3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(2):343-354. doi:10.7150/ijms.29393 This issue Cite
Research Paper
Gene Expression Profiling in Ischemic Postconditioning to Alleviate Mouse Liver Ischemia/Reperfusion Injury
1. Department of Transplant Surgery, The Third Xiangya Hospital of Central South University, Changsha 410013, China
2. Zhongnan Hospital of Wuhan University, Institute of Hepatobiliary Diseases of Wuhan University, Transplant Center of Wuhan University, Hubei Key Laboratory of Medical Technology on Transplantation, Wuhan, Hubei 430071, China
Received 2018-8-21; Accepted 2018-12-17; Published 2019-1-24
Abstract
Ischemic postconditioning (IPO) attenuates hepatic ischemia/reperfusion (I/R) injury. However, little is known about the underlying biological pathophysiology, which could be, at least in part, informed by exploring the transcriptomic changes using next-generation RNA sequencing (RNA-Seq). In this study, 18 mice (C57BL/6) were involved and randomly assigned to three groups: normal (n=6), I/R (n=6, subjected to 70% hepatic I/R), and IR+IPO (n=6, applying IPO to mice with I/R injury). We randomly selected 3 mice per group and extracted their liver tissues for next-generation RNA-Seq. We performed a bioinformatics analysis for two comparisons: normal vs. I/R and I/R vs. IR+IPO. From the analysis, 2416 differentially expressed genes (DEGs) were identified (p < 0.05 and fold change ≥ 1.5). Gene ontology (GO) analysis revealed that these genes were mainly related to cellular metabolic processes, nucleic acids and protein binding processes. The enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the DEGs were the mitogen-activated protein kinase (MAPK), IL-17 signalling pathway, regulating pluripotency of stem cells, and insulin resistance pathway. Validation of 12 selected DEGs by qRT-PCR showed that Cyr61, Atf3, Nr4a1, Gdf15, Osgin1, Egr1, Epha2, Dusp1, Dusp6, Gadd45a and Gadd45b were significantly amplified. Finally, a protein-protein interaction (PPI) network constructed to determine interactions of these 11 DEGs. In summary, by exploring gene expression profiling in regard to hepatic I/R and IPO using next-generation RNA-Seq, we suggested a few progression-related genes and pathways, providing some clues for future experimental research.
Keywords: hepatic ischemia-reperfusion injury, ischemic postconditioning, next-generation RNA-Seq, DEGs, MAPK pathway
Introduction
Hepatic ischemia/reperfusion (I/R), or the interruption of blood flow to the liver followed by subsequent reperfusion, causes an acute inflammatory response that causes cellular damage and organ dysfunction and contributes to major complications after liver transplantation or partial hepatectomy [1, 2]. The mechanism of hepatic I/R injury is complex and is controlled by multiple cytokines. Jaeschke et al. verified two obvious phases during acute liver injury after hepatic I/R [3, 4] and showed that Kupffer cells (KCs), the resident macrophages of the liver, are extremely important to the pathophysiological process of I/R-induced acute liver injury [5-7]. Once KCs are activated, pro-inflammatory cytokines including tumour necrosis factor alpha (TNF-α) and interleukin1β (IL-1β) as well as reactive oxygen species (ROS), which initiate oxidative stress, are released, subsequently promoting neutrophil infiltration into hepatic microcirculation and aggravating liver cell injury [8-10].
Currently, several pharmacological and mechanical methods have been identified that attenuate liver I/R in animal studies. For instance, melatonin, which is a molecule with notable antioxidant and anti-inflammatory properties, protects against hepatic I/R injury via Jun N-terminal kinase (JNK) pathway inhibition [11]. As a mechanical method, ischemic postconditioning (IPO), which is defined as a short series of repetitive cycles of brief reperfusion and re-occlusion applied at the onset of reperfusion after a prolonged ischemic insult, has been used to attenuate organ I/R injury in the heart [12, 13], bowel [14], kidney [15, 16], brain [17] and liver [18, 19]. Although IPO has been shown to provide protective effects against hepatic I/R injury, little is known about the underlying biological pathophysiology, which encouraged us to investigate the molecular mechanisms and pathways.
Recently, the rapid development of next-generation RNA-Seq analysis has promoted the exploration of complex diseases progression and the identification of biomarkers. For example, the RNA- Seq technique could provide high-resolution sequence information about alcoholic liver disease (ALD), through which Sun identified some new targets for the early diagnosis and therapeutic management of ALD [20]. In a previous study, Arai et al. revealed the mechanism and pathophysiology of mouse liver regeneration through gene expression profiling [21]. Altered gene expression in IPO to attenuate liver I/R injury is tightly associated with the pathophysiology and understanding IPO requires a detailed study of the transcriptomic changes that underpin this process. However, the gene expression profile during IPO attenuating hepatic I/R injury was not reported in the previous research. In this study, we explored gene expression profiles using next-generation RNA-Seq, and subsequent bioinformatics analyses were performed to assess the differentially expressed genes (DEGs) function and pathways relevant to hepatic I/R injury and IPO.
Methods and materials
Ethics Approval
This research protocol was approved by the Committee on the Ethics of Animal Experiments of the Third Xiangya Hospital and was conducted according to the Guidance for the Care and Use of Laboratory Animals of the National Institute of Health (No. LLSC (LA) 2016-030).
Animal model
A total of 20 male SPF mice (9-week-old, C57BL/6) were provided by Hunan SLAC Laboratory Animals (Hunan, China). All of the mice were housed in a standard room with ad libitum water, rodent food and a 12/12 h light/dark cycle for two weeks. After an acclimatization period, 20 mice were randomly divided into three groups: the normal (N) group (n = 6), the I/R group (n = 7, subjected to 70% hepatic I/R) and the I/R+IPO group (n = 7, applying IPO to mice with I/R injury). Two mice were excluded because of death during procedure, and each of them was from the I/R and IPO group. Finally,18 mice were included for further research and the final number per group was six. The model for partial (70%) hepatic I/R was used in accordance with previous reports [22, 23]. All mice were anaesthetized with intraperitoneal injections of sodium pentobarbital (10 mg/kg). Group N received a laparotomy without vessel blockage and the I/R group had liver ischemia induced for 1 h and then reperfusion for 4 h. The IPO group received occlusion of the porta hepatis for 1 h and was then treated with three consecutive 5-sec cycles of reperfusion followed by persistent reperfusion for 4 h. All of the mice were sacrificed, and samples (liver and blood) were collected for further analysis.
