3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(7):909-921. doi:10.7150/ijms.34245 This issue Cite
Research Paper
Possible association of arrestin domain-containing protein 3 and progression of non-alcoholic fatty liver disease
Division of Gastroenterology and Hepatology, Department of Medicine, Nihon University School of Medicine, 30-1 Oyaguchi-kamicho, Itabashi-ku, Tokyo 173-8610, Japan
#These authors equally contributed.
Received 2019-2-19; Accepted 2019-5-3; Published 2019-6-2
Abstract
The prevalence of non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) is increasing worldwide. Several effective drugs for these diseases are now in development and under clinical trials. It is important to reveal the mechanism of the development of NAFLD and NASH. We investigated the role of arrestin domain-containing protein 3 (ARRDC3), which is linked to obesity in men and regulates body mass, adiposity and energy expenditure, in the progression of NAFLD and NASH. We performed knockdown of endogenous ARRDC3 in human hepatocytes and examined the inflammasome-associated gene expression by real-time PCR-based array. We also examined the effect of conditioned medium from endogenous ARRDC3-knockdown-hepatocytes on the apoptosis of hepatic stellate cells. We observed that free acids enhanced the expression of ARRDC3 in hepatocytes. Knockdown of ARRDC3 could lead to the inhibition of inflammasome-associated gene expression in hepatocytes. We also observed that conditioned medium from endogenous ARRDC3-knockdown-hepatocytes enhances the apoptosis of hepatic stellate cells. ARRDC3 has a role in the progression of NAFLD and NASH and is one of the targets for the development of the effective treatment of NAFLD and NASH.
Keywords: ARRDC3, Hepatic Stellate Cells, Inflammasome, NASH, Steatosis
Introduction
The diagnosis rate of nonalcoholic fatty liver disease (NAFLD), including nonalcoholic steatohepatitis (NASH), continues to increase in Western and Eastern countries [1,2]. Fatty liver diseases are growing causes of cirrhosis and hepatocellular carcinoma (HCC) globally [3]. Although it has been reported that various factors are involved in the mechanism of the development of NAFLD and NASH [4], the exact mechanism is still unknown. It is important to elucidate the mechanism of the progression of NAFLD and NASH.
It has been reported that β-arrestins play an important role in metabolism [5, 6]. β-arrestins have been discovered as molecules that bind to and desensitize the activated and phosphorylated form of the G protein-coupled β2-adrenergic receptor [5]. Loss or dysfunction of β-arrestin-2 leads to the disturbance of insulin signaling [6]. β2-adrenergic receptor activation could control the antiapoptotic effects of the 27-kDa heat shock protein (HSP27) through association with β-arrestin [7]. β-arrestin dimerization regulates β2-adrenergic receptor-mitogen activated protein kinase (MAPK) signaling, cell death and proliferation [8,9]. The effects of the β2-agonists via β2-adrenergic receptors increase cAMP and interfere with gene expression of peroxisome proliferator-activated receptors (PPARs), which are transcription factors belonging to the nuclear receptor superfamily [10]. Knockdown of β-arrestin-2 also prevented the cAMP-binding protein Epac1-induced histone deacetylase 4 (HDAC4) nuclear export [11]. β2-adrenergic receptor agonists may possibly exert multiple effects including a direct-effect on liver β2-adrenergic receptors and could promote recovery from insulin-induced hypoglycemia [12].
β-arrestin-2 binds apoptosis signaling-regulating kinase 1 (ASK1), mitogen-activated protein kinase kinase 4 (MKK4), and mitogen-activated protein kinase 10 (JNK3) and promotes JNK3 activation [13]. The activation of ASK1 in hepatocytes is a key step in the progression of NASH [4, 14].
The α-arrestins are broadly expressed and include 6 mammalian members referred to as arrestin domain-containing proteins (ARRDCs) [15]. The α-arrestins also have a similar structure to β-arrestins, and these play roles in G protein-coupled receptor trafficking [15]. The α-arrestin family includes thioredoxin-interacting protein (Txnip) which has crucial functions in regulating glucose uptake and glycolytic flux through the mitochondria [16], and arrestin domain-containing protein 3 (ARRDC3), which is linked to obesity in men and regulates body mass, adiposity, and energy expenditure [16, 17]. ARRDC3 is localized in the cytoplasm and expressed in the liver.
A genome-wide association study (GWAS) identified a single nucleotide polymorphism (SNP) upstream of the ARRDC3 locus strongly associated with prognosis in early-onset breast cancer [18]. Genome-wide association analysis in East Asians also identified an SNP near the ARRDC3 gene associated with breast cancer risk [19].
In the present study, we observed the enhancement of ARRDC3 expression by the addition of oleic acids in human hepatoma cells. We have also used the siRNA targeting ARRDC3 to inhibit the expression of endogenous ARRDC3 in human hepatoma HepG2 cells and determined its effect on inflammasome pathway-associated gene expression. Furthermore, we treated human hepatic stellate cell line LX-2 with conditioned media from HepG2 cells transfected with or without ARRDC3-targeted siRNA and evaluated apoptosis of hepatic stellate cells. We have observed that the depletion of ARRDC3 in human hepatocytes resulted in the downregulation of inflammasome pathway-associated genes such as chemokine (C-X-C motief) ligand 2 (CXCL2), interleukin 6 (IL6), chemokine (C-C motief) ligand 5 (CCL5), caspase 5 (CASP5) and interferon, beta 1 (IFNB), and the enhancement of apoptosis of hepatic stellate cells treated with their conditioned media. Our results demonstrated ARRDC3 may play a role in the development of NAFLD and NASH.
Results and Discussion
Human hepatocytes express ARRDC3 mRNA
We previously observed that ARRDC3 mRNA was significantly higher expressed in the liver of NASH model rat SHRSP5/Dmcr [20] at week 4 after feeding a normal diet compared with those of the stroke-prone spontaneously hypertensive rat (SHRSP/Izm) (data not shown). SHRSP5/Dmcr or SHRSP/Izm, respectively, develops or not develops NASH at week 19 after feeding a high fat, high cholesterol-containing diet. Previous studies have demonstrated that various human cell lines express ARRDC3 [17, 21].
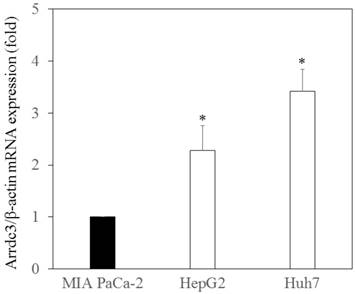
First, we examined ARRDC3 mRNA expression in the human hepatoma cell lines, HepG2 and Huh7, compared with that in human pancreatic cancer cell line MIAPaCa-2. Cellular RNA was extracted from these cell lines, and ARRDC3 mRNA levels were examined by real-time RT-PCR (Figure 1). We observed that human hepatocytes express ARRDC3 mRNA significantly higher than human pancreatic cancer cells.
