3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(13):1382-1388. doi:10.7150/ijms.21773 This issue Cite
Research Paper
Prognostic significance of high-mobility group box protein 1 genetic polymorphisms in rheumatoid arthritis disease outcome
1. Department of Orthopedics, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China;
2. Physical Education Office, Tunghai University, Taichung, Taiwan;
3. Sports Recreation and Health Management Continuing Studies, Tunghai University, Taichung, Taiwan;
4. Graduate Institute of Biomedical Science, China Medical University, Taichung, Taiwan;
5. Department of Biomedical Sciences Laboratory, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China;
6. Division of Immunology and Rheumatology, Department of Internal Medicine, China Medical University Hospital, Taichung, Taiwan;
7. Graduate Institute of Clinical Medical Science, China Medical University, Taichung, Taiwan;
8. School of Medicine, China Medical University, Taichung, Taiwan;
9. Department of Orthopedic Surgery, China Medical University Hospital, Taichung, Taiwan;
10. Department of Biotechnology, College of Health Science, Asia University, Taichung, Taiwan.
* These authors have contributed equally to this work
Received 2017-7-5; Accepted 2017-10-11; Published 2017-11-2
Abstract
Rheumatoid arthritis (RA) is a systemic inflammatory disease that causes chronic inflammation of the joints. Analysis of genetic variants offers promise for guiding treatment and improving outcomes in RA. High-mobility group box protein 1 (HMGB1) is a ubiquitous nuclear protein found in all mammal eukaryotic cells that participates in several biological functions including immune response, cell survival and apoptosis. We investigated the effects of HMGB1 gene polymorphisms on the risk of RA disease progression in a cohort of Chinese Han individuals. Four single nucleotide polymorphisms (SNPs) from the HMGB1 gene were selected and genotyped in 232 patients with RA and 353 healthy controls. We found that having one C allele in rs1360485 and one G allele in rs2249825 polymorphisms lowered the risk of RA in females. Moreover, among healthy controls, those who bore the C/G/T haplotype at SNPs rs1360485, rs2249825 and rs1412125 were at reduced risk of developing RA by 0.13-fold (p <0.05). This is the first report to examine the risk factors associated with HMGB1 SNPs in the development of RA disease in the Chinese Han population.
Keywords: HMGB1, Rheumatoid arthritis, SNP, Susceptibility.
Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by chronic inflammation and joint deformation, affecting an estimated 0.3% to 1.0% of the adult population worldwide who, if left untreated, will suffer severe disability and premature mortality [1]. Common features of RA include monocyte infiltration, inflammation, synovial swelling, pannus formation, stiffness in the joints and articular cartilage destruction [2]. The etiology of RA remains unclear. However, considerable clinical data indicate that genetic factors act as markers and contribute to the development of this disease [3]. Indeed, genetic factors may be responsible for up to 60% of overall susceptibility to RA [3]. We sought to define genetic aberrations in RA, as these may lead to a better understanding of risk prediction and guide drug discovery that may lead to a more personalized approach to treatment.
Knowing how an individual's genetic make-up regulates disease predisposition and outcome can help us better understand aspects of disease biology and facilitate diagnosis, understand the importance of tailoring treatment to the patient, and, eventually, enable a cure or the opportunity to prevent the disease. Genetic analyses of RA pathogenesis demonstrate considerable heterogeneity among individuals and populations [4, 5]. Global genome-wide association studies (GWASs) involving large patient cohorts and typically using many hundreds of thousands of single nucleotide polymorphisms (SNPs) have contributed much understanding to RA etiology [6]. These GWASs have identified a number of RA-susceptibility loci, although such studies cannot directly identify the genes or genetic variants causing the disease [6]. Previous research has found that specific SNPs in GWAS-identified susceptibility regions are associated with RA disease progression. For instance, certain SNPs in the interleukin (IL)-32, NKG2D and vascular endothelial growth factor (VEGF) genes are known to contribute to the progression of RA [7-9].