Serum enzyme and inflammation factor analyses
To assess the hepatocyte injury severity, we measured the serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels using a HITACHI 7600 Automatic Analyzer (Japan, U/L) and TNF-α and IL-1β using an Abcam ELISA kit (USA, pg/ml).
Total RNA isolation, RNA-seq library preparation and next-generation RNA-Seq
Total RNA was extracted from nine frozen mouse liver tissues (three randomly selected samples from each group) using TRIzol (Invitrogen, USA) according to the manufacturer's instructions. After quality inspection and mRNA enrichment, we used KAPA Stranded RNA-Seq Library Prep Kit (Illumina, USA) for RNA-seq library preparation, which included RNA fragmentation, random hexamer-primed first strand cDNA synthesis, dUTP-based second strand cDNA synthesis, end-repairing, A-tailing, adaptor ligation and library PCR amplification. Finally, the prepared RNA-seq libraries were qualified using an Agilent 2100 Bioanalyzer (Illumina, USA) and quantified by the qPCR absolute quantification method. Next-generation RNA-Seq was performed using the Illumina HiSeq 4000 (Illumina, USA) according to the manufacturer's instructions for 150 cycles.
Bioinformatics analysis
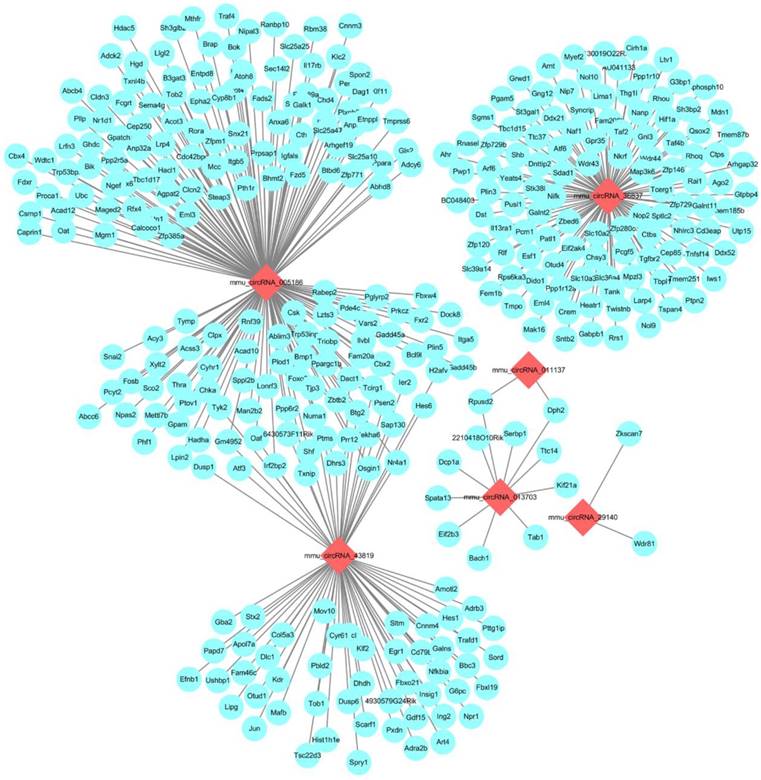
When the data were extracted, subsequent data processing was performed to use the R software Ballgown package. DEGs between the two groups were identified using fold change (FC) and p-values (FC ≥ 1.5 and p-value < 0.05). Scatter plot analysis was conducted to depict the mRNA expression distribution. Hierarchical clustering was performed to show distinguishable mRNA expression profiles among the samples. The volcano graph was created to visualize significantly dysregulated mRNAs. GO analysis was used to investigate three functionality domains: Biological Process (BP), Cellular Component (CC) and Molecular Function (MF)[24]. Pathway analysis was performed to functionally analyse and map genes to KEGG pathways. The p-value denotes the significance of the GO and KEGG pathway correlated with the various conditions (p < 0.05). The interaction of DEGs with our previous key circRNAs was determined by Coding & Noncoding Co-expression (CNC) analysis, and the CNC network was delineated by Cytoscape according to partial correlation coefficient (PCC) and p-value (PCC ≥ 0.9 and p < 0.05). A PPI network constructed to determine interactions of these DEGs by STRING analysis.
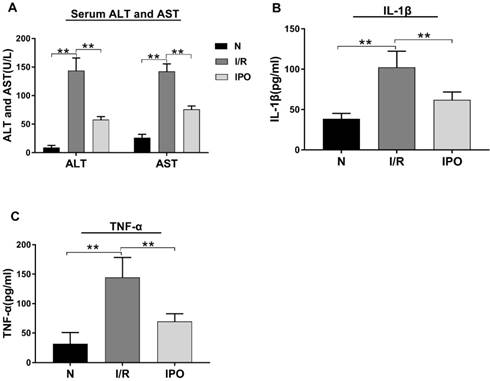
Effect of hepatic I/R injury and IPO on serum aminotransferase levels and pro-inflammatory cytokines. (A) Serum ALT and AST levels were detected by Automatic Analyzer during the I/R and IPO process. (B and C) The pro-inflammatory cytokines (IL-1β and TNF-α) were measured by ELISA kit. (n = 6 per group, *p < 0.05, **p < 0.01)
Validation of selected genes by qRT-PCR
Further qRT-PCR validation was performed with a ViiA 7 real-time PCR system (Applied Biosystems, USA) in triplicate for each sample. All of the primers were designed and synthesized by Kangchen Bio-tech (Shanghai, China). mRNA expression was defined as the threshold cycle (Ct), and GAPDH was amplified as the internal control. The relative amounts of selected mRNAs were calculated using the double-standard curve method.
Statistical analysis
All the results were expressed as the mean ± standard deviation. The data were statistically analysed and visualized with GraphPad Prism 5.0. A p-value less than 0.05 was used to indicate statistical significance.
