3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(2):189-197. doi:10.7150/ijms.30647 This issue Cite
Research Paper
Antarctic freshwater microalga, Chloromonas reticulata, suppresses inflammation and carcinogenesis
1. Department of Bioscience, Mokpo National University, Muan 58554, Republic of Korea
2. Division of Polar Life Sciences, Korea Polar Research Institute, Incheon, 21990, Republic of Korea
3. South Sea Environment Research Department, Korea Institute of Ocean Science and Technology, Geoje, 656-830, Republic of Korea
4. Department of Pharmacy, Graduate School, Sungkyunkwan University, Suwan 16419, Republic of Korea
5. Department of Polar Sciences, University of Science and Technology, Incheon 21990, Republic of Korea
*These authors contributed equally to this study
Received 2018-10-23; Accepted 2018-12-7; Published 2019-1-1
Abstract
Inflammation triggered by the innate immune system is a strategy to protect organisms from the risk of environmental infection. However, it has recently become clear that inflammation can cause a variety of human diseases, including cancer. In this study, we investigated the effects of an ethanol extract of the Antarctic freshwater microalgae, Chloromonas reticulata (ETCH), on inflammation and carcinogenesis in RAW 264.7 macrophages and HCT116 human colon cancer cells, respectively. ETCH exhibited significant anti-inflammatory activity through the dose-dependent modulation of major inflammatory markers such as COX-2, IL-6, iNOS, TNF-α, and NO production. For example, ETCH reduced LPS-induced upregulation of COX-2, IL-6, iNOS, and TNF- alpha mRNA levels, leading to a significant decrease in the levels of LPS-stimulated NO and IL-6 as well as TNF-alpha products. In contract, ETCH exhibited dose-dependent cytotoxic activity against HCT116 cells, yielding a profound reduction in the proliferation of the cancer cells. Furthermore, ETCH induced G2 phase cell cycle arrest by transcriptionally regulating of genes involved in G2 / M transition including p21 (CDKN1A), cyclin B1 (CCNB1), and CDK1; CDKN1A mRNA levels were upregulated in response to ETCH, whereas CCNB1 and CDK1 were downregulated. This study reports for the first time anti-inflammatory and anti-cancer effects of, C. reticulata and provides new insights into the molecular mechanisms of the linkage between inflammation and cancer.
Keywords: Inflammation cancer, Chloromonas reticulata, pro-inflammatory cytokines, HCT116
Introduction
Inflammation is the initial immune reaction that occurs when a foreign pathogenic agent disrupts cellular homeostasis and is an essential process for the survival of living organisms [1-3]. Chronic inflammation is likely to cause various diseases, including cancer. Therefore, if the inflammatory response is not controlled early, it may lead to the development of chronic infection, autoimmune diseases and metabolic disorders due to loss of function and homeostatic imbalance inside the living body [4-6]. Recent studies regarding increasing cancer incidence in chronic inflammatory sites have shown that inflammation and cancer have a common signaling pathway through vicious cycle linkages [7-9]. Over the past decade it has become evident that the inflammatory response is responsible for tumor development, including initiation, malignant transformation, invasion and metastasis. For example, many studies show that an inflammatory microenvironment not only promotes cancer cell growth, but also can lead to DNA damage and genomic instability leading to increased mutation rates [10-12]. The complex processes from inflammation to cancer development can occur through the elaborate modulation of major inflammatory regulatory molecules such as cytokines, NF-kB, COX-2 and iNOS. For example, cytokines produced in immune / inflammatory cells, including IL, TNF-a, and growth factors, play a modulatory role in tumorigenesis via the regulation of transcription factors such as NF-kB, STAT3, and AP-1, which act as mediators of inflammatory processes that regulate the expression of a wide range of cytokines such as IL-6, TNF-α, and chemokines [13, 14]. Recent studies have also shown that NF-kB continuously activated in various cancer types and has a role in uncontrolled cell growth, inhibition of cell death, induction of metastasis, and angiogenesis [12-14]. It has also been demonstrated that iNOS, an enzyme that induces NO production and an important regulatory molecule in both inflammation and cancer development, is overexpressed in chronic inflammatory diseases and various cancers [15, 16]. Another key regulator in the inflammatory pathway, COX-2 expression can be activated in response to various stimuli including growth factors such as proinflammatory cytokines including IL-6, TNF-α, and epidermal growth factors [17, 18]. COX-2 is also overexpressed in various types of cancer and is involved in cell proliferation, anti-apoptotic activity, angiogenesis and metastasis [19, 20]. As a result, these inflammatory substances secreted by inflammatory cells, including various cytokines, are closely related to the proliferation, survival, and metastasis of cancer cells. Inflammation in the microscopic environment of the tumor can promote tumor growth [21]. It not only aids the survival and proliferation of cancer cells, but destroys the adaptive immune response, and changes the response to hormones and therapeutic drugs. Therefore, the identification of regulatory mechanisms that mediate the common signaling pathway between inflammation and cancer and the development of drugs that interfere with these mechanisms may be a prudent approach to treat cancer by cutting off the link between inflammation and cancer.
