3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(12):1415-1422. doi:10.7150/ijms.26741 This issue Cite
Review
Role of TCF/LEF Transcription Factors in Bone Development and Osteogenesis
1. Department of Oral and Maxillofacial Surgery, School of Stomatology, Jilin University, Changchun 130021, China
2. Stomatological Hospital of Southern Medical University & Guangdong Provincial Stomatological Hospital, Guangzhou 510280, China
3. Jilin Provincial Key Laboratory of Tooth Development and Bone Remodeling, Changchun 130021, China
* These authors contributed equally to this article
Received 2018-4-18; Accepted 2018-7-29; Published 2018-9-7
Abstract
Bone formation occurs by two distinct mechanisms, namely, periosteal ossification and endochondral ossification. In both mechanisms, osteoblasts play an important role in the secretion and mineralization of bone-specific extracellular matrix. Differentiation and maturation of osteoblasts is a prerequisite to bone formation and is regulated by many factors. Recent experiments have shown that transcription factors play an important role in regulating osteoblast differentiation, proliferation, and function. Osteogenesis related transcription factors are the central targets and key mediators of the function of growth factors, such as cytokines. Transcription factors play a key role in the transformation of mesenchymal progenitor cells into functional osteoblasts. These transcription factors are closely linked with each other and in conjunction with bone-related signaling pathways form a complex network that regulates osteoblast differentiation and bone formation. In this paper, we discuss the structure of T-cell factor/lymphoid enhancer factor (TCF/LEF) and its role in embryonic skeletal development and the crosstalk with related signaling pathways and factors.
Keywords: TCF/LEF Transcription Factors, signal transduction pathway, Osteogenesis
Introduction
Bone tissue represents a complex ecosystem comprising of multiple cell types and extracellular matrix which interact with each other at certain points in time and space. The physiology and metabolism of the various elements is modulated by a variety of hormones, nerve cells, and cytokines. Physiological processes such as cell proliferation, growth, development, differentiation, senescence, apoptosis, and metabolism are regulated by a complex cell signal transduction system. The signaling systems related to bone growth and metabolism include Wnt signal transduction pathway [1, 2], osteoprotegerin (OPG)/receptor activator of nuclear factor kappaB (RANK)/RANK ligand (RANKL) signal transduction pathway [3], Transforming growth factor beta (TGF-β) signal transduction pathway [4], Mitogen-activated protein kinases (MAPK) signal transduction pathway [5], Notch signal transduction pathway [6], and Hedgehog signal transduction pathway [7]. Transcription factors are important players in these signaling pathways and play an important role in the regulation of proliferation and differentiation of osteoblasts. The transcription factors involved in bone tissue include Runx-2 (transcription factor 2) [8], Osterix (Osx) [9], Msx1/2[10], and T-cell factor/lymphoid enhancer factor (TCF/LEF) [2]. These transcription factors are interdependent and closely linked with each other to form a network in the above signaling pathways, which regulates the entire process of osteoblast proliferation and differentiation.
TCF/LEF transcription factor family is the intrinsic target of the canonical Wnt signaling pathway and is a typical transcription factor for β-catenin expressed in the nucleus. It functions by binding to specific DNA sequence that yields tractable developmental and pattern abnormalities during embryogenesis, such as aberrant bone mass homeostasis. Development of novel therapies to normalize bone mass and correct developmental defects requires a good understanding of the specific mechanism of TCF/LEFs. The TCF/LEF family, which was first discovered during a study of T lymphocytes, has been shown to regulate the expression of specific genes, such as c-myc, cylinD1, runt-related transcriptional factor 2 (Runx2), and Osx. Four isoforms of TCF/LEF have been identified in humans, namely, TCF1 (TCF7), LEF1, TCF3 (TCF7L1), and TCF4 (TCF7L2) [11-13].