High-mobility group box protein 1 (HMGB1) is a ubiquitous nuclear protein that has been discovered in mammals [10, 11]. HMGB1 contains DNA binding domains and contributes to DNA repair and the stabilization of nuclear homeostasis [12]. HMGB1 is usually localized in the cell nucleus and is secreted into the extracellular environment in response to different stimuli; either passively during cellular apoptosis or necrosis, or actively following inflammatory signals from activated immune cells or neuronal cells [13]. While previous research has confirmed the association of HMGB1 SNPs with the susceptibility and progression of other diseases, including hepatocellular carcinoma [14] and uterine cervical neoplasia [15], little is known about the association between HMGB1 SNPs, RA disease risk and progression. We therefore selected four HMGB1 SNPs to investigate the association between these SNPs and RA susceptibility in healthy controls as well as disease progression in RA patients.
Materials and Methods
Participants
We collected blood specimens from 232 patients (cases) who had been diagnosed with RA at Dongyang People's Hospital between 2014 and 2016. The control group comprised 353 healthy volunteers with no previous history of RA. We obtained written (signed) informed consent from all study participants. The study protocol was approved by the Dongyang People's Hospital Ethics Committee and Institutional Review Board (2015-YB002). Clinicopathologic data were collected for all patients from their medical records. We used a standardized questionnaire and searched the patients' electronic medical records to obtain detailed clinical data on age, sex and disease duration, as well as concurrent treatment with methotrexate, prednisolone, and tumor necrosis factor (TNF) inhibitors. At baseline, serum samples were collected from all RA patients and analyzed for the presence of anti-citrullinated protein antibodies (ACPAs), rheumatoid factor (RF), erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP). Samples were defined as ACPA-positive if anti-CCP2 titers were ≥17 IU/mL and as RF-positive if IgM RF titers were ≥30 IU/mL. Whole blood samples (3 mL) were collected from all study participants and stored at -80°C for subsequent DNA extraction.
Extraction of genomic DNA
We extracted genomic DNA from whole blood specimens using QIAamp DNA Blood Mini Kits (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Tris-EDTA (TE buffer; (10 mM Tris-HCl, 1 mM EDTA [ pH 7.8]) was used to dissolve DNA then stored at -20°C [16, 17].
SNP selection
SNPs rs2249825 (3814C/G; genomic number 31,037,903) near the exon, SNP rs1412125 (-1615T/C; genomic number 31,041,595) in the promoter region, rs1360485 (3′UTR, T/C; genomic number 31,031,884) in the 3′ untranslated region and SNP rs1045411 (2262C/T; genomic number 31,033,232) in the exon selected according to the Chinese HapMap data and previous studies [15, 18]. The minor allele frequencies of these SNPs were all ≥5 %.
DNA genotyping using real-time PCR
Genotyping for the HMGB1 SNPs was performed using the TaqMan® Single Nucleotide Polymorphism (SNP) Genotyping Assays (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions [19, 20].
Bioinformatic analysis
Genotype-Tissue Expression (GTEx) data were used to identify correlations between SNPs and levels of HMGB1 expression [21]. We conducted an investigation into expression quantitative trait loci (eQTLs), to determine the functional role of phenotype-associated SNPs.
Statistical Analyses
Chi-square analysis was used to confirm that the genotype distribution of each SNP was in Hardy-Weinberg equilibrium (HWE). A Mann-Whitney U-test and a Fisher's exact test were used to compare the significance of differences in distributions of demographic characteristics between healthy controls and RA patients. The correlations between genotype frequencies, clinicopathologic characteristics and RA risk were examined using adjusted odds ratios (AORs) with 95% confidence intervals (CIs), after controlling for other covariates. Significant differences in haplotype frequencies in cases and controls were analyzed using Haploview, according to the software package [22]. A p value of < 0.05 was considered statistically significant. Statistical analyses were performed using the Statistical Package for Social Sciences, version 9.1 (SAS Institute Inc., Cary, NC, USA).