According to recent studies, algal extracts and secondary metabolites including carotenoids, fatty acids, glycolipids, and polysaccharides have been shown to exhibit anti-inflammatory and anti-carcinogenic activities, and their pharmacological and therapeutic potentials have increasingly been highlighted [22-24]. However, the regulatory mechanisms underlying the biological activity of microalgae-derived metabolites are not yet clear. In this study, we evaluated the anti-inflammatory and cytotoxic effects of ethanol extracts from the polar microalga, Chloromonas reticulata, on mouse macrophage RAW 264.7 cells and human colon cancer HCT116 cells, respectively. Anti-inflammatory activity was determined in LPS-induced Raw 264.7 cells by measuring transcriptional levels of inflammatory regulatory genes including IL-6, TNF-α, COX-2 and iNOS. In the experiments on anticancer activity of the extracts, cancer cell proliferation and cytotoxic effects on HCT116 cells were evaluated. These results suggest that microalgae-derived extracts can be promising candidates for anti-cancer and anti-inflammatory drugs in targeted treatment against colorectal cancer.
Materials and Methods
Preparation of extract from polar microalga
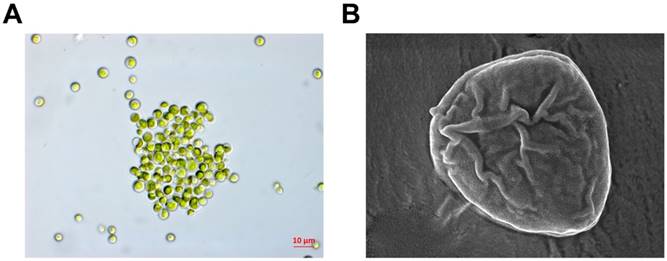
The nucleotide sequence of the genomic 18S rDNA of the Antarctic freshwater microalga, C. reticulate, which was collected near the Sejong Station (62˚ 13 'S, 58˚ 47'W), was analyzed using BLAST for sequence similarity with the NCBI GenBank database. C. reticulata samples were deposited at the Korea Polar Research Institute (#KSF100) (Figure 1). Microalgae extracts were prepared using the ethanol extraction method as described in a previous study [24].
Morphological observation using light (A) and scanning electron microscopy (B)
Cell culture
The mouse macrophage line RAW 264.7 cells and HCT116 cells used in this study were purchased from the Korean Cell Line Bank (KCLB, Seoul, Korea) and cultured in DMEM high glucose medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated FBS, 3 mM Glutamine, antibiotics (100 U/mL penicillin and 100 U/mL streptomycin) (Invitrogen, Grand Island, NY, USA) and placed in an incubator with 5% CO2 in a humidified atmosphere at 37 °C.
Determination of nitric oxide production
RAW 264.7 macrophages were stimulated with LPS from E. coli (500 ng/mL, Sigma-Aldrich, CA, USA) for 24 h after pretreatment with the ethanol extract at different concentrations (5, 10, 20, and 40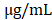 ) for 1 h, and NO production was measured in the culture solution. Griess reagent (1% sulfanilamide, 0.1% N-(1-naphathyl)-ethylenediamine dihydrochloride and 5% phosphoric acid, Promega) was added to the 96-well plate at a ratio of 1:1 and reacted for 10 min. Absorbance was measured at 540 nm using a microplate reader (Infinite 200 pro, TECAN).
) for 1 h, and NO production was measured in the culture solution. Griess reagent (1% sulfanilamide, 0.1% N-(1-naphathyl)-ethylenediamine dihydrochloride and 5% phosphoric acid, Promega) was added to the 96-well plate at a ratio of 1:1 and reacted for 10 min. Absorbance was measured at 540 nm using a microplate reader (Infinite 200 pro, TECAN).