Structural Domains of TCF/LEF
β-catenin domain
TCF/LEF contains four binding functional domains (Figure 1). These include the N-terminal β-catenin domain, a highly conserved sequence, which can combine with β-catenin. The binding process involves conformational changes in the first 50 aa via formation of an alpha helix and salt bridges with charged residues in the superhelically formed interaction groove of the central Armadillo repeat domain [14, 15]. Deletion of this domain abrogates TCF-mediated transcriptional activation, which acts as a dominant negative regulator of Wnt signaling, and was shown to result in developmental defects in Drosophila and Xenopus embryos [16-18]. TCF/LEF are largely unfolded proteins in solution and only adopt folded structures when engaged in authentic interactions [19]. This indicates that the unfolded TCF/LEF might be particularly prone to weak, biologically irrelevant interactions and underscores the importance of rigorous controls for in vitro and in vivo binding assays.
HMG DNA binding domain
Another binding functional domain of TCF/LEF is the high-mobility group (HMG) domain at the carboxy end that can bind to the promoters of target genes of the 5'-ACATCAAAG-3 'sequence (Wnt response element) in the minor groove of the DNA double helix through intermolecular affinity [18, 20, 21]. This results in a 90°-130° bending of the double-helix structure, which alters the combination of DNA and other factors to regulate gene transcription [22, 23]. The nuclear localization signal of the domain can be directly recognized by importin alpha subunits for nuclear import [24]. Moreover, the HMG domain stabilizes the interaction with DNA. The nuclear localization signal can interact with phosphate backbone motif to increase its binding capacity by a hundred-fold [22, 25].
Context-dependent regulatory domain
The context-dependent regulatory domain in TCF/LEF varies widely, with only 15%-20% identity between them. These comprise of diverse sequences and play variable roles despite having only one exon in vertebrates. This exon can be recognized by an antagonist protein [26]. Additionally, alternative splice donor and acceptor sites exist upstream and downstream, and they can interact with amino acid motifs. The functional significance of this structure is its ability to repress transcriptional activity, and this may be accomplished via recruitment of the pleiotropic repressor Groucho [27, 28].
The structural domains of TCF/LEF.
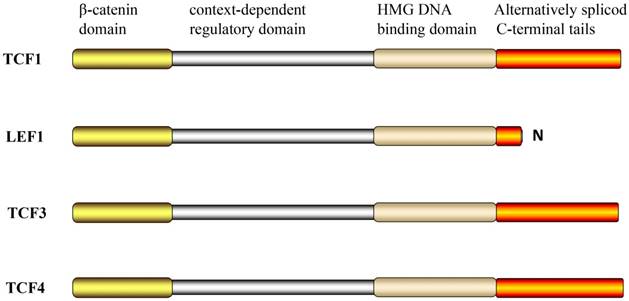
Alternatively spliced C-terminal tails
Another feature of the TCF/LEF family is that all family members have multiple C-terminuses, which are alternatively referred to as spliced C-terminal tails. Recently, the region of the TCFs C-terminal E tail near the HMG DNA binding domain was shown to contain the transcriptional activation domain (CR motif) of β-catenin. However, the LEF-1 does not have the alternative exons required for the alternative splice pattern; therefore, only the B-isomer was formed and not the E-isomer [29].
Role of TCF/LEF in embryo and skeleton development
TCF/LEF can activate transcription of downstream target genes upon activation by a variety of upstream signals and regulate biological activities, such as differentiation, proliferation, and apoptosis of osteoblasts. These play an important role in bone development, growth, and remodeling.
In the mouse embryo at 14.5 days, LEF1 was detected in the caudal, hip osteoprogenitor, and the surrounding cochlear mesenchymal cells in the bone structure [30], whereas TCF1 was detected in prechondrocytes in the mandible, palate, nasal bone, occipital bone, vertebrae, and ribs [31]. TCF4 was shown to be expressed in the mesenchymal cell region around the embryonal cartilage at 10.5 days [32], whereas TCF4 was detected in embryonal osteoblasts at 16.5 days [33].
In rats, LEF1 and TCF1 mutations result in loss of function that can lead to many malformations; the most common of these are skeletal malformations or the lack of bone elements.