Hoque et al. [22] reported that lactate negatively regulates toll-like receptor (TLR) induction of Nucleotide‑binding oligomerisation domain (NOD)-like receptor protein 3 (NLRP3) inflammasome and production of interleukin 1β (IL1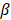 ), via β2-arrestin and the plasma membrane Gi protein coupled receptor (GPR)-81 and reduces organ injury in liver and pancreas. So, we also used human pancreatic cancer cells. As oleic acid induced steatosis and cytotoxicity on rat hepatocytes in primary culture [23], we did not use human primary hepatocytes in the present study.
), via β2-arrestin and the plasma membrane Gi protein coupled receptor (GPR)-81 and reduces organ injury in liver and pancreas. So, we also used human pancreatic cancer cells. As oleic acid induced steatosis and cytotoxicity on rat hepatocytes in primary culture [23], we did not use human primary hepatocytes in the present study.
Oleic acids enhance ARRDC3 mRNA expression
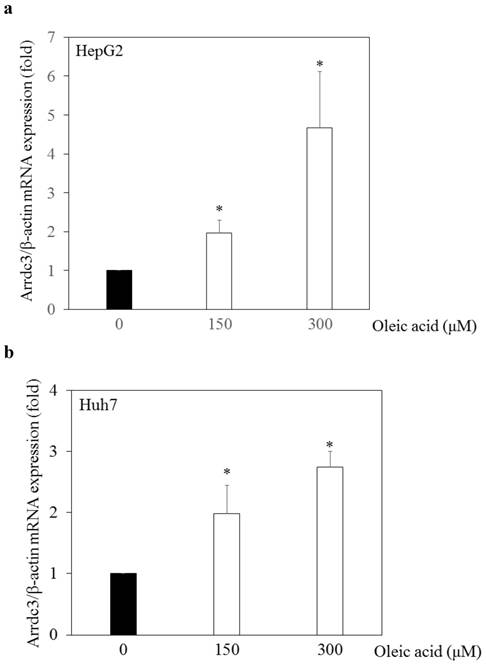
Next, we examined the effects of oleic acid, which induces steatosis in hepatocytes [24], on ARRDC3 mRNA expression in human hepatoma cell lines. We previously demonstrated that free fatty acids such as oleic acid and/or palmitic acid induced fat deposition in human hepatoma cell lines by Nile red stain [25]. We added oleic acid (0 μM, 150 μM or 300 μM) into cell culture medium of HepG2 or Huh7 cells. Twenty-four hours after the addition of oleic acid, cellular RNA was extracted and ARRDC3 mRNA levels were measured by real-time RT-PCR (Figure 2a and 2b). In both HepG2 and Huh7 cell lines, oleic acids enhanced ARRDC3 mRNA expression in a dose-dependent manner. Thus, fat deposition might be associated with ARRDC3 mRNA expression in hepatocytes.
Arrestin domain-containing protein 3 (ARRDC3) mRNA expressed in human hepatoma cells. ARRDC3 and β-actin mRNA levels were measured by real-time RT-PCR in HepG2, Huh7 and pancreatic cancer MIAPaCa-2 cells. *p < 0.05, compared with MIA PaCa-2 cells.
Effects of oleic acid on arrestin domain-containing protein 3 (ARRDC3) mRNA expression levels in human hepatoma cell lines. (a) HepG2 and (b) Huh7 cells. Real-time RT-PCR analyses of ARRDC3 and β-actin mRNA levels in HepG2 and Huh7 cells treated with or without 150 μM or 300 μM oleic acid for 24 hours. *p < 0.05, compared with 0 μM oleic acid.
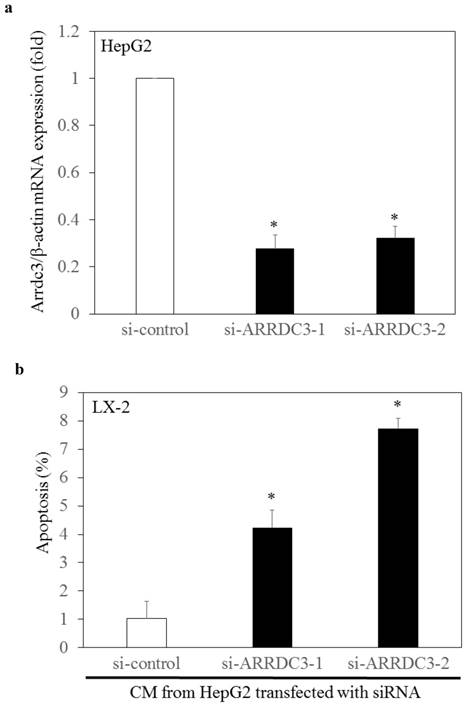
Conditioned media from endogenous arrestin domain-containing protein 3 (ARRDC3)-knockdown-HepG2 enhances apoptosis of hepatic stellate cell line LX-2. (a) ARRDC3 mRNA expression was significantly inhibited by transfection with si-ARRDC3, compared with that of si-control. si-ARRDC3-1 and si-ARRDC3-2 indicate different set of experiments. (b) Conditioned media (CM) from ARRDC3-knockdown HepG2 enhanced LX-2 cell apoptosis, compared with that of control HepG2 cells. *p < 0.05, compared with control siRNA (si-control).
Conditioned media from endogenous ARRDC3-knockdown-HepG2 enhances apoptosis of hepatic stellate cells
It is not clear whether the ARRDC3 expression in hepatocytes have any effects on human hepatic stellate cells. We investigated whether knockdown of endogenous ARRDC3 in HepG2 cells had effects on apoptosis in human hepatic stellate cell line LX-2. Forty-eight hours after transfection of siRNA into HepG2 cells, we confirmed the knockdown of ARRDC3 mRNA by real-time RT-PCR (Figure 3a). We also collected conditioned medium from HepG2 cells transfected with si-ARRDC3 or si-control, and cellular apoptosis of LX-2 cells was examined 72 hours after incubation of these media by APOPercentage apoptosis assay (Figure 3b). Cellular apoptosis of hepatic stellate cells increased after the incubation of conditioned media from ARRDC3-knockdowned HepG2 cells, compared with that from control HepG2 cells. These results suggested that upregulation of ARRDC3 in hepatocytes might inhibit hepatic stellate cell apoptosis, resulting in the progression of liver fibrosis. Although we also tried to detect apoptosis of LX-2 cells by apoptosis marker Annexin V [26], we did not see any differences more clearly (data not shown). Further studies will be needed.
Knockdown of ARRDC3 inhibits inflammasome-associated gene expression in human hepatocytes
Inflammasomes and cytokines are major players in the induction of hepatocyte apoptosis in NAFLD and NASH [4]. To further explore the mechanism, we have examined inflammasome-related gene expression profiles using real-time PCR-based focused microarrays to compare between HepG2 cells transfected with si-ARRDC3 and those with siRNA. The Inflammasome-associated gene expression between HepG2 cells transfected with si-ARRDC3 and si-control were compared using inflammasome-associated signaling target PCR array.
Out of 84 inflammasome-associated genes examined, one and 13 genes were significantly upregulated and downregulated, respectively, in HepG2 cells transfected with si-ARRDC3, compared with the si-control (p < 0.05; Table 1). Five genes (CCL5, CASP5, IL6, IFNB1 and CXCL2) were downregulated 3-fold or more. Heat shock protein 90 kDa alpha (cytosolic), class A member 1 (HSP90AA1) was the only gene that was significantly upregulated.