Enzyme-linked immunosorbent assay (ELISA)
Enzyme-linked immunosorbent assay (ELISA) was performed to determine the amounts of cytokines in the cell culture. RAW 264.7 cells were seeded into 24-well plates at the density of 4 x 105 cells/well. After 24 h of incubation, the cells were pretreated with the ethanol extract (5, 10, 20, 40 µg/mL) and then treated with 0.5 µg/mL LPS after 1 h. After 18 h, the cell culture was obtained and used for cytokine measurement. The concentrations of TNF-α and IL-6 in the supernatants of RAW 264.7 cell cultures were determined using an ELISA kit, according to the manufacturer's instructions (BioLegend, Inc.). Briefly, the culture was diluted to the appropriate concentration, and 50 μL of the supernatant was added to a 96-well plate coated with cytokines overnight at 4 °C. After washing three times with the washing buffer, 100 μL of biotinylated antibody reagent was added to each well, allowed to react at room temperature for 1 h, washed three times, treated with 100 μL of streptavidine-HRP solution and incubated for 1 h at room temperature. After the reaction, the cells were washed three times with the washing buffer. Following reaction with 100 μL of di (2-ethylhexyl) -2, 4, 5-trimethoxy benzalmalonate (TMB) substrate for 5-30 min, 100 μL of the stop solution was added and absorbance was measured at 450 nm.
Cytotoxicity assay
An MTS assay was performed to determine the cytotoxic effect of the ethanol extract on RAW 264.7 cells and HCT116 cells. The cells were seeded into 3 × 105cells / well on a 96-well plate. After incubation for 24 h, 10% MTT solution was added to the cell culture medium, and it was incubated at 37 °C for 2 h. The absorbance was measured using a microplate reader (Thermo Scientific Inc., San Diego, CA, USA) at 570 nm.
Colony-forming assay
The HCT116 cell line was treated with the ethanol extract and a colony formation assay was performed to measure cell viability. Cells were plated at 1 × 103 cells/mL in a 6-well plate. The cells were treated with 0, 12.5, 25, 50, and 100 µg/mL ethanol extracts for 24 h and washed in fresh medium. They were then incubated at 37 °C in a 5% CO2 incubator for 7-8 days. The colony formed was fixed with 10% formalin and counted by staining with 0.01% crystal violet as described in a previous study [25]. Cell viability was expressed as 100% of the control value.
Quantitative real-time polymerase chain reaction analysis (RT-PCR)
RAW 264.7 cells were seeded at a density of 1 × 105 cells / well in a 6- well plate and incubated for 24 h to stabilize the cells. The cells were treated with LPS (500 ng/mL) for 30 min, followed by incubation for 24 h. The cells were washed twice with PBS, and total RNA was extracted using Tripure Isolation Reagent (Roche). Next, 5 μg of total RNA was synthesized with cDNA using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). RT-PCR was performed by monitoring the increase in the amount of SYBR-Green in real time using a Rotor-Gene 6500 RT-PCR (Corbett Research, Sydney, Australia). The primer sequences used are indicated in Table S1. The RT-PCR thermal cycling conditions were as follows: denaturation at 95 °C for 10 sec, annealing at 60 °C for 15 sec and elongation at 72 °C for 60 sec. RT-PCR data were collected using the Rotor-Gene 6500 detection system.
Immunoblot analysis
The expression levels of iNOS and COX-2 proteins were analyzed using Western blot to confirm the anti-inflammatory effects of the ethanol extract. The cells were lysed using a buffer containing 2 mM phenyl-methylsulfonyl fluoride, protease inhibitors (cOmpleteTM, Roche), 1 mM Na3VO4, 50 mM NaF and 10 mM EDTA. The cell lysate was centrifuged at 15,000 × g for 30 min at 4 °C to obtain a supernatant containing protein and quantified using the Bradford method. Next, 20 μg of the quantified protein was electrophoresed on 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (BIO-RAD, Hercules, CA, USA). The membrane was blocked with T-TBS (0.1% Tween 20 + TBS) solution containing 5% bovine serum albumin (BSA) for 2 h at room temperature. The primary antibody against iNOS (1/1000 dilution), COX-2 (1/1000 dilution), and GAPDH (1/1000 dilution) was exposed to a secondary antibody, horseradish peroxidase-conjugated anti-rabbit IgG (1/5000 dilution), and the membrane was reacted with ECL detection reagents (Millipore, Billerica, MA, USA).
Cell cycle analysis
To evaluate cell cycle arrest induced by the ethanol extract in HCT116 cells, flow cytometry assay was performed. The cells were seeded at 1 × 106 cells/well into a 6-well plate and cultured for 24 h. Then, the ethanol extract was added at concentrations of 25, 50, and 100 μg/mL. The cells were cultured for 24 h, and the suspended cells and trypsin-treated cells were collected and centrifuged at 2,000 rpm for 5 min at 4 °C. The supernatant was removed and further centrifuged at 5,000 rpm for 5 min at 4 °C. Again, the supernatant was removed and 1 mL 70% ethanol was added to fix the cells at 4 °C. Next, staining was done using cell cycle reagent (20 μg/mL propidium iodide, 0.1 mM EDTA, 10 µg/mL RNase (from bovine pancreas, and DNase free) and 1% triton X-100 prepared in PBS) for 30 min in dark at 4 °C. Finally, a flow cytometer (Beckman Coulter EPICS XL) was used to analyze the cell cycle.