Results
All study participants were of Chinese Han ethnicity (Table 1). Between-group differences were significant for age and gender (both p < 0.001). The majority of RA patients were female (83.6%), with a mean age of 54 years, and an average RA duration of 52 months. At baseline, 60.8% were using TNF inhibitors, 45.7% were receiving methotrexate (45.7%) and 46.1% were receiving prednisolone. The majority were RF-positive (85.3%) and ACPA-positive (78.9%). To reduce possible interference of confounding variables, AORs with 95% CIs were estimated by multiple logistic regression models after controlling for age in each comparison.
Genotype distributions for SNPs rs1360485, rs1045411, rs2249825 and rs1412125 are presented in Table 2. In the healthy controls, all genotypic frequencies were in HWE (p > 0.05). In both healthy controls and RA patients, the highest distribution frequencies for the rs1360485, rs1045411, rs2249825 and rs1412125 genes were, respectively, homozygous for T/T, homozygous for C/C, homozygous for C/C, and homozygous for T/T (Table 2). The incidence of these polymorphisms did not differ significantly between RA patients and healthy controls (Table 2).
However, among females, controls with one C allele in the rs1360485 were 65% less likely (AOR 0.65; 95% CI: 0.44-0.97) and those with one G allele in the rs2249825 were 58% less likely (AOR 0.58; 95% CI: 0.36-0.94) to develop RA in the future, compared with RA patients (Table 3). These results indicate a protective effect of HMGB1 gene polymorphisms in females.
HMGB1 genotypes in RA patients were evaluated to clarify the role of HMGB1 rs1360485 and rs2249825 polymorphisms in regard to RF and ACPA clinical status, RA drug consumption, as well as ESR and CRP values. No significant associations were found between any polymorphisms and clinical status, RA drug use, clinical pathologic markers or genotypes in RA patients (Table 4).
Characteristics of the study population
| Variables | Control (n=353) N (%) | Case (n=232) N (%) | OR | (95% CI) | P Value | |
|---|---|---|---|---|---|---|
| Age, year (mean±SD) | 45.03 ± 17.39 | 54.38 ± 11.60 | < 0.0001 | |||
| < 50a | 214 (60.62) | 78 (33.62) | 1 | |||
| ≥ 50a | 139 (39.38) | 154 (66.38) | 3.04 | (2.15-4.30) | < 0.0001 | |
| Gender | ||||||
| female | 167 (47.3) | 194 (83.6) | 1 | |||
| male | 186 (52.7) | 38 (16.4) | 0.176 | (0.12-0.26) | < 0.0001 | |
| RF | ||||||
| Negative | ND | 34 (14.67) | ||||
| Positive | ND | 198 (85.3) | ||||
| ACPA | ||||||
| Negative | ND | 49 (21.1) | ||||
| Positive | ND | 183 (78.9) | ||||
| Anti-TNF | ||||||
| Use | ND | 141 (60.8) | ||||
| Non-use | ND | 91 (39.2) | ||||
| Methotrexate | ||||||
| Use | ND | 106 (45.7) | ||||
| Non-use | ND | 126 (54.3) | ||||
| Prednisolone | ||||||
| Use | ND | 107 (46.1) | ||||
| Non-use | ND | 125 (53.9) | ||||
| ESR (mean±SD) | 30.65 ± 25.06 | |||||
| < 20b | ND | 107 (46.1) | ||||
| ≥ 20b | ND | 125 (53.9) | ||||
| CRP (mean±SD) | 22.64 ± 80.39 | |||||
| < 8c | ND | 140 (61.2) | ||||
| ≥ 8c | ND | 90 (38.8) | ||||
| RA duration, month (mean±SD) | ND | 52.45 ± 70.40 | ||||
N = number; OR = odds ratio; CI = confidence interval; RF = rheumatoid factor; ND = not determined; ACPA = anti-citrullinated protein antibodies; TNF = tumor necrosis factor; ESR = erythrocyte sedimentation rate; CRP = C-reactive protein; RA = rheumatoid arthritis.
a Median age for entire group b normal value of ESR; C normal value of CRP.