TCF1 knockout mice showed a slight decrease in bone mineral density at one month after birth [33], however, this decrease was not as severe as that observed after osteoblast-specific gene β-catenin deletion, presumably, because of the role of TCF4 in osteoblasts. Moreover, the number and function of osteoclasts were increased, while the number and function of osteoblasts remained unchanged [34]. However, the bone resorption was accelerated because of the reduced amount of osteoprotegerin [35].
LEF-1 knockout mice are smaller than normal littermates, display numerous defects, (such as the lack of teeth, body hair, and beard) in tissues formed by epithelial and mesenchymal interactions, and die within two weeks after birth [36]. LEF1-/- female mice showed reduced number of osteoblasts and decreased trabecular bone mass [37], whereas the male mice did not exhibit any of these defects. LEF-1 was expressed in the primary cranial osteoblasts and MC3T3 precursor cell lines; however, its expression was lower than that of other TCFs. The expression of LEF-1 showed a gradual decrease until it was undetectable on the ninth day of MC3T3 cells culture in osteoblast culture medium. This indicated that LEF-1 was in a downregulated state during the phase of osteoblast terminal differentiation [15]. Inhibition of LEF-1 in MC3T3-E1 was shown to render the LEF-1 short hairpin RNA differentiation rate faster than that in the control group. Therefore, matrix mineralization and the expression of cell marker genes [alkaline phosphatase (ALP), OC, and sialoprotein] occur three to four days in advance [15]. LEF-1 overexpression was shown to inhibit the expression and differentiation of osteoblast markers, which suggests that LEF-1 inhibits the terminal osteogenesis of osteoblasts [38].
Animals that lack both LEF1 and TCF7, similar to wnt3a-/-, did not develop limbs and died at the embryonal age of approximately 10.5 days [39].
Given that TCF-3 is required for early anteroposterior patterning, TCF3-depletion results in embryonic death [40].
As compared to their normal siblings, TCF4-deficient mice were normal in size and appearance; however, the developmental defects of intestinal epithelial cells and crypt cells led to their death shortly after birth [32]. Primary osteoblasts from Lrp5-/- mice lose LEF1, but not TCF4 expression, after 10 days of in vitro culture, which indicates that LEF1 and TCF4 may have distinct functions in osteoblasts [41].
In adults, expression of TCF/LEF transcription factors is generally limited to the mitotically active cells in renewable tissues such as the lymphoid follicles, skin, hair follicles, colon, intestines, testes, and tumors [42-44], as well as bone tissue. TCF/LEF activity is increased in the bone tissues undergoing remodeling and in areas showing osteoblastic proliferation [45]. The expression level of TCF/LEF was shown to be downregulated in the differentiated cells and absent at the end of proliferation [46]. TCF1 and TCF4 can be detected in adult primary osteoblasts.
TCF/LEF crosstalk in signaling pathways in osteogenesis
Recent studies have shown that multiple factors involved in the Wnt signaling pathway play important roles in the regulation of osteoblast proliferation, differentiation, and apoptosis in bone. Gene mutation of some factors in this pathway may lead to abnormal bone formation. For example, β-catenin inactivation in mesenchymal stem cells can inhibit osteogenic differentiation. sFRP-deficient mice exhibited increased Wnt activation and bone mass, along with significant increase in the expression of Runx2 and its target genes (e.g., OCN). TCF/LEF, as an intracellular transcription factor in the canonical Wnt signaling pathway, initiates the transcription of downstream target genes by recruiting β-catenin into the nucleus and binding with DNA and β-catenin. As an intermediate factor, TCF/LEF is also involved in the crosstalk between the Wnt/β-catenin pathway and other signaling pathways (such as the β-TGF/BMP and FGF signal pathways) to regulate the transcription of related signaling proteins (Runx2, Smad, OPG, and estrogen); thus it plays an important role in bone tissue development and remodeling (Figure 2).