Effects of knockdown of endogenous arrestin domain-containing protein 3 (ARRDC3) on inflammasome-associated gene expression in human HepG2 cells. Changes of gene expression in HepG2 cells transfected with si-ARRDC3, compared with si-control.
| Gene Symbol | Pathways | si-ARRDC3 vs. si-control | p-values |
|---|---|---|---|
| HSP90B1 | Inflammasomes (Negative regulation) | -1.57 | 0.000089 |
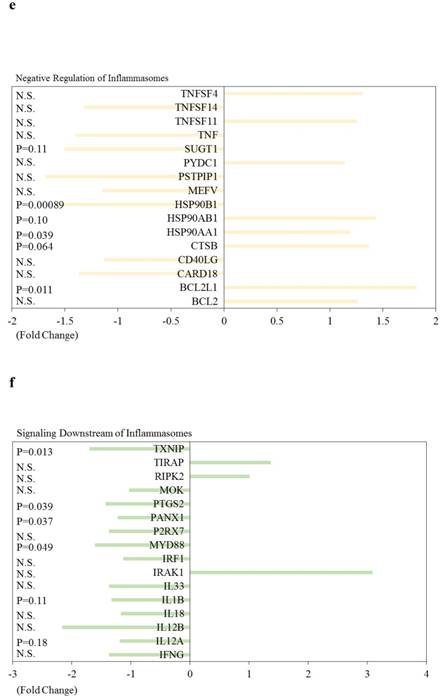
| BIRC3 | Signaling Downstream of NOD-Like Receptors | -1.90 | 0.0011 |
| CXCL2 | Signaling Downstream of NOD-Like Receptors | -3.69 | 0.0011 |
| IL6 | Signaling Downstream of NOD-Like Receptors | -6.90 | 0.0017 |
| CCL5 | Signaling Downstream of NOD-Like Receptors | -10.56 | 0.0069 |
| CASP1 | Inflammasomes (IPAF/NLRP1/NLRP3) | -1.51 | 0.0085 |
| CASP5 | Inflammasomes (NLRP1) | -10.06 | 0.010 |
| TXNIP | Signaling Downstream of Inflammasomes | -1.70 | 0.013 |
| MAP3K7 | Signaling Downstream of NOD-Like Receptors | -1.30 | 0.021 |
| PANX1 | Signaling Downstream of Inflammasomes | -1.22 | 0.037 |
| HSP90AA1 | Inflammasomes (Negative regulation) | 1.19 | 0.039 |
| PTGS2 | Signaling Downstream of Inflammasomes | -1.43 | 0.039 |
| MYD88 | Signaling Downstream of Inflammasomes | -1.61 | 0.049 |
| IFNB1 | Signaling Downstream of NOD-Like Receptors | -4.61 | 0.050 |
HSP90B1, heat shock protein 90 beta family member 1; BIRC3, baculoviral IAP repeat containing 3; CXCL2, C-X-C motif chemokine ligand 2; IL6, interleukin 6; CCL5, C-C motif chemokine ligand 5; CASP1, caspase 1; CASP5, caspase 5; TXNIP, thioredoxin interacting protein; MAP3K7, mitogen-activated protein kinase kinase kinase 7; PANX1, pannexin 1; HSP90AA1, heat shock protein 90 alpha family class A member 1; PTGS2, prostaglandin-endoperoxide synthase 2; MYD88, myeloid differentiation primary response 88; IFNB1, interferon beta 1; IPAF (NLRC4), NLR family CARD domain containing 4; NLRP1, NLR family pyrin domain containing 1; NLRP3, NLR family pyrin domain containing 3.
Expression levels of endoplasmic reticulum molecule Heat shock protein 90 kDa beta (Grp94), member 1 (HSP90B1) were significantly up-regulated in the livers of zebrafish larvae fed high fat with or without high cholesterol diets [27]. Baculoviral IAP repeat containing 3 (BIRC3), a severe hypoxia-activated gene, was significantly increased in simple hepatic steatosis compared with the controls [28]. A Western-type cholesterol-containing diet significantly induced hepatic expression of CXCL2 [29]. IL6 levels were increased in NASH and correlated with GP130 expression [30]. Steatosis induced CCL5/RANTES was associated with early-stage liver fibrosis in the progression of NAFLD [31]. NLRP3 inflammasome, pro-IL1β, active-CASP1 and IL1β activation occurs in NAFLD [32].
Elevation of ceramide levels was associated with activation of CASP5 and the subsequent cleavage of HuR and apoptotic cell death in the liver [33]. The reactive oxygen species (ROS)-thioredoxin interacting protein (TXNIP) pathway mediates hepatocellular NOD-like receptor (NLR) family pyrin domain containing 3 (NLRP3) inflammasome activation, inflammation and lipid accumulation in fructose-induced NAFLD [34]. Mitogen-activated protein kinase kinase kinase 7 (MAP3K7) induced adipocyte differentiation through peroxisome proliferator-activated receptor gamma (PPARγ) signaling [35].
Pannexin 1 (PANX1)-dependent pathophysiological extracellular ATP release in lipoapoptosis is capable of stimulating migration of human monocytes in chronic liver injury induced by free fatty acids [36]. HSP90AA1 is one of the nine critical genes related to the pathogenesis of hepatocellular carcinoma [37]. Prostaglandin-endoperoxide synthase 2 (PTGS2) and myeloid differentiation primary response gene 88 (Myd88) are also associated with NAFLD and NASH [38, 39]. Mitochondrial damage in steatohepatitis extends to mitochondrial antiviral-signaling protein MAVS, an adapter of helicase receptors, resulting in inefficient type I IFN and inflammatory cytokine response [40]. Thus, it is possible that ARRDC3 might be involved in the inflammasome-associated pathways involved in the pathogenesis of NAFLD and NASH.
We performed further pathway analysis. Effects of knockdown of ARRDC3 on inflammasome-associated pathways in human hepatocytes are shown in Figure 4. Most of inflammasome-associated genes were downregulated in HepG2 cells transfected with si-ARRDC3, compared with the si-control. However, among negative regulation molecules of inflammasomes, HSP90AA1 was significantly upregulated and B-cell CLL/lymphoma 2 (BCL2)-like 1 (BCL2L1), cathepsin B (CTSB), heat shock protein 90 kDa alpha, class B member 1 (HSP90AB) tended to be upregulated.