Statistical analysis
Values are presented as mean ± SEM of three independent biological experiments. Statistically significant differences between each treated group and the control group were determined using one-way analysis of variance followed by Student's t-test; p values < 0.05 were considered to indicate a statistically significant difference.
Results
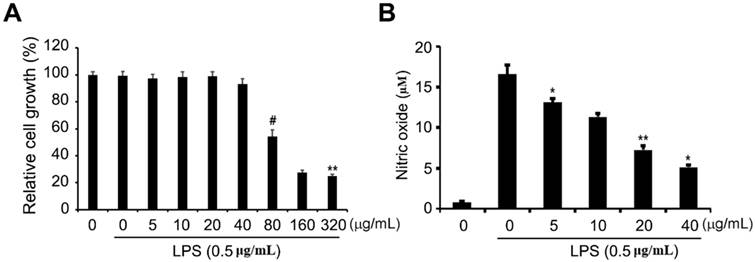
Effects of ethanol extract on RAW 264.7 cell viability and NO production
To determine the cytotoxic effects of the ethanol extract on RAW 264.7 cells, the cells were treated with different concentrations of the extract ranging from 5 to 320 μg/mL and then incubated with 1 μg/mL LPS for 24 h, after which the cellular proliferation assay (MTS) was performed. As shown in Fig 2A, RAW 264.7 cell proliferation was not significantly affected by treatment with the ethanol extract of up to 40 μg/mL compared to that of cells without LPS treatment. LPS stimulates toll-like receptor 4 (TLR4) in macrophages or monocytes and pro-inflammatory cytokines, including tumor necrosis factor-α (TNF-α), interleukin oxalate (NO), and eicosanoid [26, 27]. These inflammatory mediators are known to cause lethal consequences to the host if acute or excessive reactions occur, and they play an important role in bio-defense by producing NO and cytokine early in the inflammatory reaction. Therefore, we wondered whether the ethanol extract could exhibit an inhibitory effect on inflammation. Griess reagent assay was performed to determine the level of NO, which is one of the major pro-inflammatory mediators in LPS-induced RAW 264.7 cells exposed to non-cytotoxic concentrations of ethanol extract (5 to 40 μg/mL). We observed that ethanol extract led to a significant dose-dependent decrease in LPS-accelerated NO production (Fig. 2B), suggesting that the ethanol extract has anti-inflammatory activity in LPS-induced inflammation.
Effects of ETCH on cellular proliferation and NO production in LPS-induced RAW 264.7 macrophages. After 24 h of treatment, cell viability was evaluated using an MTT assay (A) and NO production was determined by Griess reaction (B). Statistical significance was determined by Student's t-test. The results are shown as mean ± SD values of three experiments. *p < 0.05, **p < 0.01, and #p < 0.001.
Effects of ETCH on the expression levels of proinflammatory regulators. The Transcriptional (A) and translational (B) expression levels of iNOS and COX-2 genes in response to ETCH treatment. The data have been normalized using an internal control, GAPDH. Statistical significance was determined by Student's t-test. Results are shown as mean ± SD values of three experiments; *p < 0.05, **p < 0.01, and #p < 0.001.
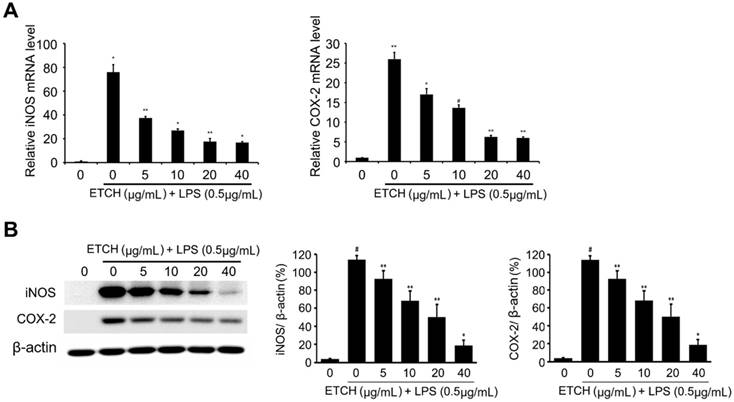
Effect of ethanol extract on LPS-induced iNOS/COX-2 expression
iNOS is an enzyme that makes a primary contribution to the synthesis of NO and produces NO by converting intracellular L-arginine into L-citrulline [28, 29]. The excess NO produced in this way induces the expression of inflammatory cytokines and causes an inflammatory reaction that results in tissue damage, gene mutations, and neuronal cell damage. In addition, COX-2, which is produced via pro-oxidant or pro-inflammatory stimuli by the activation of MEKK-1 and NF-κB, plays a pivotal role in inflammatory response as it increases prostaglandin synthesis. To investigate the relevance of iNOS and COX-2 in inhibitory mechanisms of NO production, both mRNA and protein expression levels of iNOS and COX-2 were examined using qRT-PCR and immunoblot analysis. According to our data, the expression levels of iNOS and COX-2 were strongly induced during LPS treatment at both the transcriptional and translational levels, whereas the expression level of iNOS induced by LPS was significantly decreased in a concentration-dependent manner in the experimental group treated with different concentrations of the ethanol extract ranging from 5 to 40 μg/mL (Fig. 3). Taken together, these data suggest that the ethanol extract C. reticulata, has strong anti-inflammatory effects because it inhibits NO production via the suppression of COX-2 and iNOS expression.