Runx2
Runx2, an osteoblast-specific transcription factor, can combine with core-binding factor β, which can promote the expression of osteoblast-specific genes, such as ALP, OCN, and Col I. Cbfα1/Runx2 plays an important role in the process of mesenchymal stem cell differentiation into osteoblast, which can inhibit their differentiation into adipocytes and chondrocytes [47, 48]. Furthermore, in vitro experiments have shown that Runx2 promotes the activity of ALP and the expression of bone matrix protein gene in immature mesenchymal stem cells and osteoblasts [49, 50]. Cbfα1/Runx2-deficient mice do not manifest intramembranous and endochondral ossification. Osteoblasts can express the two proteins, Cbfα1 and Runx-2 [51]; however, the expression levels of these genes vary at different times during differentiation. The promoter of Runx2 contains the consensus sequence of TCF close to the regulatory site of Runx2, and the β-catenin-TCF/LEF dimer can bind to the Runx2 promoter and promote its transcriptional activity.
LEF1 is a known inhibitor of Cbfα1. The DNA binding sites for LEF1 and Runx2 in osteocalcin are adjacent to each other. Mutation of the DNA binding site in LEF1 was shown to increase the osteocalcin promoter activity, and LEF1 was shown to inhibit the regulation of osteocalcin by Runx2 in osteoblast lineages [52, 53].
Runx2 has been shown to regulate the expression of TCF/LEF family genes [54]. In osteoblasts and chondrocytes, Runx2 can strongly promote the expression of TCF1 and LEF1, whereas the expressions of TCF1 and TCF4 were found to be decreased in Runx2-/- endochondral skeletons. The expressions of TCF1, LEF1, TCF4, and Runx2/ were reduced in Runx2-/- calvaria. Runx2 can enhance reporter activity of TCF1 promoter [55]. In addition to the expression patterns of TCF/LEF family genes and Runx2, researchers found that RUNX2 regulates TCF1 expression at least among the TCF/LEF family genes. RUNX2 can regulate LEF1△N p2 promoter, which is located in the intron between exons 3 and 4 of LEF1. Also, the overexpression of LEF1△N induces the expression of osteoblast differentiation genes (osteocalcin and Col1a1) in differentiating osteoblasts [56].
The crosstalk of TCF/LEF between the signaling pathways.
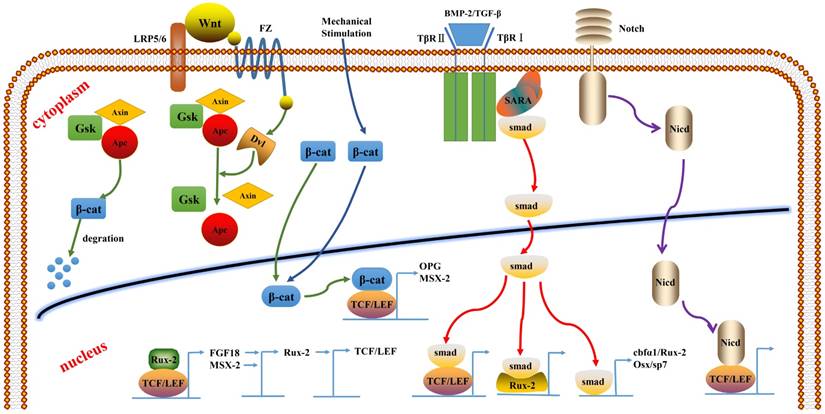
BMP-2/TGFβ/Smad signaling pathway
The BMP/TGFβ/Smad signaling pathway plays an important role in bone development. BMP-2 is an acidic glycoprotein that is widely expressed in the bone matrix. It is a member of the TGF-β superfamily and can induce osteoblast differentiation and promote bone formation as one of the important extracellular signal molecules in bone tissue. BMP-2 can transduce and regulate the transcription of osteogenic gene by activating Smad signals and, thereby, plays a role in osteogenesis. BMP-2 was found to upregulate the expression of 66 genes, including Smad6, Smad7, Msx2, and 13 other related transcription factors.