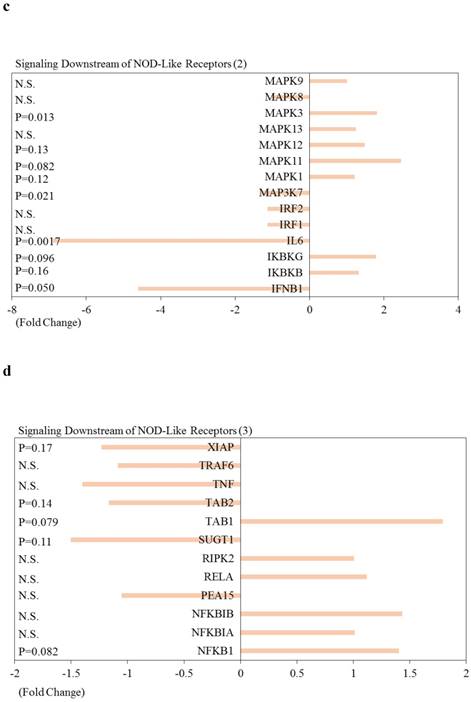
Effects of knockdown of endogenous arrestin domain-containing protein 3 (ARRDC3) on inflammasome-associated pathways in human HepG2 cells. Changes of gene expression in HepG2 cells transfected with si-ARRDC3, compared with si-control. (a) Absent in melanoma 2 (AIM2), (b) Ice protease-activating factor (IPAF), (c) Nucleotide‑binding oligomerisation domain (NOD)‑like receptor protein 1 (NLRP1), (d) NOD-like receptor family pyrin domain containing 3 (NLRP3), (e) Negative regulation of inflammasomes, (f) Signaling downstream of inflammasomes. P, p-values. N.S., not statistically significant difference.
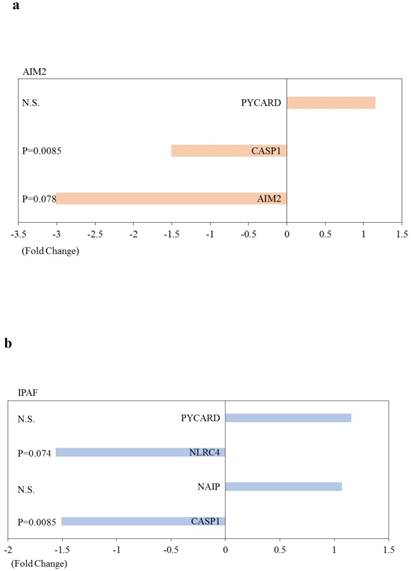
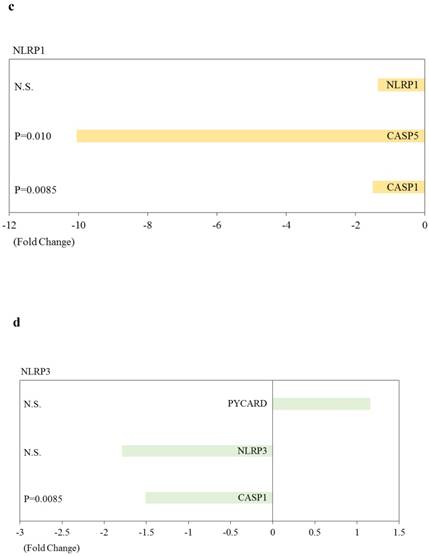
We performed further pathway analysis. Effects of knockdown of ARRDC3 on inflammasome-associated pathways in human hepatocytes are shown in Figure 4. Most of inflammasome-associated genes were downregulated in HepG2 cells transfected with si-ARRDC3, compared with the si-control. However, among negative regulation molecules of inflammasomes, HSP90AA1 was significantly upregulated and B-cell CLL/lymphoma 2 (BCL2)-like 1 (BCL2L1), cathepsin B (CTSB), heat shock protein 90 kDa alpha, class B member 1 (HSP90AB) tended to be upregulated.
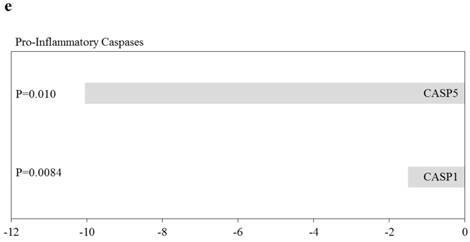
Effects of knockdown of ARRDC3 on Nucleotide‑binding oligomerisation domain (NOD)‑like receptor-associated pathways and pro-inflammatory caspases in human hepatocytes are shown in Figure 5. Among NOD-like receptor-related molecules, NLR family, CARD domain containing 4 (NLRC4) and NLR family, pyrin domain containing 9 (NLRP9) tended to be downregulated, and NLR family member X1 (NLRX1) and NOD1 tended to be upregulated (Figure 5). Of interest, among Signaling downstream of NOD‑like receptor-related molecules, Fas-associated via death domain (FADD), inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKBKB), inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma (IKBKG), Mitogen-activated protein kinase 1 (MAPK1), MAPK3, MAPK11, MAPK12, nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NFKB1) and transforming growth factor (TGF)-beta activated kinase 1/MAP3K7 binding protein 1 (TAB1) tended to be upregulated (Figure 5b-5d). Two inflammatory caspases were significantly downregulated in HepG2 cells transfected with si-ARRDC3, compared with the si-control (Figure 5e).
In the present study, we demonstrated that free fatty acids induced ARRDC3 mRNA expression in hepatocytes and that upregulation of ARRDC3 in hepatocytes is associated with inhibition of hepatic stellate cell apoptosis, which may lead to the progression of liver fibrosis. We also demonstrated that ARRDC3 is strongly associated with inflammasome-associated gene expression. These results indicate that ARRDC3 plays a role in the progression of NAFLD and NASH.
A previous study [17] has shown that ARRDC3 deficiency in mice protects against obesity. ARRDC3 is a gene required for β2-adrenergic receptor regulation and colocalizes with β2-adrenergic receptors [41]. ARRDC3 also plays an important role in neural precursor development downregulated protein 4 (NEDD4)-mediated ubiquitination and endocytosis of activated β2-adrenergic receptors and subsequent β2-adrenergic receptor degradation [41]. Shi et al. [42] reported that abrogation of β2-adrenergic receptors is known to modulate hepatic lipid accumulation and glucose tolerance in aging mice. Of interest, in the present study, we found an association between lipid accumulation and ARRDC3 expression in hepatocytes (Figure 1).
Two E3 ligases NEDD4 and NEDD4l, which are known to regulate membrane protein internalization and degradation via the endocytic pathway [43], are the proteins responsible for transmembrane BAX inhibitor motif-containing 1 (TMBIM1) ubiquitination [44]. TMBIM1 is an effective suppressor of steatohepatitis and a previously unknown regulator of the multivesicular body (MVB)-lysosomal pathway via targeting of the lysosomal degradation of TLR4 [44].
Effects of knockdown of endogenous arrestin domain-containing protein 3 (ARRDC3) on Nucleotide‑binding oligomerisation domain (NOD)‑like receptor-associated pathways and pro-inflammatory caspases in human HepG2 cells. Changes of gene expression in HepG2 cells transfected with si-ARRDC3, compared with si-control. (a) NOD‑like receptors, (b), (c), (d) Signaling downstream of NOD‑like receptors, (e) Pro-inflammatory caspases. P, p-values. N.S., not statistically significant difference.