Effects of the ethanol extract on LPS-induced pro-inflammatory cytokine production
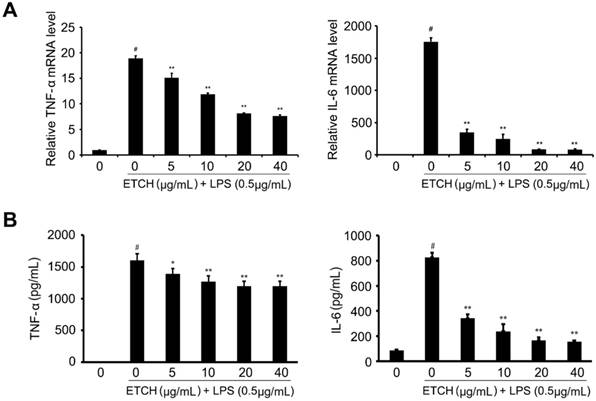
To investigate the effects of the ethanol extract on the expression of various proinflammatory and inflammatory cytokines induced by LPS, such as TNF-α and IL-6, we examined the mRNA expression levels of cytokines. After pretreatment with the ethanol extract for 24 h, the expression levels of TNF-α and IL-6 mRNAs in LPS-stimulated cells were determined by qRT-PCR. We found that TNF-α mRNA levels gradually decreased in a concentration-dependent manner compared to that of the LPS-treated group without ethanol extract, whereas IL-6 was significantly reduced even at the low concentration of 5 μg/mL, suggesting that ethanol extract contained a large amount of physiologically active substances capable of reducing the mRNA levels of inflammation-related genes (Fig. 4). In addition, LPS-induced increases in TNF-α and IL-6 production were also decreased in response to treatment of ethanol extract; IL-6 production was significantly decreased in a concentration-dependent manner, whereas TNF-α was slightly affected despite a significant decrease in its mRNA levels.
Cytotoxic effects of ethanol extract on human colon cancer cells
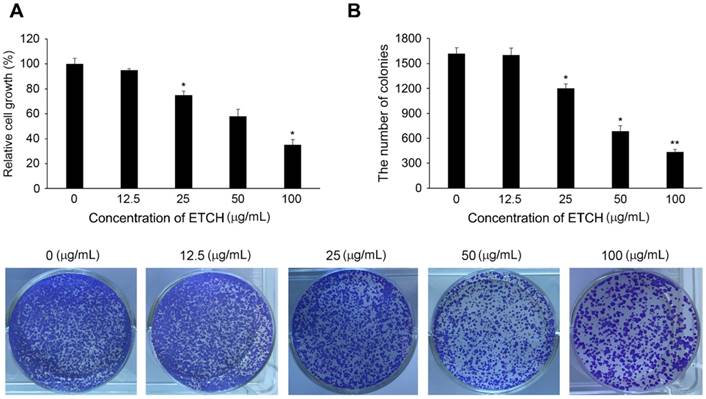
If acute inflammation is not resolved and progresses to chronic inflammation, inflammatory cells produce various inflammation inducers such as TNF-α and interleukin-6 (IL-6), which can damage or mutate DNA and initiate tumorigenesis. To better understand the effect of ethanol extract on the linkage between inflammation and cancer, we investigated its anti-cancer activity by determining its growth inhibitory regulation of human colon cancer cells, HCT116. In this study, HCT116 cells were treated with various concentrations of the extract (0, 12.5, 25, 50, and 100 μg/mL) and incubated for 24 h. As shown in Figure 5A, cellular proliferation in HCT116 was found to be inhibited in a concentration-dependent manner: the survival rate was 92.2% at the concentration of 12.5 μg/mL, 78.5% at the concentration of 25 μg/mL, 58.8% at the concentration of 50 μg/mL, and 35.4% at the concentration of 100 μg/mL. In addition, the cancer cells treated with the extract showed significant suppression of colony formation unlike controls (Fig. 5B). These results suggest that the ethanol extract has high anti-cancer activity against colon cancer cells and may act as an effective inhibitor of cancer development caused by chronic inflammation.