BMP can activate its downstream signal transduction upon binding to its receptor. Subsequently, the Smad-specific transcription factor complex assembles and translocates to the nucleus. The Smad complex interacts with other DNA components and other transcription factors, such as Runx2 and LEF1, to regulate gene expression. Smad4 or TGFβ-specific Smad3 interacts with LEF1 and activates gene expression upon stimulation by BMP2 and TGFβ signaling [57]. Through this process, the BMP/TGFβ and Wnt signaling pathways are interlaced with each other, and Smad and TCF/LEF proteins interact to regulate the expression of target genes. Some experiments have shown that BMPs act upstream of Wnts, while others have shown that BMPs action is downstream or parallel to Wnts. A study of osteocytes showed that BMP2 was located in the upstream of the Wnt signal. We found that β-catenin and BMP-2 promote the differentiation of mesenchymal stem cells into osteoblasts and inhibit their differentiation into adipocytes, and that they also can promote bone formation in vivo. The osteoinductivity of β-catenin requires interaction between osteoblasts and other specific factors, such as BMP-2, and the process partly depends on the TCF/LEF activity [58]. ΔN-LEF1 variants are upregulated in response to BMP2 signals through RUNX2-dependent, but SMAD4-independent activation of the P2 alternative promoter [56].
Smad proteins, such as Smad1, 5, and 8, specifically bind to BMPs and undergo phosphorylation. Subsequently, these translocate to the nucleus and activate osteoblast-specific transcription factors, such as Cbfα1/Runx2 and Osx/Sp7, which induces differentiation of MSCs into osteoblasts. A study also found that both smad4 and LEF1 are involved in the activation of β-catenin transcriptional complexes. β-catenin and LEF/TCF form complexes with Smad4, which promotes the entry of Smad4 into the nucleus, while the active translocation of Smad into the nucleus is accelerated by BMPs and TGFβ. The osteocalcin gene promoter contains the smad binding site, which regulates the expression of the genes together with TCF/LEF. The Smad DNA homologous sequence can also bind to TCF4/β-catenin.
Wnt/β-catenin signal pathway
In the absence of Wnt signaling, the β-catenin in the cytoplasm that has not been phosphorylated and accumulated to a certain extent enters the nucleus and interacts with TCF/LEF to activate the transcription of target genes, causing osteoblast differentiation and proliferation.
The Wnt/β-catenin signal pathway may activate the expression of Runx2 gene through TCF1 in the same way as that in the BMP/TGF-β signaling pathway [59]. β-catenin mutation enhances the LEF1-mediated inhibition of Runx2. In turn, Runx2 can induce the expression of TCF1, which upregulates the expression of osteoblast-associated genes, such as Col-1, ALP, and OCN, thereby controlling osteoblast differentiation and bone development [60]. The Wnt/β-catenin/TCF1/Runx2 signaling pathway was shown to enhance the activity of ALP in undifferentiated BMSCs and to promote osteogenic differentiation and proliferation of BMSCs. The Wnt/β-catenin signaling pathway can inhibit terminal differentiation of the osteoblast cell line.
The SFRP1 knockout mice exhibited high bone mass and significantly increased expression of TCF1, Runx2, and OC. TCF1 overexpression increased Runx2 activity by two to five times and increased osteoblast mRNA expression by seven to eight times. When SFRP1 inhibits the Wnt canonical signal pathway, the above expression disappears. This finding suggests that the mechanism of Wnt/β-catenin signaling pathway may be the same as that of the BMP signaling pathway, that is, via activation of TCF-Runx2-mediated regulation of osteoblast differentiation and bone development. LEF1 was found to have an activation site of Runx2, and it requires the key transcription factors, such as Runx2, Osx, and Dbc5, of osteogenic differentiation for β-catenin to promote osteogenesis. The complex interaction between Runx2 and TCF/LEF regulates the downstream target genes. β-catenin signal was also found to upregulate the Runx2 level in mesenchymal stem cells [59], whereas silencing of β-catenin gene was found to downregulate Runx2 expression and inhibit osteogenic differentiation[61, 62]. However, β-catenin and TCF/LEF factors in Wnt have been reported to have a complex association with Smad4 or Smad1 in BMP9 [63, 64]. Yet, crosstalk between signals may be more complex than expected, and LEF-1 has been reported to inhibit the osteocalcin promoter by interacting with Runx2. Another study reported that canonical Wnt signaling can directly stimulate Runx2 expression.