Results
IPO attenuated liver I/R
The effect of hepatic I/R injury and IPO was evaluated be assessing the serum levels of ALT, AST, IL-1β and TNF-α. The ALT, AST, IL-1 β and TNF-α serum levels were significantly increased in the I/R group compared with those in the N group. However, these values decreased significantly in the IPO group compared with those in the I/R group (Figure 1), indicating that IPO attenuated I/R injury successfully.
mRNA expression patterns during IPO protection against hepatic I/R injury via next-generation RNA-Seq analysis
Next-generation RNA-Seq showed that 2,416 of 22,249 genes were differentially expressed overall (p < 0.05 and FC ≥ 1.5). Of these, we identified that 320 and 567 genes were up-regulated and down-regulated, respectively, between the N and I/R group. Additionally, 853 and 676 genes were up-regulated and down-regulated, respectively, in the IPO group compared with their expression in the I/R group. Scatter plot graph analysis was conducted to depict the gene expression distribution (Figure 2A). Hierarchical clustering analysis evaluated these 2416 significantly expressed genes, which were indicated by p < 0.05 and an FC ≥ 1.5 between the N, I/R and IPO groups (Figure 2B). Each column represents the expression pattern of one sample, and high and low expression levels are indicated by the “red” and “green” lines, respectively. The volcano graph was created to visualize significant DEGs (Figure 2C). CNC analysis integrated these DEGs and our previously verified six circRNAs with the hepatic I/R injury and IPO data. Additionally, 380 DEGs had roles establishing the regulation network, as depicted in Figure 3.
GO and KEGG pathway analysis of differentially expressed genes
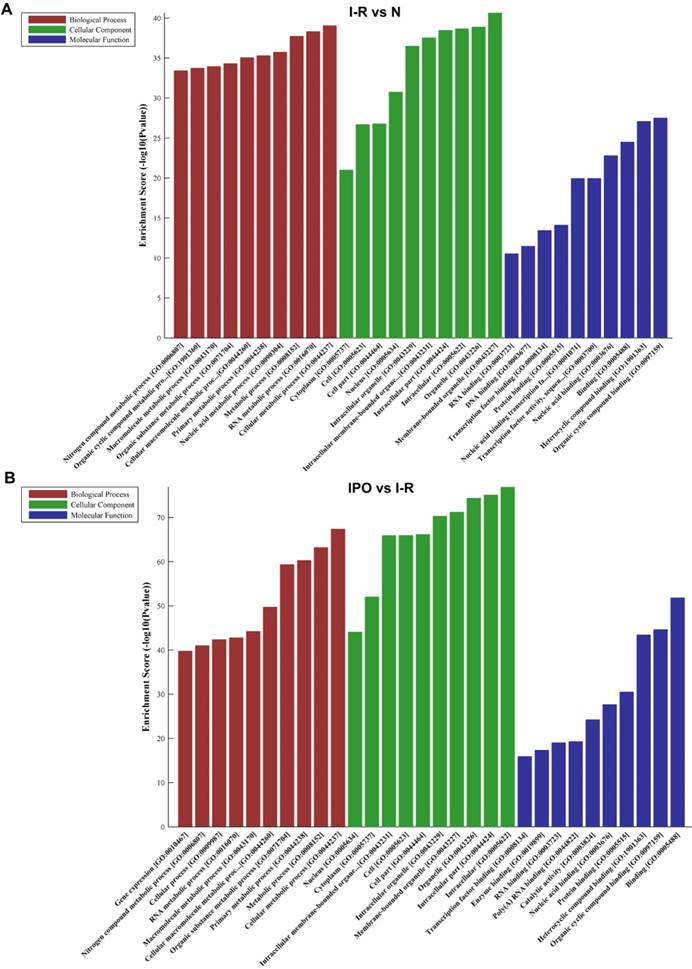
The top 10 GO terms from the BP, CC, and MF domains in the compared groups are ranked according to enrichment score and by p-value (Figure 4).
Bioinformatics analysis of mRNA expression patterns during IPO and hepatic I/R injury by Next-generation RNA-Seq. A. Scatter plot graph analysis was conducted to exhibit all the mRNA expression distribution. The dashed lines represent the default significant fold change (1.5) in the scatter plot analysis. B. Hierarchical clustering was used to evaluate the 2416 DEGs when comparing with each of normal, I/R and IPO group samples. One sample expression pattern was represented by each column and high and low expression was indicated by the “red” and “green” line, respectively. C. The volcano graph was performed to show significantly DEGs in a visible way and the vertical green lines correspond to 1.5-fold up- and down-regulation and the horizontal line represents the p-value (0.05)
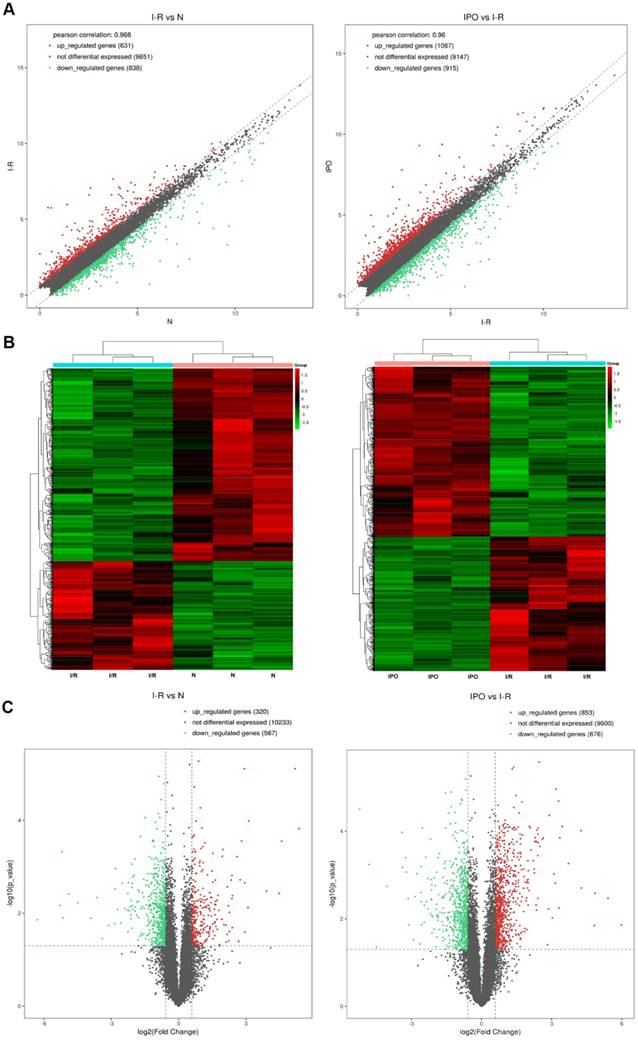
The CNC network analysis. The network consists of 6 circRNAs (red) and 380 mRNAs (blue)
In the BP domain, the most meaningful enriched GO terms were related to nucleic acid and cellular metabolic processes and included RNA metabolic process (GO:0016070), Nucleic acid metabolic process (GO:0090304), Gene expression (GO:0010467), Cellular macromolecule metabolic process (GO:0044260) and Cellular metabolic process (GO:0044237).