Effects of ETCH on the expression and production of proinflammatory cytokines. Transcriptional levels (A) and production (B) of cytokines, TNF-α and IL-6. Statistical significance was determined by Student's t-test. Results are shown as mean ± SD values of three experiments. *p < 0.05, **p < 0.01, and #p < 0.001.
Cytotoxic effects of ETCH on the growth of human colon cancer cells, HCT116. Inhibition of cellular proliferation (A) and colony formation (B) in response to ETCH treatment. ). Statistical significance was determined by Student's t-test. The results are shown as mean ± SD values of three experiments. *p < 0.05, and **p < 0.01.
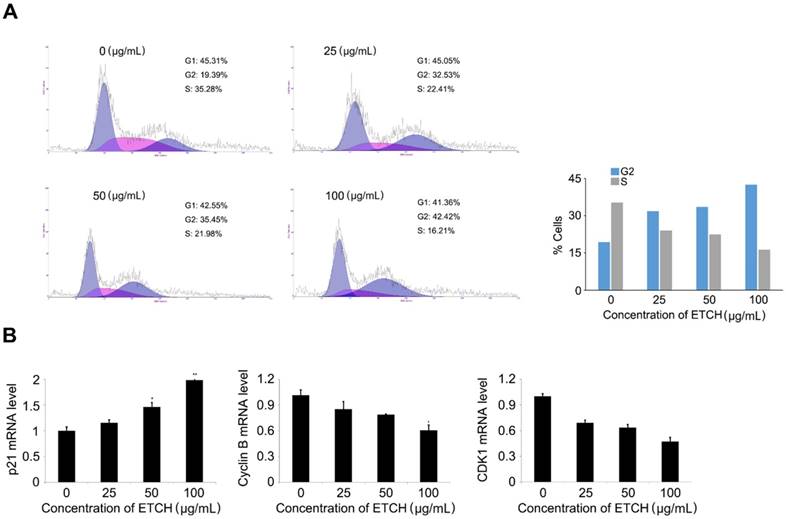
Induction of cell cycle arrest (A) and expression pattern of the cell cycle-regulated genes in response to ETCH treatment (B). Statistical significance was determined by Student's t-test. Results are shown as mean ± SD values of three experiments. *p < 0.05, **p < 0.01.
Induction of cell cycle arrest in ethanol extract-treated cancer cells
To better understand the mechanism of the anticancer activity of the extracts, the effect of the extract on the cell cycle distribution of HCT116 cells was investigated. HCT116 cells were treated with various concentrations of the extract (0-100 μg/mL), cultured for 24 h, and then analyzed by flow cytometry. We observed that the extract increased cell populations in the G0 / G1 phase in a concentration-dependent manner, while reducing the cell population in the S phase, leading to G2 arrest. Of the control cells, 19.36% and 35.28% were in the G2 phase and S phase, respectively. However, the percentage of extract-treated cells in the G2 phase (38.61%) gradually increased in response to increasing doses of the extract (19.36% at 0 μg/mL, 32.53% at 25 μg/mL, 35.45% at 50 μg/mL and 42.42% at 100 μg/mL). The cell population in the S phase was decreased (35.28% at 0 μg/mL, 22.42% at 25 μg/mL, 21.98% at 50 μg/mL and 16.21 μg/mL) (Fig. 6A). Furthermore, G2 arrest in response to the extract treatment was supported by the expression pattern of cell-cycle-associated genes; expression levels of cyclin B1 and CDK1 mRNA, whereas are regulators associated with G2 transition, were gradually decreased, while the level of p21 mRNA, an inhibitor of G2 transition, was increased (Fig. 6B). These data imply that, at least in part, G2 arrest can contribute to inhibitory regulation of extract-mediated cellular proliferation through the modulation of marker genes associated with G2 transition, such as p21, cyclin B1, and CDK1.