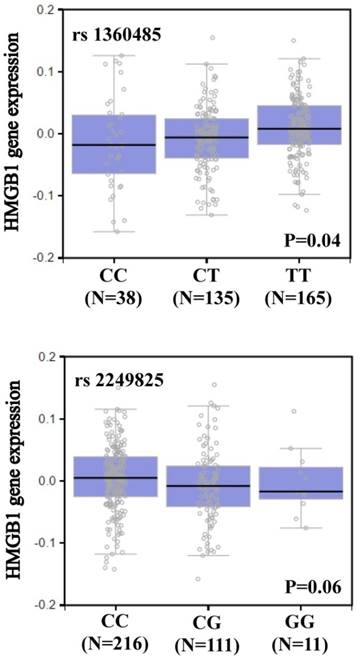
We searched the GTEx database to investigate whether rs1360485 and rs2249825 were associated with HMGB1 expression. Individuals carrying a genotype with the variant C at rs1360485 showed a trend for reduced expression of HMGB1, compared with the wild-type homozygous genotypes (p < 0.05, Figure 1). In addition, those with a genotype with the variant G at rs2249825 had a slightly reduced expression of HMGB1 (p = 0.06, Figure 1).
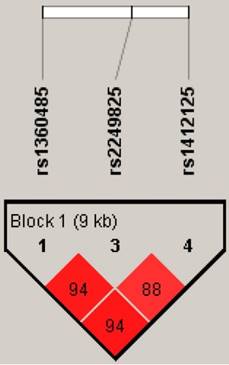
An analysis of distribution frequencies of HMGB1 rs1360485, rs2249825 rs1412125 haplotypes revealed that the most common haplotype in healthy controls was T/C/T (69.12%), which was therefore selected as the reference. The C/G/T HMGB1 haplotype significantly reduced the risk for developing RA by 0.13-fold (95% CI: 0.03-0.5, p <0.05) (Table 5). The reconstructed linkage disequilibrium plot of the four SNPs is shown in Figure 2. We found a haploblock in which rs1360485 and rs2249825 showed 94% linkage disequilibrium. In addition, rs1360485 and rs1412125 as well as rs2249825 and rs1412125 expressed 94% and 88% linkage disequilibrium, respectively (Figure 2).
HMGB1 displays a significant eQTL association with rs1360485 and rs2249825 genotypes in whole blood (GTEx data set).
Discussion
RA is a complex immune-related disease, with many genetic components and environmental factors implicated in its development. Although the availability of novel therapies in the last few years have enabled an increasing number of patients with RA to achieve a state where they have only mild or no symptoms [23], a proportion of patients remains treatment-refractory and continues to experience progressive joint destruction, functional impairment and increased mortality. In such patients, the limitations of current RA therapies emphasize the importance of continuing to investigate the pathogenesis of RA disease and identifying new therapeutic targets. We therefore sought to determine whether genetic polymorphisms in HMGB1-related genes contribute to a higher susceptibility for RA. To the best of our knowledge, our study is the first to examine the distribution of the rs1360485, rs1045411, rs2249825 and rs1412125 SNPs and their possible association with RA development. We also investigated the associations of these HMGB1 SNPs with clinical status, RA drug therapies and clinical pathologic markers, leading to a susceptibility to RA. In analyses adjusting for confounding factors, females who had one C allele in rs1360485 or one G allele in rs2249825 were at reduced risk of developing RA, by 0.65-fold (95% CI: 0.44-0.97; p < 0.05) and 0.58-fold (95% CI: 0.36-0.94; p < 0.05), respectively. However, concentration of HMGB1 in serum were unavailable because patients had only recently enrolled in the study. Osteoarthritis (OA) is a chronic inflammatory joints disorder associated with degradation of joints [24, 25]. Nevertheless, we did not find any report to examine HMGB1 SNP with OA development. Furthermore, the role of HMGB1 SNP in OA patients should also be evaluated.