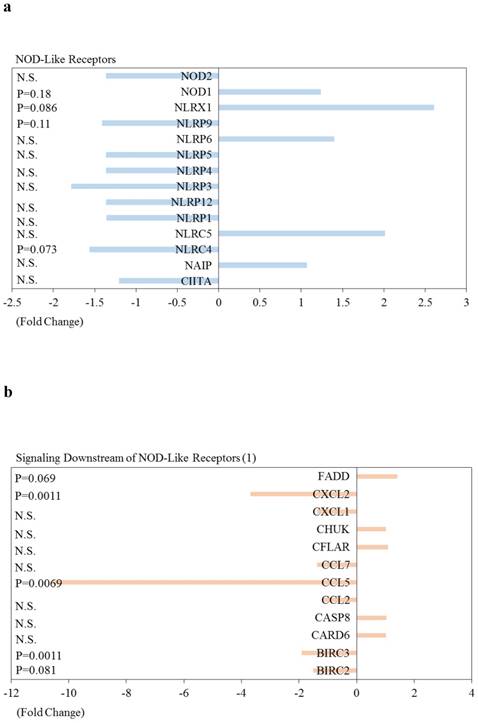
We also observed that knockdown of ARRDC3 in human hepatocytes down-regulates inflammasome-associated gene expression (Table 1). It has been reported that activation of inflammasomes plays a role in the development of NAFLD and NASH [27-40, 44]. The association between ARRDC3 and inflammasome-related pathways may have a role in the development of NAFLD and NASH. Further studies will be needed to clarify this point.
Cell death is very important in the progression of NAFLD and NASH [4]. β-adrenergic receptor stimulation clearly induced the expression of v-raf-leukemia viral oncogene 1 (RAF-1) [45]. Inhibition of the pro-apoptotic function of ASK1 by RAF-1 may be the reason for maintaining survival [46]. Inhibition of the ASK1 pathway through the suppression of ARRDC3 may provide a novel mechanism in the management of NAFLD and NASH.
The number of patients with NAFLD and NAS has been increasing in the USA, Europe and Asian countries [3, 4]. NAFLD and NASH can lead to advanced liver diseases including cirrhosis and HCC [3]. Selonsertib which is a serine/threonine kinase inhibitor and targets ASK1 is now in phase III clinical trial for the treatment of NASH [47]. In phase II clinical trials of this drug, according to magnetic resonance (MR) elastography and biopsies at baseline and week 24, 33% (18/54) had fibrosis improvement (≥1-stage reduction) after undergoing 24 weeks of treatment with the study drug [48]. According to MR imaging-estimated proton density fat fraction and biopsies at baseline and week 24, a ≥1-grade reduction in steatosis was observed in 28% (18/65) [48]. A combination therapy of anti-inflammatory and anti-fibrotic intervention could be effective for NAFLD and NASH. ASK1 pathway plays a role in both inflammation and fibrosis of NAFLD and NASH [4, 49, 50].
Materials and Methods
Cell lines and reagents
Human hepatoma cell lines (HepG2 and Huh7), hepatic stellate cell line LX-2 and human pancreatic cancer MIAPaCa-2 cells were maintained in Roswell Park Memorial Institute medium (RPMI 1640) (Sigma, St. Louis, MO, USA) supplemented with 1-10% fetal bovine serum, penicillin (100 U/mL) and streptomycin (100 μg/mL) at 5% CO2 and 37°C. HepG2, Huh7 and MIAPaCa-2 cells were purchased from the Japanese Collection of Research Bioresources Cell Bank (Ibaraki, Osaka, Japan) [26, 51]. LX-2 cells, spontaneously immortalized cells, were kindly provided by Prof. Scott L. Friedman, Mount Sinai Medical School, NY, USA [52]. Oleic acid-albumin from bovine serum was purchased from Sigma.
Incubation of human hepatoma cell lines with oleic acids
Before 24 hours of treatment with oleic acids, HepG2 and Huh7 cells were seeded in 6-well plates at a density of 0.5 x 106 cells/well. Cells were washed with PBS and incubated with or without 150 μM or 300 μM oleic acids in RPMI with 10% fetal bovine serum for 24 hours.
RNA extraction, cDNA synthesis and real-time reverse transcription-PCR (RT-PCR)
Cellular RNA was isolated from cells by using the RNeasy Mini Kit (Qiagen, Tokyo, Japan). cDNA synthesis was performed by using PrimeScript RT reagent (Perfect Real Time) (Takara Bio, Otsu, Shiga, Japan) with random hexamers and oligo dT primers on GeneAmp PCR system 5700 (Applied Biosystems, Foster, CA, USA). PCR amplification was performed on cDNA templates using primers specific for ARRDC3 (sense primer [5'-ATCCCAGTGTGATGTGACGA-3'] and antisense primer [5'-TTTGCAACAGAATCGGAAAA-3']) and for actin-beta (sense primer [5'-CAGCCATGTACGTTGCTATCCAGG-3']) and antisense primer [5'-AGGTCCAGACGCAGGATGGCATG-3']). For RNA quantification, real-time PCR was performed by using Power SYBR Green Master Mix (Thermo Fisher Scientific, Tokyo, Japan) with a 7500 Fast real-time PCR system (Applied Biosystems) as described previously [53]. The actin housekeeping gene was used for normalization, and data were analyzed by the comparative threshold cycle method. Relative quantification of gene expression using the 2-ΔΔCt method correlated with absolute gene quantification obtained by standard curve [53]. Each real-time PCR assay was performed in triplicate.
Transfection of small interfering RNA (siRNA)
To transiently knockdown ARRDC3, approximately 0.5×105 cells were seeded in 35 mm-plates (Iwaki Glass, Tokyo, Japan) 24 hours prior to transfection. Cells were transfected with 50 nM each of siRNA specific for ARRDC3 (si-ARRDC3) or control siRNA (si-control), using Effectene transfection reagent (Qiagen) according to the manufacturer's protocol [53]. After 48 hours of transfection, cellular RNA and conditioned medium were collected.
Detection of apoptosis of LX-2 cells
After 72 hours of incubation with conditioned media from HepG2 cells transfected with si-ARRDC3 or si-control, the APOPercentage apoptosis assay (Biocolor, Belfast, Northern Ireland) was used to evaluate apoptosis of LX-2 cells following the manufacturer's instruction. Transfer and exposure of phosphatidylserine to the exterior surface of the membrane have been linked to the onset of apoptosis. Phosphatidylserine transmembrane movement results in uptake of APOPercentage dye by apoptosis-committed cells. Purple-red stained cells were identified as apoptotic cells by light microscopy [26].
Inflammasomes-associated signaling target PCR array
HepG2 cells were transfected with 50 nM each of si-ARRDC3 or si-control. After 48 hours of transfection, cellular RNA was extracted from both cells using the RNeasy Mini Kit (Qiagen). cDNA was synthesized with an RT2 First Strand cDNA Kit (Qiagen) according to the manufacturer's protocol. To examine the expression of 84 inflammasome-associated genes, a human inflammasomes RT2 Prolifer PCR array (Qiagen) was performed with the SYBR Green real-time PCR-based method on 7500 Fast real-time PCR system (Applied Biosystems)[20]. The cycling program was as follows: 95°C for 10 minutes for 1 cycle, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were analyzed using RT2 Profiler PCR Array Data Analysis software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Gene expression was normalized to 5 internal control genes (beta-actin, beta-2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, hypoxantine phosphoribosyltransferase 1 and ribosomal protein, large, P0) to determine the fold change in gene expression by 2-ddCT (comparative cycle threshold) method.