Discussion
Inflammatory manifests due to various causes such as physical or chemical stimulation including bacterial infection and is one of the mechanisms for repairing or regeneration of the damaged tissues [1-3]. However, according to both molecular biology studies and clinical or epidemiological studies, chronic inflammation is involved in a variety of carcinogenic processes such as induction of genetic instability, and metaplastic variation and aberrant gene expression, induction of proliferation, apoptosis resistance, and angiogenesis [3-5]. In fact, various inflammatory mediators including cytokines, chemokines, cyclooxygenase-2 (COX-2), prostaglandin (PG), and nitric oxide synthase (NOS) have been implicated in tumor initiation, progression, and metastasis. It was recently reported that physiologically active pharmaceuticals derived from microalgae including anti-tumor, anti-bacterial, anti-fungal, and neuroactive substances are being produced. For example, tubercidin, a nitrogen compound extracted from Tolypothrix byssoidea, has been shown to be active in vitro against P-388 lymphocytic leukemia [22]. L-asparaginase extracted from Chlamydomonas inhibits the growth of lymphoid sarcoma in rat models [30]. Here, we investigated the anti-inflammatory and anti-cancer effects of ethanol extracts from C. reticulata in RAW 264.7 and HCT116 cells, respectively. The MTS assay was first performed on RAW 264.7 cells to define a non-cytotoxic concentration range. The ethanol extract was found to markedly reduce the expression of major regulatory factors of inflammation such as iNOS and COX-2 at the both transcriptional and translational levels, and to consequently inhibit NO production. In addition, ETCH significantly inhibited HCT116 cell proliferation, at least in part by arresting G2 phase transition. In general, macrophages are distributed evenly throughout the human body and play a role in both inflammatory and immune responses. LPS stimulates TLR4 to activate NF-κB, TNF-α, IL-6, and various other inflammatory factors such as NO and PGs [26-28]. An appropriate concentration of NO is physiologically necessary for smooth muscle relaxation, platelet aggregation inhibition, immune regulation, vasodilation and neurotransmission. However, excess NO production lead to various chronic inflammatory diseases. As another important inflammatory factor, PGE2, produced by COX-2, increases the permeability of blood vessels and may cause fever and swelling during the inflammation process [17-19]. In addition, cytokines such as TNF-α, IL-1β, and IL-6 are mainly produced by activated macrophages and play a role in early inflammatory responses against microorganisms. Therefore, inhibition of overexpression of iNOS, COX-2, and inflammatory cytokines, precursors of NO and PGE2 and inflammatory factors, in activated macrophages may be an important factor in controlling the inflammatory response. In this study, RAW 264.7 cells were treated with ETCH and treated with LPS to induce inflammation, followed by NO and their precursors, iNOS, COX-2 expression, and inflammatory cytokines such as TNF-α and IL-6. Based on our data, ETCH effectively reduced the production of LPS-induced inflammatory factors and precursors and inflammatory cytokines and regulated the expression levels of inflammatory regulators such as COX-2, IL-6, iNOS, and TNF-α at the transcriptional or translational level, leading to increased production of inflammatory NO and inflammatory cytokines IL-6 and TNF-α. However, both mRNA and protein levels of IL-6 were significantly inhibited by ETCH treatment, but TNF-α production was slightly decreased as compared to its mRNA level. This discrepancy between TNF-α mRNA and production levels suggests that the expression levels of TNF-α mRNA not significantly affect its biosynthesis in macrophages. On the other hand, harsh environments such as those of the Antarctic region would have required organisms including microalgae to develop specific self-defense mechanisms through the synthesis of specific secondary metabolites necessary for environmental adaptation. Therefore, the ETCH used in this study may contain bioactive molecules with potent anti-inflammatory and anti-cancer effects. In addition, the identification of physiologically active compounds that mediate common signaling pathways between inflammation and cancer and the development of drugs that interfere with the function of key regulatory genes can be an effective approach to cancer treatment by eliminating the link between inflammation and cancer. In a further study, the biological efficacy of the microalgae extract will be clarified, and the potential for its commercial use in functional food materials and anti-cancer drugs will be demonstrated.
Supplementary Material
Supplementary table.