The most enriched GO CC terms primarily focused on the cell, such as Organelle (GO:0043226), Membrane-bounded organelle (GO:0043227), Intracellular organelle (GO:0043229), Cytoplasm (GO:0005737) and Nucleus (GO:0005634).
As for MF terms, nucleic acid and protein binding were very important in the GO terms ranked by enrichment score. Represented terms were Nucleic acid binding (GO:0003676), DNA binding (GO:0003677), RNA binding (GO:0003723), Transcription factor binding (GO:0008134), Transcription factor activity and sequence-specific DNA binding (GO:0003700) and Protein binding (GO:0005515).
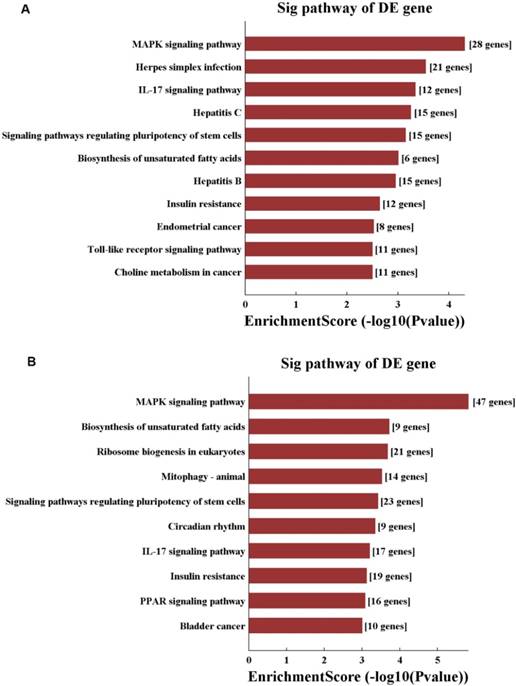
Moreover, KEGG pathway analysis was performed, and pathways were selected and ranked by p-value. Overall, 125 pathways were connected to in hepatic I/R injury and IPO. The top 10 pathways in the compared groups (N vs. I/R and I/R vs. I/R+IPO) were listed according to enrichment score and were ranked by p-value (Figure 5). Identical pathways in both sets were the MAPK signalling pathway, the IL-17 signalling pathway, regulating pluripotency of stem cells, and the insulin resistance pathway.
GO analysis of DEGs with top 10 Enrichment score under the theme of BP, CC and MF in N vs. I /R (A) and I/R vs. IPO (B) group
Validation of selected DEGs by qRT-PCR
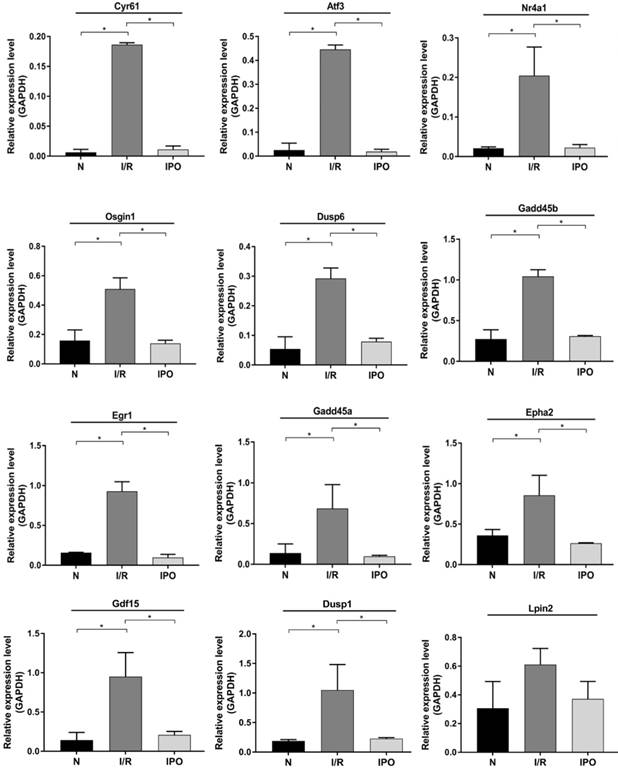
Twelve DEGs were selected based on a combination of p-value, FC, PCC and Fragments per Kilobase of transcript per million mapped reads (FPKM) (Table 1). All of the primers were designed and synthesized by Kangchen Bio-tech (Table 2). The results confirmed that consistent with the RNA-Seq results, 11 genes were significantly amplified by qRT-PCR including Cyr61, Atf3, Nr4a1, Gdf15, Osgin1, Egr1, Epha2, Dusp1, Dusp6, Gadd45a and Gadd45b (Figure 6).
KEGG pathway analysis of N vs. I/R (A) and I/R vs. IPO (B) group with top 10 Enrichment score