Statistical analysis
All experiments were repeated at least three times independently, and all statistical analyses were performed using DA Stats software (O. Nagata, Nifty Serve: PAF01644). Statistical analyses were performed using a 2-tailed Student t-test or Welch t-test for paired data.
Conclusion
Recent studies demonstrated that ARRDC3 also play roles in human cancer signaling [54, 55]. We identified ARRDC3 as an important positive regulator in NAFLD and NASH. Targeting ARRDC3 may be a good strategy to develop a novel therapeutic method against NAFLD and NASH.
Abbreviations
NAFLD: Non-alcoholic fatty liver disease; NASH: Non-alcoholic steatohepatitis; ARRDC3: Arrestin domain-containing protein 3; AIM2: Absent in melanoma 2; BCL2: B-cell CLL/lymphoma 2; BCL2L1: BCL2-like 1; BIRC2: Baculoviral inhibitor of apoptosis (IAP) repeat containing 2; BIRC3: Baculoviral IAP repeat containing 3; CARD18: Caspase recruitment domain family, member 18; CARD6: Caspase recruitment domain family, member 6; CASP1: Caspase 1, apoptosis-related cysteine peptidase; CASP5: Caspase 5, apoptosis-related cysteine peptidase; CASP8: Caspase 8, apoptosis-related cysteine peptidase; CCL2: Chemokine (C-C motief) ligand 2; CCL5: Chemokine (C-C motief) ligand 5; CCL7: Chemokine (C-C motief) ligand 7; CD40LG: CD40 ligand; CFLAR: CASP8 and FADD-like apoptosis regulator; CHUK: Conserved helix-loop-helix ubiquitous kinase; CIITA: Class II, major histocompatibility complex, transactivator; CTSB: Cathepsin B; CXCL1: Chemokine (C-X-C motief) ligand 1; CXCL2: Chemokine (C-X-C motief) ligand 2; FADD: Fas-associated via death domain; HSP90AA1: Heat shock protein 90 kDa alpha, class A member 1; HSP90AB1: Heat shock protein 90 kDa alpha, class B member 1; HSP90B1: Heat shock protein 90 kDa beta (Grp94), member 1; IFNB1: Interferon, beta 1, fibroblast; IFNG: Interferon, gamma; IKBKB: Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta; IKBKG: Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma; IL12A: Interleukin 12A; IL12B: Interleukin 12B; IL18: Interleukin 18; IL1B: Interleukin 1, beta; IL33: Interleukin 33; IL6: Interleukin 6; IRAK1: Interleukin-1 receptor-associated kinase 1; IRF1: Interferon regulatory factor 1; IRF2: Interferon regulatory factor 2; MAP3K7: Mitogen-activated protein kinase kinase kinase 7; MAPK1: Mitogen-activated protein kinase 1; MAPK11: Mitogen-activated protein kinase 11; MAPK12: Mitogen-activated protein kinase 12; MAPK13: Mitogen-activated protein kinase 13; MAPK3: Mitogen-activated protein kinase 3; MAPK8: Mitogen-activated protein kinase 8; MAPK9: Mitogen-activated protein kinase 9; MEFV: Mediterranean fever; MYD88: Myeloid differentiation primary response gene (88); NAIP: NOD-like receptor (NLR) family, apoptosis inhibitory protein; NFKB1: Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1; NFKB1A: Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha; NFKB1B: Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta; NLRC4: NLR family, CARD domain containing 4; NLRC5: NLR family, CARD domain containing 5; NLRP1: NLR family, pyrin domain containing 1; NLRP12: NLR family, pyrin domain containing 12; NLRP3: NLR family, pyrin domain containing 3; NLRP4: NLR family, pyrin domain containing 4; NLRP5: NLR family, pyrin domain containing 5; NLRP6: NLR family, pyrin domain containing 6; NLRP9: NLR family, pyrin domain containing 9; NLRX1: NLR family member X1; NOD1: Nucleotide-binding oligomerization domain containing 1; NOD2: Nucleotide-binding oligomerization domain containing 2; P2RX7: Purinergic receptor P2X, ligand-gated ion channel, 7; PANX1: Pannexin 1; PEA15: Phosphoprotein enriched in astrocytes 15; PSTPIP1: Proline-serine-threonine phosphatase interacting protein 1; PTGS2: Prostaglandin-endoperoxide synthase 2; PYCARD: PYD and CARD domain containing; PYDC1: PYD (pyrin domain) containing 1; MOK: Renal tumor antigen; RELA: V-rel reticuloendotheliosis viral oncogene homolog A (avian); RIPK2: Receptor-interacting serine-threonine kinase 2; SUGT1: SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae); TAB1: TGF-beta activated kinase1/MAP3K7 binding protein 1; TAB2: TGF-beta activated kinase1/MAP3K7 binding protein 2; TIRAP: Toll-interleukin 1 receptor (TIR) domain containing adaptor protein; TNF: Tumor necrosis factor; TNFSF11: Tumor necrosis factor (ligand) superfamily, member 11; TNFSF14: Tumor necrosis factor (ligand) superfamily, member 14; TNFSF4: Tumor necrosis factor (ligand) superfamily, member 4; TRAF6: TNF receptor-associated factor 6; TXNIP: Thioredoxin interacting protein; XIAP: X-linked inhibitor of apoptosis.
Acknowledgements
The authors thanks to Prof. Scott L. Friedman, Mount Sinai Medical School, NY, USA for providing us LX-2 cells.
Funding
This work was supported by JSPS KAKENHI GRANT Number JP17K09404 (to T.K.).