OPG
A cellular and molecular level study found that β-catenin combined with TCF can promote OPG expression in osteoblasts and inhibit osteoclast differentiation; this suggests that β-catenin and TCF in the canonical Wnt signaling pathway is involved in the regulation of bone formation and inhibition of bone resorption. The overexpression of β-catenin can indirectly affect the osteoclasts and decrease the OPG. Inhibition of LEF-1 expression in MC3T3 cells resulted in 80% less expression of OPG as compared to that in the control group [65]. LEF1 and ETs family members work together in the promotion of osteopontin in the Wnt pathway [29].
Msx-2
A study revealed that β-catenin and BMP2 can synergistically upregulate the transcription of Msx-2 gene. TCF/LEF can transactivate the Msx-2 promoter, and Msx-2 may be the upstream regulator of Runx2, which plays an important role in bone formation [54].
FGF 18
Recent evidence suggests that Runx2 and TCF/LEF complex can regulate the expression of FGF18, a direct target gene for the Wnt/β-catenin signaling pathway, by binding to a specific site on the FGF18 promoter. In turn, FGF can induce Runx2 expression and activate Runx2 in the osteoblast lineage; which is a regulatory process for osteogenesis [50].
Notch signal pathway
Notch receptors are transmembrane proteins that promote epithelial-mesenchymal transition and regulate cell proliferation and differentiation. By binding to the ligand, notch undergoes protein cleavage to produce a notch intracellular domain (NICD), which translocates to the nucleus and regulates gene expression by interacting with specific transcription factors. In the osteoblast gene expression, NICD damages cell differentiation and blocks the expression of TCF/LEF target promoter. NICD interacts with the highly conserved HMG region of LEF1 and therefore has a direct effect on Wnt-induced gene expression [66].
Estrogen
Estrogen deficiency disrupts the balance of osteoblastic and osteoclastic activity, reduces bone mineral density, and induces osteoporosis [67]. One of the effects of estrogen on osteoblasts and osteoclasts is the inhibition of apoptosis. TCF1 and TCF4 can bind to estrogen receptor (ER) and regulate their transcriptional activity. Thus, TCF1 stimulates ligand-dependent ER activity, whereas TCF4 antagonizes ER/E2 function.
Mechanical stimulation
Mechanical stimulation can cause rapid accumulation of β-catenin in the cytoplasm that moves to the nucleus and regulates the expression of target gene in consort with TCF/LEF [68]. Experimental results show that TCF1 mRNA and LEF1 mRNA expressions in the exercise group were significantly higher than those in the quiet group. Exercise is believed to increase the probability of complex formation between β-catenin and TCF/LEF and increase the expression of downstream target genes. Authors also found that LEF1 may be more sensitive to exercise stimulation than TCF1.