12 DEGs were screened for validation by qRT-PCR.
| Gene Name | FCa | p-value | N_FPKMb | I/R_FPKM | IPO_FPKM | PCCc |
|---|---|---|---|---|---|---|
| Cyr61 | 23.8 | 0.0003 | 1.4 | 6 | 1.9 | 0.9524 |
| Atf3 | 22.1 | 0.0037 | 2.6 | 7.1 | 2.3 | 0.9454 |
| Nr4a1 | 8.7 | 0.0001 | 1.4 | 4.5 | 1.3 | 0.916 |
| Gdf15 | 8.5 | 0.0076 | 4 | 7.1 | 4.9 | 0.9253 |
| Osgin1 | 7.7 | 0.0256 | 3.7 | 6.6 | 3.2 | 0.9132 |
| Dusp6 | 5.2 | 0.0204 | 3.4 | 5.8 | 3.7 | 0.9368 |
| Gadd45b | 5.2 | 0.0015 | 4.2 | 6.5 | 4.1 | 0.9634 |
| Dusp1 | 5.2 | 0.0048 | 4.5 | 6.9 | 4.6 | 0.9421 |
| Egr1 | 4.5 | 0.0285 | 4.2 | 6.4 | 3.3 | 0.9077 |
| Gadd45a | 4.3 | 0.0181 | 3.5 | 5.6 | 3 | 0.9304 |
| Lpin2 | 3.3 | 0.0136 | 4.2 | 5.9 | 4.4 | 0.9178 |
| Epha2 | 2.7 | 0.0194 | 3 | 4.4 | 2.5 | 0.9393 |
aFC: Fold change. bGroup FPKM: Fragments per Kilobase of transcript per million mapped reads. cPCC: partial correlation coefficient of CNC
The primers sequence used in this study
| Gene name | Primer | product length (bp) |
|---|---|---|
| GAPDH | F:5' CACTGAGCAAGAGAGGCCCTAT 3' R:5' GCAGCGAACTTTATTGATGGTATT 3' | 144 |
| Cyr61 | F:5'CGAGTTACCAATGACAACCCAG 3' R :5' TGCAGCACCGGCCATCTA 3' | 223 |
| Atf3 | F:5' GGCGGCGAGAAAGAAATA 3' R :5' ATTCTGAGCCCGGACGAT 3' | 206 |
| Nr4a1 | F:5' TACCAATCTTCTCACTTCCCTC 3' R :5' GCCCACTTTCGGATAACG 3' | 180 |
| Gdf15 | F:5' AGAACCAAGTCCTGACCCAG 3' R:5'AATCTCACCTCTGGACTGAGTAT 3' | 51 |
| Osgin1 | F:5' GCAGAGGTCTCCGCAACA 3' R :5' CGGTAGTAGTGGGCGATGT 3' | 55 |
| Egr1 | F:5' GAGCGAACAACCCTATGAG 3' R :5' GTCGTTTGGCTGGGATAA 3' | 102 |
| Lpin2 | F:5' ACAGGACAATAGGAAGGAGGAG 3' R :5' AGGGTAGGTGGTTTCTAATGG 3' | 220 |
| Epha2 | F:5' AGGGAGAAGGATGGTGAGTT 3' R :5' CTTCCAGCACACGCGAC 3' | 184 |
| Dusp6 | F:5' CCCAATCTGTTTGAGAATGCG 3' R :5' ACGGTGACAGAGCGGCTGA 3' | 179 |
| Dusp1 | F:5' GCAGCAAACAGTCCACCC 3' R :5' CCGAGAAGCGTGATAGGC 3' | 167 |
| Gadd45b | F:5' ACCCTGATCCAGTCGTTCT 3' R :5' GGACCCATTGGTTATTGC 3' | 232 |
| Gadd45a | F:5' TGTGCTGGTGACGAACCC 3' R :5' ACCCACTGATCCATGTAGCG 3' | 99 |
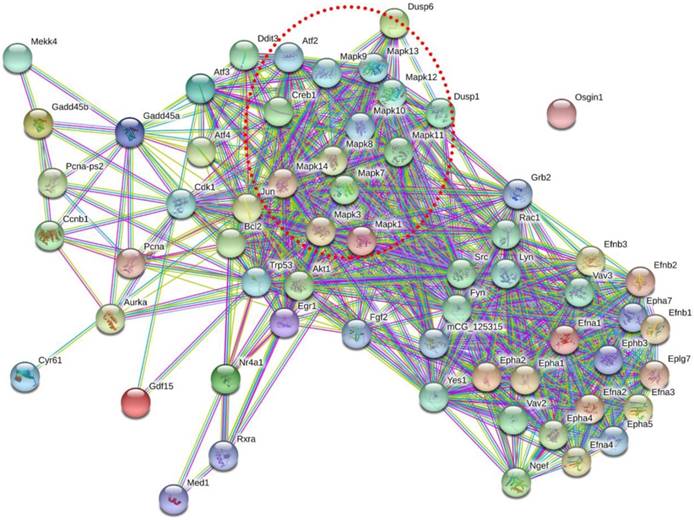
To determine how these 11 DEGs interact with each other, we identified potential PPI network for these DEGs (Figure 7). Signal-net analysis integrated these 11 genes using STRING analysis and 61 nodes were involved in the establishment of the gene regulation network, with 636 edges. From the PPI network, we found that MAPK gene family made a significant contribution to the interactions of these DEGs, which indicated the importance of MAPK pathway.
Discussion
We used next-generation RNA-Seq to explore gene expression profiling in regard to hepatic I/R and IPO. In this study, we identified 2416 DEGs that have potential to be novel regulators and might, at least in part, elucidate the pathophysiological mechanism of IPO in attenuating hepatic I/R injury. Through the use of bioinformatics analysis, we found that the most enriched BP and MF terms for DEGs were almost all related to intracellular nucleic acid and protein metabolic and binding processes, indicating that hepatocyte necrosis and proliferation play a crucial role in hepatic I/R injury and IPO-induced protection. Our findings agree with a previous report stating that cell necrosis and apoptosis caused by damaged ATP biosynthesis contributes substantially to inflammation in the hepatic reperfusion period [25].
Validation of selected DEGs by qRT-PCR. 12 DEGs were validated using qRT-PCR among 3 groups. And 11 of them were significantly amplified and consistent with the RNA-Sequencing results. *p < 0.05
Protein-protein interaction network of these 11 DEGs. Nodes represented genes. Purple lines represented experimental evidence; yellow lines represented text-mining evidence; light lines represented database evidence; The red dashed frame labelled the MAPK gene family.