Competing Interests
Tatsuo Kanda and Mitsuhiko Moriyama received research grants from AbbVie, Eisai, Daiichi-Sankyo, Shionogi, Mitsubishi-Tanabe Pharma, Astellas, Ono Pharma and Takeda Pharma. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
1. Younossi Z, Stepanova M, Ong JP. et al. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clin Gastroenterol Hepatol. 2019;17:748-55 DOI: 10.1016/j.cgh.2018.05.057. PMID: 29908364
2. Tateishi R, Okanoue T, Fujiwara N. et al. Clinical characteristics, treatment, and prognosis of non-B, non-C hepatocellular carcinoma: a large retrospective multicenter cohort study. J Gastroenterol. 2015;50:350-60 DOI: 10.1007/s00535-014-0973-8. PMID: 24929638
3. Estes C, Anstee QM, Arias-Loste MT. et al. Modeling NAFLD Disease Burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J Hepatol. 2018;69:896-904 DOI: 10.1016/j.jhep.2018.05.036. PMID: 29886156
4. Kanda T, Matsuoka S, Yamazaki M. et al. Apoptosis and non-alcoholic fatty liver diseases. World J Gastroenterol. 2018;24:2661-72 DOI: 10.3748/wjg.v24.i25.2661. PMID: 29991872
5. Lefkowitz RJ, Rajagopal K, Whalen EJ. New roles for beta-arrestins in cell signaling: not just for seven-transmembrane receptors. Mol Cell. 2006;24:643-52 DOI: 10.1016/j.molcel.2006.11.007. PMID: 17157248
6. Luan B, Zhao J, Wu H. et al. Deficiency of a beta-arrestin-2 signal complex contributes to insulin resistance. Nature. 2009;457:1146-9 DOI: 10.1038/nature07617. PMID: 19122674
7. Rojanathammanee L, Harmon EB, Grisanti LA. et al. The 27-kDa heat shock protein confers cytoprotective effects through a beta 2-adrenergic receptor agonist-initiated complex with beta-arrestin. Mol Pharmacol. 2009;75:855-65 DOI: 10.1124/mol.108.053397. PMID: 19176359
8. Xu TR, Baillie GS, Bhari N. et al. Mutations of beta-arrestin 2 that limit self-association also interfere with interactions with the beta2-adrenoceptor and the ERK1/2 MAPKs: implications for beta2-adrenoceptor signalling via the ERK1/2 MAPKs. Biochem J. 2008;413:51-60 DOI: 10.1042/BJ20080685. PMID: 18435604
9. Boularan C, Scott MG, Bourougaa K. et al. beta-arrestin 2 oligomerization controls the Mdm2-dependent inhibition of p53. Proc Natl Acad Sci USA. 2007;104:18061-6 DOI: 10.1073/pnas.0705550104. PMID: 17984062
10. Fuster G, Busquets S, Ametller E. et al. Are peroxisome proliferator-activated receptors involved in skeletal muscle wasting during experimental cancer cachexia? Role of beta2-adrenergic agonists. Cancer Res. 2007;67:6512-9 DOI: 10.1158/0008-5472.CAN-07-0231. PMID: 17616713
11. Berthouze-Duquesnes M, Lucas A, Saulière A. et al. Specific interactions between Epac1, β-arrestin2 and PDE4D5 regulate β-adrenergic receptor subtype differential effects on cardiac hypertrophic signaling. Cell Signal. 2013;25:970-80 DOI: 10.1016/j.cellsig.2012.12.007. PMID: 23266473
12. Szepietowska B, Zhu W, Sherwin RS. β2-Adrenergic receptor agonist administration promotes counter-regulatory responses and recovery from hypoglycaemia in rats. Diabetologia. 2013;56:2517-23 DOI: 10.1007/s00125-013-3009-7. PMID: 23933834
13. Breitman M, Kook S, Gimenez LE. et al. Silent scaffolds: inhibition OF c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653-64 DOI: 10.1074/jbc.M112.358192. PMID: 22523077
14. Zhang P, Wang PX, Zhao LP. et al. The deubiquitinating enzyme TNFAIP3 mediates inactivation of hepatic ASK1 and ameliorates nonalcoholic steatohepatitis. Nat Med. 2018;24:84-94 DOI: 10.1038/nm.4453. PMID: 29227477
15. Kang DS, Tian X, Benovic JL. Role of β-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr Opin Cell Biol. 2014;27:63-71 DOI: 10.1016/j.ceb.2013.11.005. PMID: 24680432
16. Patwari P, Lee RT. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab. 2012;23:216-22 DOI: 10.1016/j.tem.2012.03.003. PMID: 22520962
17. Patwari P, Emilsson V, Schadt EE. et al. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metab. 2011;14:671-83 DOI: 10.1016/j.cmet.2011.08.011. PMID: 21982743
18. Rafiq S, Tapper W, Collins A. et al. Identification of inherited genetic variations influencing prognosis in early-onset breast cancer. Cancer Res. 2013;73:1883-91 DOI: 10.1158/0008-5472.CAN-12-3377. PMID: 23319801
19. Cai Q, Zhang B, Sung H. et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat Genet. 2014;46:886-90 DOI: 10.1038/ng.3041. PMID: 25038754
20. Higuchi T, Moriyama M, Fukushima A. et al. Association of mRNA expression of iron metabolism-associated genes and progression of non-alcoholic steatohepatitis in rats. Oncotarget. 2018;9:26183-94 DOI: 10.18632/oncotarget.25488. PMID: 29899851
21. Wang D, Yang PN, Chen J. et al. Promoter hypermethylation may be an important mechanism of the transcriptional inactivation of ARRDC3, GATA5, and ELP3 in invasive ductal breast carcinoma. Mol Cell Biochem. 2014;396:67-77 DOI: 10.1007/s11010-014-2143-y. PMID: 25148870
22. Hoque R, Farooq A, Ghani A. et al. Lactate reduces liver and pancreatic injury in Toll-like receptor- and inflammasome-mediated inflammation via GPR81-mediated suppression of innate immunity. Gastroenterology. 2014;146:1763-74 DOI: 10.1053/j.gastro.2014.03.014. PMID: 24657625
23. Moravcová A, Červinková Z, Kučera O. et al. The effect of oleic and palmitic acid on induction of steatosis and cytotoxicity on rat hepatocytes in primary culture. Physiol Res. 2015;64(Suppl 5):S627-36 PMID: 26674288
24. Ricchi M, Odoardi MR, Carulli L. et al. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol. 2009;24:830-40 DOI: 10.1111/j.1440-1746.2008.05733.x. PMID: 19207680
25. Nwe Win N, Kanda T, Nakamura M. et al. Free fatty acids or high-concentration glucose enhances hepatitis A virus replication in association with a reduction in glucose-regulated protein 78 expression. Biochem Biophys Res Commun. 2017;483:694-9 DOI: 10.1016/j.bbrc.2016.12.080. PMID: 27986562
26. Sasaki R, Kanda T, Nakamura M. et al. Possible Involvement of Hepatitis B Virus Infection of Hepatocytes in the Attenuation of Apoptosis in Hepatic Stellate Cells. PLoS One. 2016;11:e0146314. DOI: 10.1371/journal.pone.0146314. PMID: 26731332
27. Dai W, Wang K, Zheng X. et al. High fat plus high cholesterol diet lead to hepatic steatosis in zebrafish larvae: a novel model for screening anti-hepatic steatosis drugs. Nutr Metab. (Lond.). 2015;12:42. DOI: 10.1186/s12986-015-0036-z. PMID: 26583037
28. Sookoian S, Gianotti TF, Rosselli MS. et al. Liver transcriptional profile of atherosclerosis-related genes in human nonalcoholic fatty liver disease. Atherosclerosis. 2011;218:378-85 DOI: 10.1016/j.atherosclerosis.2011.05.014. PMID: 21664615