Acknowledgements
This study was supported by the research projects (PE19180) of Korea Polar Research Institute, the Republic of Korea and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1C1B5085764).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Petersen HJ, Smith AM. The role of the innate immune system in granulomatous disorders. Front Immunol. 2013;4:120
2. Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol Cell. 2014;54:281-288
3. Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859-870
4. Straub RH, Schradin G. Chronic inflammatory systemic diseases. Evol Med Public Health. 2016;2016:37-51
5. Okin D, Medzhitov R. Evolution of inflammatory diseases. Curr Biol. 2012;22:R733-R740
6. Chimenti MS, Triggianese P, Conigliaro P, Candi E, Melino G, Perricone R. The interplay between inflammation and metabolism in rheumatoid arthritis. Cell Death Dis. 2015;6:e1887
7. Hunter P. The inflammation theory of disease. EMBO Rep. 2012;13:968-970
8. Hasturk H, Kantarci A, Van Dyke TE. Oral inflammatory diseases and systemic inflammation: role of the macrophage. Front Immunol. 2012;3:118
9. Godkin A, Smith KA. Chronic infections with viruses or parasites: breaking bad to make good. Immunology. 2017;150:389-396
10. Kim JB, Han AR, Park EY, Kim JY, Cho W, Lee J. et al. Inhibition of LPS-induced iNOS, COX-2 and cytokines expression by poncirin through the NF-kappaB inactivation in RAW 264.7 macrophage cells. Biol Pharm Bull. 2007;30:2345-2351
11. Chun KS, Cha HH, Shin JW, Na HK, Park KK, Chung WY. et al. Nitric oxide induces expression of cyclooxygenase-2 in mouse skin through activation of NF-kappaB. Carcinogenesis. 2004;25:445-454
12. Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86
13. Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2:823-830
14. Park MH, Hong JT. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5:15
15. Vannini F, Kashfi K, Nath N. The dual role of iNOS in cancer. Redox Biol. 2015;6:334-343
16. Kostourou V, Cartwright JE, Johnstone AP, Boult JK, Cullis ER, Whitley G, Robinson SP. The role of tumour-derived iNOS in tumour progression and angiogenesis. Br J Cancer. 2011;104:83-90
17. Hua F, Li CH, Wang H, Xu HG. Relationship between expression of COX-2, NF-κB, IL-6 and autoimmune-type recurrent miscarriage. Asian Pac J Trop Med. 2013;6:990-994
18. Harizi H, Norbert G. Inhibitionof IL-6, TNF-alpha, and cyclooxygenase-2 protein expression by prostaglandin E2-induced IL-10 in bone marrow-derived dendritic cells. Cell Immunol. 2004;228:99-109
19. Liu B, Qu L, Yan S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. 2015;15:106
20. Pang LY, Hurst EA, Argyle DJ. Cyclooxygenase-2: A role in cancer stem cell survival and reproduction of cancer cells during therapy. Stem Cells Int. 2016;2016:2048731
21. Mostofa AG, Punganuru SR, Madala HR, Al-Obaide M, Srivenugopal KS. The process and regulatory components of inflammation in brain oncogenesis. Biomolecules. 2017;7:E34
22. Rodríguez-Luna A, Talero E, Terencio MDC, González-Rodríguez ML, Rabasco AM, de Los Reyes C, Motilva V, Ávila-Román J. Topical Application of Glycolipids from Isochrysis galbana Prevents Epidermal Hyperplasia in Mice. Mar Drugs. 2017;16:pii E2
23. Gutiérrez-Pliego LE, Martinez-Carrillo BE, Reséndiz-Albor AA, Arciniega-Martínez IM, Escoto-Herrera JA, Rosales-Gómez CA, Valdés-Ramos R. Effect of supplementation with n-3 fatty acids extracted from microalgae on inflammation biomarkers from two different strains of mice. J Lipids. 2018:4765358
24. Talero E, García-Mauriño S, Ávila-Román J, Rodríguez-Luna A, Alcaide A, Motilva V. Bioactive compounds isolated from microalgae in chronic inflammation and cancer. Mar Drugs. 2015;13:6152-6209
25. Mijatovic T, Houzet L, Defrance P, Droogmans L, Huez G, Kruys V. Tumor necrosis factor-alpha mRNA remains unstable and hypoadenylated upon stimulation of macrophages by lipopolysaccharides. Eur J Biochem. 2000;267:6004-6012
26. Leon CG, Tory R, Jia J, Sivak O, Wasan KM. Discovery and development of toll-like receptor 4 (TLR4) antagonists: a new paradigm for treating sepsis and other disease. Pharm Res. 2008;25:1751-1761
27. Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145-151
28. MaAdam E, Haboubi HN, Forrester G, Eltahir Z, Spencer-Harty S, Davies C, Griffiths AP, Baxter JN, Jenkins GJ. Inducible nitric oxide synthase (iNOS) and nitric oxide (NO) are important mediators of reflux-induced cell signaling in esophageal cells. Carcinogenesis. 2012;33:2035-2043
29. Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639-653
30. Izadpanah Qeshmi F, Homaei A, Fernandes P, Javadpour S. Marine microbial L-asparaginase: biochemistry, molecular approaches and application in tumor therapy and in food industry. Microbiol Res. 2018;208:99-112
Author contact
![]() Corresponding author: Email: sangheekimre.kr, Tel: 82-32-760-5515 Fax: 82-32-760-5509
Corresponding author: Email: sangheekimre.kr, Tel: 82-32-760-5515 Fax: 82-32-760-5509

 Global reach, higher impact
Global reach, higher impact