Conclusion
Based on various experimental data, it is evident that the roles of TCF/LEF transcription factors in embryonic development are time- and position-specific, which indicates that they have different functions in embryonic development. An abnormality in one of the transcription factors may lead to embryonal malformations or abnormalities, especially of the bone tissues, and may even be fatal. Osteogenesis and differentiation of osteoblasts is regulated synergistically by a complex regulatory network consisting of several key signaling pathways (such as BMP, Wnt, Notch, and FGF), which interact with each other and with TCF/LEF by interacting directly or indirectly with key transcription factors, such as Runx2 or Osx. Additionally, abnormal expression of one of the TCF/LEF transcription factors may be partially compensated by other factors. TCF/LEF transcription factors not only receive β-catenin signal and then start the downstream target gene transcription to participate in the classic Wnt signal pathway, but are also involved in crosstalk with other signal pathways; these signals can influence and control each other directly or indirectly. Thereby, TCF/LEF transcription factors play a key role in osteogenesis and bone growth and transformation. However, the specific functional mechanisms and interactions of the transcription factors are still unclear. These questions are subjects of investigations that are being actively pursued to better understand bone physiology and pathology.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Baron R, Rawadi G. Targeting the Wnt/beta-catenin pathway to regulate bone formation in the adult skeleton. Endocrinology. 2007;148:2635-43
2. Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202-9
3. Theoleyre S, Wittrant Y, Tat SK, Fortun Y, Redini F, Heymann D. The molecular triad OPG/RANK/RANKL: involvement in the orchestration of pathophysiological bone remodeling. Cytokine Growth Factor Rev. 2004;15:457-75
4. Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8:272-88
5. Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D. et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120:2457-73
6. Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T. et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306-14
7. St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072-86
8. Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. Biofactors. 2010;36:25-32
9. Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H. et al. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107:12919-24
10. Roybal PG, Wu NL, Sun J, Ting MC, Schafer CA, Maxson RE. Inactivation of Msx1 and Msx2 in neural crest reveals an unexpected role in suppressing heterotopic bone formation in the head. Dev Biol. 2010;343:28-39
11. Veien ES, Grierson MJ, Saund RS, Dorsky RI. Expression pattern of zebrafish tcf7 suggests unexplored domains of Wnt/beta-catenin activity. Dev Dyn. 2005;233:233-9
12. Waterman ML. Lymphoid enhancer factor/T cell factor expression in colorectal cancer. Cancer Metastasis Rev. 2004;23:41-52
13. Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303-11
14. Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048-52
15. Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885-96
16. Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R. et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638-42
17. Molenaar M, Roose J, Peterson J, Venanzi S, Clevers H, Destree O. Differential expression of the HMG box transcription factors XTcf-3 and XLef-1 during early xenopus development. Mech Dev. 1998;75:151-4
18. van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J. et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789-99
19. Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364-71
20. van Beest M, Dooijes D, van De Wetering M, Kjaerulff S, Bonvin A, Nielsen O. et al. Sequence-specific high mobility group box factors recognize 10-12-base pair minor groove motifs. J Biol Chem. 2000;275:27266-73
21. Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH. et al. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352-63
22. van de Wetering M, Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992;11:3039-44
23. Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185-95
24. Prieve MG, Guttridge KL, Munguia J, Waterman ML. Differential importin-alpha recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol Cell Biol. 1998;18:4819-32
25. Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567-78
26. Ghogomu SM, van Venrooy S, Ritthaler M, Wedlich D, Gradl D. HIC-5 is a novel repressor of lymphoid enhancer factor/T-cell factor-driven transcription. J Biol Chem. 2006;281:1755-64
27. Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H. et al. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604-8
28. Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P. et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608-12
29. Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492-504
30. Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19-39
31. Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F. et al. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439-48
32. Cho EA, Dressler GR. TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech Dev. 1998;77:9-18
33. Glass DA 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H. et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751-64