In this study, we found the same top 10 significantly enriched pathways between N vs. I/R and I/R vs. I/R+IPO, which were the MAPK pathway, the IL-17 pathway, regulating pluripotency of stem cells, and insulin resistance pathway. The MAPK signalling pathway primarily consists of an extracellular signal‑regulated kinase that regulates numerous cellular activities, including proliferation, differentiation, survival, death and transformation. Signalling activated upon hepatic I/R injury includes members of the MAPK family [26] and, as mentioned in a recent study, propylene glycol alginate sodium sulphate pre-conditioning, which attenuated hepatic I/R injury by focusing on the MAPK pathway [27]. IL-17 is a pro-inflammatory cytokine with a key role recruiting neutrophils and macrophages to sites of inflammation, subsequently causing damage after hepatic I/R injury [28]. Furthermore, Patrizia et al. demonstrated that interferon regulatory factor 3 deficiency enhances hepatic I/R injury by mediating the IL-17 pathway [29]. Pluripotent stem cells (PSCs), which are induced from mesenchymal stem cells (MSCs), have been utilized for basic research because of their high proliferation rate and engraftment capacity [30]. Several reports investigated the pivotal role of PSCs on I/R injury. For instance, glutathione peroxidase 3 delivered in human-induced PSCs (hiPSCs) attenuated hepatic I/R injury by inhibiting hepatic senescence and extracellular vesicles released from MSCs, which protect against murine renal and hepatic I/R injury [31-33]. Insulin is an important hormone that reduces plasma glucose in vivo and is regulated by insulin signalling. Although a previous report indicated that hepatic I/R injury regulates insulin signalling during the early reperfusion phase, the mechanism of insulin resistance in hepatic I/R injury remains unclear. The above results agree with previous evidence and reported mechanisms, highlighting the ability and accuracy of RNA-Seq analysis. In the meantime, we suggest that IPO might protect against hepatic I/R injury by regulating the four predicted pathways.
Data from selected DEGs verification experiments revealed 11 significantly changed genes following qRT-PCR amplification. The expression trend for the 11 qRT-PCR genes was consistent with the RNA-Seq data. Cyr61, which is a gene with one of the largest fold changes in this study, belongs to the CNN protein family and regulates complex cellular activities such as cell adhesion, proliferation and apoptosis [34]. Bian et al. reported that Cyr61 expression in hepatocytes was involved in the hepatic pro-inflammatory response and macrophage infiltration in murine non-alcoholic fatty liver disease [35], which agrees with the results of this study. Furthermore, Atf3, which is a member of the ATF/cyclic AMP-responsive element binding protein transcription factor family that represses inflammatory gene expression in multiple diseases [36], was also significantly up-regulated. Several previous reports demonstrated that I/R can significantly increase Atf3 expression during the reperfusion phase in the kidney, heart and brain [37-39]. As far as we know, regarding the potential mechanisms involved in the IPO, several studies have postulated that IPO decreases the burst production of pro-inflammatory mediators [23], modulates the hepatocytes apoptotic cascade [10], and improves liver regeneration [40], which showed good agreement with our data. Taken together, we suggest Cyr61 and Atf3 may serve a vital role in the development of IPO attenuating hepatic I/R injury.
Furthermore, six amplified DEGs (Dusp1/6, Gadd45a/b, Egr1 and Epha2) were significantly enriched in the predicted MAPK signalling pathway, emphasizing the importance of this pathway in the hepatic I/R and IPO process. Dusp 1/6 is a member of the Dusp protein family, which dephosphorylates the threonine/ serine and tyrosine residues of their substrates [41]. Tongda Xu revealed that in myocardial I/R injury, inhibition of Dusp2- mediated c-JNK dephosphorylation and activation of Dusp4/16-mediated extracellular regulated protein kinases1/2 (ERK1/2) phosphorylation exerted an anti-apoptotic role [42]. Furthermore, Gadd45b and Egr1 appeared to be pivotal factors preventing apoptosis and autophagy during cerebral I/R injury [43, 44]. And targeting Epha2 receptors might be a novel anticancer strategy because of the critical role Epha signalling plays in tumour growth and metastasis [45]. The functions of six DEGs were mainly associated with apoptosis and autophagy, which was in line with MAPK pathway's role. At the same time, PPI network indicated that MAPK pathway played a significant part in these DEGs interactions. Several studies also have reported that IPO inhibits apoptosis after renal and liver I/R injury [16, 46]. Our data and previous evidences have suggested that these DEGs and MAPK pathway makes a contribution to IPO attenuating liver I/R injury. For other amplified DEGs, Chao et al. reported that Nr4a1 deletion altered systemic glucose metabolism and caused insulin resistance after deletion in mice [47]. The main connection of Gdf15 in liver disease has been with non-alcoholic steatohepatitis and hepatic fibrosis [48, 49]. In reviewing the literature, no evidence was discovered associating liver disease with Osgin1 or Lpin2 expression. However, the potential function and role of these DEGs in the pathophysiology of hepatic I/R injury and IPO require further exploration.
Conclusion
To the best of our knowledge, this study is the first to explore gene expression profiling with regard to hepatic I/R and IPO using next-generation RNA-Seq. We suggested a few progression-related genes and pathways, such as Cyr61, Atf3, MAPK pathway and IL-17 pathway and so on, providing some clues for future experimental research. Further validations, particularly in human tissues, may provide more comprehensive understanding of the underlying biological pathophysiology surrounding ischemic postconditioning attenuating mouse liver I/R injury.
Abbreviations
IPO: Ischemic postconditioning; I/R: ischemia/ reperfusion; DEGs: differentially expressed genes; GO: Gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MAPK: mitogen-activated protein kinase; PPI: Protein-protein interaction; KCs: Kupffer cells; TNF-α: tumor necrosis factor alpha; IL-1β: interleukin1β; ROS: reactive oxygen species; ALD: Alcoholic liver disease; ALT: alanine aminotransferase; AST: aspartate aminotransferase; FC: fold change; BP: Biological Process; CC: Cellular Component; MF: Molecular Function; CNC: Coding & Noncoding Co-expression; PCC: Partial correlation coefficient. FPKM: Fragments per Kilobase of transcript per million mapped reads; PSC: Pluripotent stem cell; hiPSC: human-induced PSC; MSC: mesenchymal stem cell; AMP: Adenosine monophosphate; JNK: c-Jun N-terminal kinase; ERK1/2: Extracellular regulated protein kinases 1/2.
Acknowledgements
We would like to thank Kangchen Bio-tech for their technical support with the next-generation RNA-Seq. The work was supported by the National Natural Science Foundation of China (grant number U1403222).
Authors' contributions
PZ and YN performed the animal experiments and wrote the manuscript. PZ, YM, KC and YN analyzed the data. QY designed the study and contributed experimental materials. All authors read and approved the final version of the manuscript.
Data Availability
The next-generation RNA-Seq, GO and KEGG analysis data used to support the findings of this study are available from the corresponding author on reasonable request. or GEO database (accession number: GSE117066).
Competing Interests
The authors have declared that no competing interest exists.