HMGB1 pairwise linkage disequilibrium patterns. Schematic presentation of the HMGB1 haploblock, indicating the locations of the SNP polymorphism and the pairwise linkage disequilibrium measures D'. The measure of D' of SNP is established according to the fraction, where a value of over 80 represents high D'
HMGB1 regulates several functions with inside or outside of cell, for example immune response, DNA repair, chromatin stabilization, cell apoptosis and gene transcription HMGB1 may play a role in the development of RA and OA [26, 27], although the evidence is uncertain as to any correlation between HMGB1 polymorphisms and development of RA disease. In this study, we examined four HMGB1 SNPs in 232 RA patients and in 353 healthy controls. We did not find significant differences in the incidence of RA patients with these polymorphisms compared to that of healthy controls. However, we found that in the female presence of one C allele in rs1360485 and one G allele in rs2249825 were both associated with significantly lower risks for the development of RA disease. The polymorphisms in the 3ʹ-flanking region of a gene can control gene expression [28]. Data from the GTEx database demonstrated that variant C at rs1360485 and variant G at rs2249825 showed a trend for reduced expression of HMGB1, compared with the wild-type T and C homozygous genotypes.
Distribution frequencies of HMGB1 genotypes and allele frequencies of four tag SNPs in cases and controls.
| Variables | Control(n=353) N (%) | Case(n=232) N (%) | OR | (95% CI) | P Value | AORa | (95% CI) | P Value | |
|---|---|---|---|---|---|---|---|---|---|
| rs1360485 | |||||||||
| TT | 218 (61.8) | 152 (65.5) | 1 | 1 | |||||
| CT | 177 (33.1) | 75 (32.3) | 0.92 | (0.64-1.31) | 0.64 | 1.04 | (0.69-1.57) | 0.85 | |
| CC | 18 (5.1) | 5 (2.2) | 0.4 | (0.15-1.10) | 0.07 | 0.41 | (0.13-1.27) | 0.12 | |
| TT | 218 (61.76) | 152 (62.52) | 1 | 1 | |||||
| CT+CC | 135 (38.24) | 80 (34.48) | 0.85 | (0.60-1.20) | 0.36 | 0.81 | (0.64-1.42) | 0.92 | |
| T | 553 (78.33) | 379 (81.68) | 1 | 1 | |||||
| C | 153 (21.67) | 85 (18.32) | 0.81 | (0.60-1.09) | 0.16 | 0.88 | (0.63-1.63) | 0.45 | |
| rs1045411 | |||||||||
| CC | 233 (66.0) | 146 (62.9) | 1 | 1 | |||||
| CT | 109 (30.9) | 82 (35.4) | 1.2 | (0.84-1.71) | 0.32 | 1.43 | (0.95-2.15) | 0.09 | |
| TT | 11 (3.1) | 4 (1.7) | 0.58 | (0.18-1.86) | 0.36 | 0.54 | (0.14-2.10) | 0.37 | |
| CC | 233 (66.01) | 146 (62.93) | 1 | 1 | |||||
| CT+TT | 120 (33.99) | 86 (37.07) | 1.14 | (0.81-1.62) | 0.45 | 0.15 | (0.90-2.00) | 1.34 | |
| C | 575 (81.44) | 375 (80.60) | 1 | 1 | |||||
| T | 131 (18.56) | 90 (19.40) | 1.06 | (0.78-1.42) | 0.72 | 1.18 | (0.84-1.66) | 0.34 | |
| rs2249825 | |||||||||
| CC | 264 (74.8) | 180 (77.6) | 1 | 1 | |||||
| CG | 79 (22.4) | 50 (21.5) | 0.93 | (0.62-1.39) | 0.72 | 0.97 | (0.61-1.54) | 0.86 | |
| GG | 10 (2.8) | 2 (0.9) | 0.29 | (0.06-1.36) | 0.12 | 0.26 | (0.05-1.44) | 0.12 | |
| CC | 264 (74.79) | 180 (77.59) | 1 | 1 | |||||
| CG+GG | 89 (25.21) | 52 (22.41) | 0.86 | (0.58-1.27) | 0.44 | 0.6 | (0.57-1.39) | 0.89 | |
| C | 607 (85.98) | 410 (88.36) | 1 | 1 | |||||
| G | 99 (14.02) | 54 (11.64) | 0.81 | (0.57-1.15) | 0.24 | 0.83 | (0.55-1.24) | 0.36 | |
| rs1412125 | |||||||||
| TT | 197 (55.8) | 138 (59.5) | 1 | 1 | |||||
| CT | 139 (39.4) | 80 (34.5) | 0.82 | (058-1.17) | 0.27 | 0.86 | (0.57-1.28) | 0.45 | |
| CC | 17 (4.8) | 14 (6.0) | 1.18 | (0.56-2.46) | 0.67 | 1.78 | (0.73-4.31) | 0.2 | |
| TT | 197 (55.81) | 138 (59.48) | 1 | 1 | |||||
| CT+CC | 156 (44.19) | 94 (40.52) | 0.86 | (0.62-1.24) | 0.38 | 0.72 | (0.63-1.37) | 0.93 | |
| T | 533 (75.50) | 356 (76.72) | 1 | 1 | |||||
| C | 173 (24.50) | 108 (23.28) | 0.93 | (0.71-1.23) | 0.63 | 1.04 | (0.75-1.42) | 0.83 | |
a Logistic regression with adjustment for age, gender.