34. Glass DA 2nd, Karsenty G. In vivo analysis of Wnt signaling in bone. Endocrinology. 2007;148:2630-4
35. Glass DA 2nd, Karsenty G. Canonical Wnt signaling in osteoblasts is required for osteoclast differentiation. Ann N Y Acad Sci. 2006;1068:117-30
36. van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L. et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691-703
37. Noh T, Gabet Y, Cogan J, Shi Y, Tank A, Sasaki T. et al. Lef1 haploinsufficient mice display a low turnover and low bone mass phenotype in a gender- and age-specific manner. PLoS One. 2009;4:e5438
38. Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J Exp Med. 2004;200:797-803
39. Galceran J, Farinas I, Depew MJ, Clevers H, Grosschedl R. Wnt3a-/-like phenotype and limb deficiency in Lef1(-/-)Tcf1(-/-) mice. Genes Dev. 1999;13:709-17
40. Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV. et al. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263-74
41. Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA 2nd. et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303-14
42. Alonso L, Fuchs E. Stem cells in the skin: waste not, Wnt not. Genes Dev. 2003;17:1189-200
43. Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311-20
44. Staal FJ, Clevers HC. Wnt signaling in the thymus. Curr Opin Immunol. 2003;15:204-8
45. Qi H, Aguiar DJ, Williams SM, La Pean A, Pan W, Verfaillie CM. Identification of genes responsible for osteoblast differentiation from human mesodermal progenitor cells. Proc Natl Acad Sci U S A. 2003;100:3305-10
46. Shibamoto S, Winer J, Williams M, Polakis P. A blockade in Wnt signaling is activated following the differentiation of F9 teratocarcinoma cells. Exp Cell Res. 2004;292:11-20
47. Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233-9
48. Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K. et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952-63
49. Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747-54
50. Reinhold MI, Naski MC. Direct interactions of Runx2 and canonical Wnt signaling induce FGF18. J Biol Chem. 2007;282:3653-63
51. Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429-33
52. Kahler RA, Westendorf JJ. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J Biol Chem. 2003;278:11937-44
53. Zhong N, Gersch RP, Hadjiargyrou M. Wnt signaling activation during bone regeneration and the role of Dishevelled in chondrocyte proliferation and differentiation. Bone. 2006;39:5-16
54. Komori T. Signaling networks in RUNX2-dependent bone development. J Cell Biochem. 2011;112:750-5
55. Mikasa M, Rokutanda S, Komori H, Ito K, Tsang YS, Date Y. et al. Regulation of Tcf7 by Runx2 in chondrocyte maturation and proliferation. J Bone Miner Metab. 2011;29:291-9
56. Hoeppner LH, Secreto F, Jensen ED, Li X, Kahler RA, Westendorf JJ. Runx2 and bone morphogenic protein 2 regulate the expression of an alternative Lef1 transcript during osteoblast maturation. J Cell Physiol. 2009;221:480-9
57. Rawadi G, Vayssiere B, Dunn F, Baron R, Roman-Roman S. BMP-2 controls alkaline phosphatase expression and osteoblast mineralization by a Wnt autocrine loop. J Bone Miner Res. 2003;18:1842-53
58. Mbalaviele G, Sheikh S, Stains JP, Salazar VS, Cheng SL, Chen D. et al. Beta-catenin and BMP-2 synergize to promote osteoblast differentiation and new bone formation. J Cell Biochem. 2005;94:403-18
59. Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS. et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132-40
60. Zhang M, Yan Y, Lim YB, Tang D, Xie R, Chen A. et al. BMP-2 modulates beta-catenin signaling through stimulation of Lrp5 expression and inhibition of beta-TrCP expression in osteoblasts. J Cell Biochem. 2009;108:896-905
61. Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042-5
62. van Bezooijen RL, Bronckers AL, Gortzak RA, Hogendoorn PC, van der Wee-Pals L, Balemans W. et al. Sclerostin in mineralized matrices and van Buchem disease. J Dent Res. 2009;88:569-74
63. Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H. et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature. 2000;403:781-5
64. Hussein SM, Duff EK, Sirard C. Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J Biol Chem. 2003;278:48805-14
65. Drumm M, Teletchea S, Kozelka J. Recognition complex between the HMG domain of LEF-1 and its cognate DNA studied by molecular dynamics simulations with explicit solvation. J Biomol Struct Dyn. 2005;23:1-11
66. Sciaudone M, Gazzerro E, Priest L, Delany AM, Canalis E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology. 2003;144:5631-9
67. Manolagas SC, Kousteni S, Jilka RL. Sex steroids and bone. Recent Prog Horm Res. 2002;57:385-409
68. Case N, Ma M, Sen B, Xie Z, Gross TS, Rubin J. Beta-catenin levels influence rapid mechanical responses in osteoblasts. J Biol Chem. 2008;283:29196-205
Author contact
![]() Corresponding author: Prof. Bing Han, Department of Oral and Maxillofacial Surgery, School of Stomatology, Jilin University, 1500 Tsinghua Road, Changchun 130021, China. Tel: +86 0431 85579316 Email: hbingedu.cn
Corresponding author: Prof. Bing Han, Department of Oral and Maxillofacial Surgery, School of Stomatology, Jilin University, 1500 Tsinghua Road, Changchun 130021, China. Tel: +86 0431 85579316 Email: hbingedu.cn

 Global reach, higher impact
Global reach, higher impact