References
1. de Rougemont O, Lehmann K, Clavien PA. Preconditioning, organ preservation, and postconditioning to prevent ischemia-reperfusion injury to the liver. Liver Transplantation. 2009;15:1172-82
2. Boros P, Bromberg J. New cellular and molecular immune pathways in ischemia/reperfusion injury. American Journal of Transplantation. 2006;6:652-8
3. Jaeschke H, Bautista AP, Spolarics Z, Spitzer JJ. Superoxide generation by Kupffer cells and priming of neutrophils during reperfusion after hepatic ischemia. Free radical research communications. 1991;15:277-84
4. Jaeschke H. Reactive oxygen and ischemia/reperfusion injury of the liver. Chemico-biological interactions. 1991;79:115-36
5. Akai S, Uematsu Y, Tsuneyama K, Oda S, Yokoi T. Kupffer cell-mediated exacerbation of methimazole-induced acute liver injury in rats. Journal of Applied Toxicology. 2016;36:702-15
6. Liu Y, Ji H, Zhang Y, Shen X, Gao F, He X. et al. Recipient T cell TIM-3 and hepatocyte galectin-9 signalling protects mouse liver transplants against ischemia-reperfusion injury. Journal of hepatology. 2015;62:563-72
7. Huang J, Shen X-D, Yue S, Zhu J, Gao F, Zhai Y. et al. Adoptive Transfer of Heme Oxygenase-1 (HO-1)-Modified Macrophages Rescues the Nuclear Factor Erythroid 2-Related Factor (Nrf2) Antiinflammatory Phenotype in Liver Ischemia/Reperfusion Injury. Molecular Medicine. 2014;20:448
8. Feng M, Wang Q, Zhang F, Lu L. Ex vivo induced regulatory T cells regulate inflammatory response of Kupffer cells by TGF-beta and attenuate liver ischemia reperfusion injury. International immunopharmacology. 2012;12:189-96
9. Kin H, Wang N-P, Mykytenko J, Reeves J, Deneve J, Jiang R. et al. INHIBITION OF MYOCARDIAL APOPTOSIS BY POSTCONDITIONING IS ASSOCIATED WITH ATTENUATION OF OXIDATIVE STRESS-MEDIATED NUCLEAR FACTOR-κκB TRANSLOCATION AND TNFαα RELEASE. Shock. 2008;29:761-8
10. Yoon S-Y, Kim CY, Han HJ, Lee KO, Song T-J. Protective effect of ischemic postconditioning against hepatic ischemic reperfusion injury in rat liver. Annals of surgical treatment and research. 2015;88:241-5
11. Uehara T, Bennett B, Sakata ST, Satoh Y, Bilter GK, Westwick JK. et al. JNK mediates hepatic ischemia reperfusion injury. Journal of hepatology. 2005;42:850-9
12. Penna C, Tullio F, Moro F, Folino A, Merlino A, Pagliaro P. Effects of a protocol of ischemic postconditioning and/or captopril in hearts of normotensive and hypertensive rats. Basic research in cardiology. 2010;105:181-92
13. Zhuo C, Wang Y, Wang X, Wang Y, Chen Y. Cardioprotection by ischemic postconditioning is abolished in depressed rats: role of Akt and signal transducer and activator of transcription-3. Molecular and cellular biochemistry. 2011;346:39-47
14. Santos CHMd, Gomes OM, Pontes JCDV, Miiji LNO, Bispo MAF. The ischemic preconditioning and postconditioning effect on the intestinal mucosa of rats undergoing mesenteric ischemia/reperfusion procedure. Acta cirurgica brasileira. 2008;23:22-8
15. Liu X, Chen H, Zhan B, Xing B, Zhou J, Zhu H. et al. Attenuation of reperfusion injury by renal ischemic postconditioning: the role of NO. Biochemical and biophysical research communications. 2007;359:628-34
16. Chen H, Xing B, Liu X, Zhan B, Zhou J, Zhu H. et al. Ischemic postconditioning inhibits apoptosis after renal ischemia/reperfusion injury in rat. Transplant International. 2008;21:364-71
17. Rehni AK, Singh N. Role of phosphoinositide 3-kinase in ischemic postconditioning-induced attenuation of cerebral ischemia-evoked behavioral deficits in mice. Pharmacological reports. 2007;59:192
18. Santos CHMd, Pontes JCDV, Miiji LNO, Nakamura DI, Galhardo CAV, Aguena SM. Postconditioning effect in the hepatic ischemia and reperfusion in rats. Acta cirurgica brasileira. 2010;25:163-8
19. Teixeira ARF, Molan NT, Kubrusly MS, Bellodi-Privato M, Coelho AM, Leite KR. et al. Postconditioning ameliorates lipid peroxidation in liver ischemia-reperfusion injury in rats. Acta cirurgica brasileira. 2009;24:52-6
20. Sun J, Li B, Sun A, Zhao K, Ma Y, Zhao J. et al. Comprehensive analysis of aberrantly expressed profiles of messenger RNA in alcoholic liver disease. Journal of cellular biochemistry. 2018
21. Arai M, Yokosuka O, Chiba T, Imazeki F, Kato M, Hashida J. et al. Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. Journal of Biological Chemistry. 2003
22. Izuishi K, Tsung A, Hossain MA, Fujiwara M, Wakabayashi H, Masaki T. et al. Ischemic preconditioning of the murine liver protects through the Akt kinase pathway. Hepatology. 2006;44:573-80
23. Guo JY, Yang T, Sun XG, Zhou NY, Li FS, Long D. et al. Ischemic postconditioning attenuates liver warm ischemia-reperfusion injury through Akt-eNOS-NO-HIF pathway. Journal of biomedical science. 2011;18:79
24. Xie C, Li B, Xu Y, Ji D, Chen C. Characterization of the global transcriptome for Pyropia haitanensis (Bangiales, Rhodophyta) and development of cSSR markers. BMC genomics. 2013;14:107
25. Sammut IA, Burton K, Balogun E, Sarathchandra P, Brooks KJ, Bates TE. et al. Time-dependent impairment of mitochondrial function after storage and transplantation of rabbit kidneys. Transplantation. 2000;69:1265-75
26. Li J, Wang F, Xia Y, Dai W, Chen K, Li S. et al. Astaxanthin pretreatment attenuates hepatic ischemia reperfusion-induced apoptosis and autophagy via the ROS/MAPK pathway in mice. Marine drugs. 2015;13:3368-87
