocyte and macrophage colony-stimulating factors by modified low-density lipoproteins. Nature. 1990;344:254-7
5. Liu Y, Zhou D, Zhang Z, Song Y, Zhang D, Zhao T. et al. Effects of genetic variants on lipid parameters and dyslipidemia in a Chinese population. Journal of lipid research. 2011;52:354-60
6. Tousoulis D, Charakida M, Stefanadis C. Endothelial function and inflammation in coronary artery disease. Heart. 2006;92:441-4
7. Tietge UJ. Hyperlipidemia and cardiovascular disease: inflammation, dyslipidemia, and atherosclerosis. Current opinion in lipidology. 2014;25:94-5
8. Ridker PM. Hyperlipidemia as an instigator of inflammation: inaugurating new approaches to vascular prevention. Journal of the American Heart Association. 2012;1:3-5
9. Gracie JA, Robertson SE, McInnes IB. Interleukin-18. Journal of leukocyte biology. 2003;73:213-24
10. Yamanishi K, Maeda S, Kuwahara-Otani S, Watanabe Y, Yoshida M, Ikubo K. et al. Interleukin-18-deficient mice develop dyslipidemia resulting in nonalcoholic fatty liver disease and steatohepatitis. Transl Res. 2016;173:101-14 e7
11. Hivert MF, Sun Q, Shrader P, Mantzoros CS, Meigs JB, Hu FB. Circulating IL-18 and the risk of type 2 diabetes in women. Diabetologia. 2009;52:2101-8
12. Troseid M, Seljeflot I, Hjerkinn EM, Arnesen H. Interleukin-18 is a strong predictor of cardiovascular events in elderly men with the metabolic syndrome: synergistic effect of inflammation and hyperglycemia. Diabetes care. 2009;32:486-92
13. Dezayee ZM. Interleukin-18 can predict pre-clinical atherosclerosis and poor glycemic control in type 2 diabetes mellitus. International journal of applied & basic medical research. 2011;1:109-12
14. Hernesniemi JA, Karhunen PJ, Oksala N, Kahonen M, Levula M, Rontu R. et al. Interleukin 18 gene promoter polymorphism: a link between hypertension and pre-hospital sudden cardiac death: the Helsinki Sudden Death Study. European heart journal. 2009;30:2939-46
15. Jefferis BJ, Papacosta O, Owen CG, Wannamethee SG, Humphries SE, Woodward M. et al. Interleukin 18 and coronary heart disease: prospective study and systematic review. Atherosclerosis. 2011;217:227-33
16. Ramji DP, Davies TS. Cytokines in atherosclerosis: Key players in all stages of disease and promising therapeutic targets. Cytokine & growth factor reviews. 2015;26:673-85
17. G RK, K MS, G KK, Kurapati M, M S, T MA. et al. Evaluation of Hs-CRP levels and interleukin 18 (-137G/C) promoter polymorphism in risk prediction of coronary artery disease in first degree relatives. PloS one. 2015;10:e0120359
18. Thompson SR, Humphries SE. Interleukin-18 genetics and inflammatory disease susceptibility. Genes and immunity. 2007;8:91-9
19. Hernesniemi JA, Karhunen PJ, Rontu R, Ilveskoski E, Kajander O, Goebeler S. et al. Interleukin-18 promoter polymorphism associates with the occurrence of sudden cardiac death among Caucasian males: the Helsinki Sudden Death Study. Atherosclerosis. 2008;196:643-9
20. Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112:146-52
21. Koch W, Wolferstetter H, Schatke A, Schomig A, Kastrati A. Interleukin 18 gene variation and risk of acute myocardial infarction. Cytokine. 2011;56:786-91
22. Alizargar J, Bai CH. Factors associated with carotid Intima media thickness and carotid plaque score in community-dwelling and non-diabetic individuals. BMC cardiovascular disorders. 2018;18:21
23. Hsieh YC, Seshadri S, Chung WT, Hsieh FI, Hsu YH, Lin HJ. et al. Association between genetic variant on chromosome 12p13 and stroke survival and recurrence: a one year prospective study in Taiwan. Journal of biomedical science. 2012;19:1
24. Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA. et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv Biobank. 2015;13:311-9
25. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-5
26. Arimitsu J, Hirano T, Higa S, Kawai M, Naka T, Ogata A. et al. IL-18 gene polymorphisms affect IL-18 production capability by monocytes. Biochemical and biophysical research communications. 2006;342:1413-6
27. Thompson SR, Novick D, Stock CJ, Sanders J, Brull D, Cooper J. et al. Free Interleukin (IL)-18 levels, and the impact of IL18 and IL18BP genetic variation, in CHD patients and healthy men. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:2743-9
28. Opstad TB, Pettersen AA, Arnesen H, Seljeflot I. Circulating levels of IL-18 are significantly influenced by the IL-18 +183 A/G polymorphism in coronary artery disease patients with diabetes type 2 and the metabolic syndrome: an observational study. Cardiovascular diabetology. 2011;10:110
29. Zheng J, Chen T, Lin H. IL-10, IL-18 Gene Polymorphisms Might Influence Predisposition to Coronary Artery Disease in East Asians: A Meta-Analysis. Immunological investigations. 2021;50:37-46
30. O'Brien LC, Mezzaroma E, Van Tassell BW, Marchetti C, Carbone S, Abbate A. et al. Interleukin-18 as a therapeutic target in acute myocardial infarction and heart failure. Mol Med. 2014;20:221-9
Author contact
![]() Corresponding author: Chyi-Huey Bai, Ph. D., Department of Public Health, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan. TEL: 02-6620-2589 #16013; Email: baichedu.tw.
Corresponding author: Chyi-Huey Bai, Ph. D., Department of Public Health, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan. TEL: 02-6620-2589 #16013; Email: baichedu.tw.

 Global reach, higher impact
Global reach, higher impact