29. Morrison MC, Liang W, Mulder P. et al. Mirtoselect, an anthocyanin-rich bilberry extract, attenuates non-alcoholic steatohepatitis and associated fibrosis in ApoE(∗)3Leiden mice. J Hepatol. 2015;62:1180-6 DOI: 10.1016/j.jhep.2014.12.011. PMID: 25514555
30. Min HK, Mirshahi F, Verdianelli A. et al. Activation of the GP130-STAT3 axis and its potential implications in nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol. 2015;308:G794-803 DOI: 10.1152/ajpgi.00390.2014. PMID: 25747354
31. Li BH, He FP, Yang X. et al. Steatosis induced CCL5 contributes to early-stage liver fibrosis in nonalcoholic fatty liver disease progress. Transl Res. 2017;180:103-17.e4 DOI: 10.1016/j.trsl.2016.08.006. PMID: 27639593
32. Mridha AR, Wree A, Robertson AAB. et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66:1037-46 DOI: 10.1016/j.jhep.2017.01.022. PMID: 28167322
33. Zhu Q, Lin L, Cheng Q. et al. The role of acid sphingomyelinase and caspase 5 in hypoxia-induced HuR cleavage and subsequent apoptosis in hepatocytes. Biochim Biophys Acta. 2012;1821:1453-61 DOI: 10.1016/j.bbalip.2012.08.005. PMID: 22906436
34. Zhang X, Zhang JH, Chen XY. et al. Reactive oxygen species-induced TXNIP drives fructose-mediated hepatic inflammation and lipid accumulation through NLRP3 inflammasome activation. Antioxid Redox Signal. 2015;22:848-70 DOI: 10.1089/ars.2014.5868. PMID: 25602171
35. Zhang Y, O'Keefe RJ, Jonason JH. BMP-TAK1 (MAP3K7) Induces Adipocyte Differentiation Through PPARγ Signaling. J Cell Biochem. 2017;118:204-10 DOI: 10.1002/jcb.25626. PMID: 27293199
36. Xiao F, Waldrop SL, Bronk SF. et al. Lipoapoptosis induced by saturated free fatty acids stimulates monocyte migration: a novel role for Pannexin1 in liver cells. Purinergic Signal. 2015;11:347-59 DOI: 10.1007/s11302-015-9456-5. PMID: 26054298
37. Yang MR, Zhang Y, Wu XX, Chen W. Critical genes of hepatocellular carcinoma revealed by network and module analysis of RNA-seq data. Eur Rev Med Pharmacol Sci. 2016;20:4248-56 PMID: 27831650
38. Marcolin E, San-Miguel B, Vallejo D. et al. Quercetin treatment ameliorates inflammation and fibrosis in mice with nonalcoholic steatohepatitis. J Nutr. 2012;142:1821-8 DOI: 10.3945/jn.112.165274. PMID: 22915297
39. Yang L, Miura K, Zhang B. et al. TRIF Differentially Regulates Hepatic Steatosis and Inflammation/Fibrosis in Mice. Cell Mol. Gastroenterol. Hepatol. 2017:3 469-83. DOI: 10.1016/j.jcmgh.2016.12.004. PMID: 28462384
40. Csak T, Dolganiuc A, Kodys K. et al. Mitochondrial antiviral signaling protein defect links impaired antiviral response and liver injury in steatohepatitis in mice. Hepatology. 2011;53:1917-31 DOI: 10.1002/hep.24301. PMID: 21425308
41. Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 2010;11:605-11 DOI: 10.1038/embor.2010.80. PMID: 20559325
42. Shi Y, Shu ZJ, Xue X. et al. β2-Adrenergic receptor ablation modulates hepatic lipid accumulation and glucose tolerance in aging mice. Exp Gerontol. 2016;78:32-8 DOI: 10.1016/j.exger.2016.03.005. PMID: 26952573
43. Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398-409 DOI: 10.1038/nrm2690. PMID: 19436320
44. Zhao GN, Zhang P, Gong J. et al. Tmbim1 is a multivesicular body regulator that protects against non-alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat Med. 2017;23:742-52 DOI: 10.1038/nm.4334. PMID: 28481357
45. Safi SZ, Qvist R, Ong G. et al. Stimulation of β-adrenergic receptors plays a protective role via increased expression of RAF-1 and PDX-1 in hyperglycemic rat pancreatic islet (RIN-m5F) cells. Arch Med Sci. 2017;13:470-80 DOI: 10.5114/aoms.2016.64131. PMID: 28261303
46. Chen J, Fujii K, Zhang L. et al. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci USA. 2001;98:7783-8 DOI: 10.1073/pnas.141224398. PMID: 11427728
47. Ji N, Yang Y, Cai CY. et al. Selonsertib (GS-4997), an ASK1 inhibitor, antagonizes multidrug resistance in ABCB1- and ABCG2-overexpressing cancer cells. Cancer Lett. 2019;440-441:82-93 DOI: 10.1016/j.canlet.2018.10.007. PMID: 30315846
48. Jayakumar S, Middleton MS, Lawitz EJ. et al. Longitudinal correlations between MRE, MRI-PDFF, and liver histology in patients with non-alcoholic steatohepatitis: Analysis of data from a phase II trial of selonsertib. J Hepatol. 2019;70:133-41 DOI: 10.1016/j.jhep.2018.09.024. PMID: 30291868
49. Tacke F, Weiskirchen R. An update on the recent advances in antifibrotic therapy. Expert Rev Gastroenterol Hepatol. 2018;12:1143-52 DOI: 10.1080/17474124.2018.1530110. PMID: 30261763
50. Sumida Y, Yoneda M. Current and future pharmacological therapies for NAFLD/NASH. J Gastroenterol. 2018;53:362-76 DOI: 10.1007/s00535-017-1415-1. PMID: 29247356
51. Sasaki R, Kanda T, Wu S. et al. Association between hepatitis B virus and MHC class I polypeptide-related chain A in human hepatocytes derived from human-mouse chimeric mouse liver. Biochem Biophys Res Commun. 2015;464:1192-5 DOI: 10.1016/j.bbrc.2015.07.102. PMID: 26212443
52. Xu L, Hui AY, Albanis E. et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142-51 DOI: 10.1136/gut.2004.042127. PMID: 15591520
53. Wu S, Kanda T, Imazeki F. et al. Hepatitis B virus e antigen downregulates cytokine production in human hepatoma cell lines. Viral Immunol. 2010;23:467-76 DOI: 10.1089/vim.2010.0042. PMID: 20883161
54. Arakaki AKS, Pan WA, Trejo J. GPCRs in Cancer: Protease-Activated Receptors, Endocytic Adaptors and Signaling. Int J Mol Sci. 2018:19 DOI: 10.3390/ijms19071886. PMID: 29954076
55. Takeuchi F, Kukimoto I, Li Z. et al. Genome-wide association study of cervical cancer suggests a role for ARRDC3 gene in human papillomavirus infection. Hum Mol Genet. 2019:28 341-8. DOI: 10.1093/hmg/ddy390. PMID: 30412241
Author contact
![]() Corresponding author: Tatsuo Kanda, M.D., Ph.D., Associate Professor, Division of Gastroenterology and Hepatology, Department of Medicine, Nihon University School of Medicine, 30-1 Oyaguchi-kamicho, Itabashi-ku, Tokyo 173-8610, Japan. E-mail: kanda.tatsuoac.jp; Phone: +81-3-3972-8111; Fax: +81-3-3956-8496
Corresponding author: Tatsuo Kanda, M.D., Ph.D., Associate Professor, Division of Gastroenterology and Hepatology, Department of Medicine, Nihon University School of Medicine, 30-1 Oyaguchi-kamicho, Itabashi-ku, Tokyo 173-8610, Japan. E-mail: kanda.tatsuoac.jp; Phone: +81-3-3972-8111; Fax: +81-3-3956-8496

 Global reach, higher impact
Global reach, higher impact