27. Xu S, Niu P, Chen K, Xia Y, Yu Q, Liu N. et al. The liver protection of propylene glycol alginate sodium sulfate preconditioning against ischemia reperfusion injury: focusing MAPK pathway activity. Scientific reports. 2017;7:15175
28. Kono H, Fujii H, Ogiku M, Hosomura N, Amemiya H, Tsuchiya M. et al. Role of IL-17A in neutrophil recruitment and hepatic injury after warm ischemia-reperfusion mice. The Journal of Immunology. 2011;187:4818-25
29. Loi P, Yuan Q, Torres D, Delbauve S, Laute MA, Lalmand MC. et al. Interferon regulatory factor 3 deficiency leads to interleukin-17-mediated liver ischemia-reperfusion injury. Hepatology. 2013;57:351-61
30. Abu-Amara M, Yang SY, Tapuria N, Fuller B, Davidson B, Seifalian A. Liver ischemia/reperfusion injury: processes in inflammatory networks—a review. Liver Transpl. 2010;16:1016-32
31. Qi X, Ng KT-P, Lian Q, Li CX, Geng W, Ling CC. et al. Glutathione Peroxidase 3 Delivered by hiPSC-MSCs Ameliorated Hepatic IR Injury via Inhibition of Hepatic Senescence. Theranostics. 2018;8:212
32. Yuan X, Li D, Chen X, Han C, Xu L, Huang T. et al. Extracellular vesicles from human-induced pluripotent stem cell-derived mesenchymal stromal cells (hiPSC-MSCs) protect against renal ischemia/reperfusion injury via delivering specificity protein (SP1) and transcriptional activating of sphingosine kinase 1 and inhibiting necroptosis. Cell death & disease. 2017;8:3200
33. Haga H, Yan IK, Takahashi K, Matsuda A, Patel T. Extracellular Vesicles from Bone Marrow-Derived Mesenchymal Stem Cells Improve Survival from Lethal Hepatic Failure in Mice. Stem cells translational medicine. 2017;6:1262-72
34. Jun J-I, Kim K-H, Lau LF. The matricellular protein CCN1 mediates neutrophil efferocytosis in cutaneous wound healing. Nature communications. 2015;6:7386
35. Bian Z, Peng Y, You Z, Wang Q, Miao Q, Liu Y. et al. CCN1 expression in hepatocytes contributes to macrophage infiltration in nonalcoholic fatty liver disease in mice. Journal of lipid research. 2013;54:44-54
36. Chen B, Wolfgang CD, Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Molecular and cellular biology. 1996;16:1157-68
37. Yoshida T, Sugiura H, Mitobe M, Tsuchiya K, Shirota S, Nishimura S. et al. ATF3 protects against renal ischemia-reperfusion injury. Journal of the American Society of Nephrology. 2008;19:217-24
38. Kim M-Y, Seo EJ, Lee DH, Kim EJ, Kim HS, Cho H-Y. et al. Gadd45β is a novel mediator of cardiomyocyte apoptosis induced by ischaemia/hypoxia. Cardiovascular research. 2010;87:119-26
39. Wang L, Deng S, Lu Y, Zhang Y, Yang L, Guan Y. et al. Increased inflammation and brain injury after transient focal cerebral ischemia in activating transcription factor 3 knockout mice. Neuroscience. 2012;220:100-8
40. Young SB, Pires AR, Boaventura GT, Ferreira AM, Martinho JM, Galhardo MA. Effect of ischemic preconditioning and postconditioning on liver regeneration in prepubertal rats. Transplantation proceedings. 2014;46:1867-71
41. Farooq A, Zhou M-M. Structure and regulation of MAPK phosphatases. Cellular signalling. 2004;16:769-79
42. Xu T, Wu X, Chen Q, Zhu S, Liu Y, Pan D. et al. The anti-apoptotic and cardioprotective effects of salvianolic acid a on rat cardiomyocytes following ischemia/reperfusion by DUSP-mediated regulation of the ERK1/2/JNK pathway. PloS one. 2014;9:e102292
43. He G, Xu W, Tong L, Li S, Su S, Tan X. et al. Gadd45b prevents autophagy and apoptosis against rat cerebral neuron oxygen-glucose deprivation/reperfusion injury. Apoptosis. 2016;21:390-403
44. Wang A, Zhang H, Liang Z, Xu K, Qiu W, Tian Y. et al. U0126 attenuates ischemia/reperfusion-induced apoptosis and autophagy in myocardium through MEK/ERK/EGR-1 pathway. European journal of pharmacology. 2016;788:280-5
45. Dobrzanski P, Hunter K, Jones-Bolin S, Chang H, Robinson C, Pritchard S. et al. Antiangiogenic and antitumor efficacy of EphA2 receptor antagonist. Cancer research. 2004;64:910-9
46. Sun K, Liu Z-S, Sun Q. Role of mitochondria in cell apoptosis during hepatic ischemia-reperfusion injury and protective effect of ischemic postconditioning. World journal of gastroenterology: WJG. 2004;10:1934
47. Chao LC, Wroblewski K, Zhang Z, Pei L, Vergnes L, Ilkayeva OR. et al. Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes. 2009;58:2788-96
48. Chung HK, Kim JT, Kim H-W, Kwon M, Kim SY, Shong M. et al. GDF15 deficiency exacerbates chronic alcohol-and carbon tetrachloride-induced liver injury. Scientific reports. 2017;7:17238
49. Kim KH, Kim SH, Han DH, Jo YS, Lee Y-h, Lee M-S. Growth differentiation factor 15 ameliorates nonalcoholic steatohepatitis and related metabolic disorders in mice. Scientific reports. 2018;8:6789
Author contact
![]() Corresponding authors: Qifa Ye. E-mail address: yqf_chinacom; Mail address: No.138 Tongzipo Road, Changsha, Hunan, China; Telephone number: +8615116256469 and Ying Niu. E-mail address: niuying1com; Mail address: No.138 Tongzipo Road, Changsha, Hunan, China; Telephone number: +8613975195016
Corresponding authors: Qifa Ye. E-mail address: yqf_chinacom; Mail address: No.138 Tongzipo Road, Changsha, Hunan, China; Telephone number: +8615116256469 and Ying Niu. E-mail address: niuying1com; Mail address: No.138 Tongzipo Road, Changsha, Hunan, China; Telephone number: +8613975195016

 Global reach, higher impact
Global reach, higher impact