b Hardy-Weinberg equilibrium test.
Association of HMGB1 alleles and 4 tag SNPs with RA disease in the female study participants.
| Variables | Control(n=167) N (%) | Case(n=194) N (%) | OR | (95% CI) | P Value | AORa | (95% CI) | P Value | |
|---|---|---|---|---|---|---|---|---|---|
| Age, year (mean±SD) | 41.95 ± 14.86 | 52.98 ± 11.55 | < 0.0001 | ||||||
| rs1360485 | |||||||||
| T | 253 (75.75) | 323 (83.25) | 1 | 1 | |||||
| C | 81 (24.25) | 65 (16.75) | 0.63 | (0.44-0.91) | 0.01 | 0.65 | (0.44-0.97) | 0.04 | |
| rs1045411 | |||||||||
| C | 261 (78.14) | 320 (82.47) | 1 | 1 | |||||
| T | 73 (21.86) | 68 (17.53) | 0.76 | (0.53-1.10) | 0.14 | 0.86 | (0.57-1.28) | 0.45 | |
| rs2249825 | |||||||||
| C | 280 (83.83) | 348 (89.69) | 1 | 1 | |||||
| G | 54 (16.17) | 40 (10.31) | 0.6 | (0.38-0.92) | 0.02 | 0.58 | (0.36-0.94) | 0.03 | |
| rs1412125 | |||||||||
| T | 250 (74.85) | 302 (77.84) | 1 | 1 | |||||
| C | 84 (25.15) | 86 (22.16) | 0.85 | (0.60-1.20) | 0.35 | 0.92 | (0.63-1.34) | 0.66 | |
a Logistic regression with adjustment for age.
Genotype frequencies of HMGB1 rs1360485/rs2249825 polymorphisms in RA patients, stratified by RA markers status.
| Variables | N (%) | N (%) | OR | (95% CI) | P Value | AORa | (95% CI) | P Value | |
|---|---|---|---|---|---|---|---|---|---|
| rs1360485 | TT (n=152) | CT+CC (n=80) | |||||||
| RF | Negative | 25 (16.45) | 9 (11.25) | 1 | 1 | ||||
| Positive | 127 (83.55) | 71 (88.75) | 1.55 | (0.69-3.51) | 0.29 | 1.57 | (0.69-3.59) | 0.29 | |
| ACPA | Negative | 34 (22.37) | 15 (18.75) | 1 | 1 | ||||
| Positive | 118 (77.63) | 65 (81.25) | 1.25 | (0.63-2.46) | 0.52 | 1.32 | (0.66-2.64) | 0.44 | |
| Anti-TNF | Negative | 93 (61.18) | 48 (60.00) | 1 | 1 | ||||
| Positive | 59 (38.82) | 32 (40.00) | 1.05 | (0.60-1.83) | 0.86 | 0.95 | (0.54-1.69) | 0.87 | |
| Methotrexate | Negative | 69 (45.39) | 37 (46.25) | 1 | 1 | ||||
| Positive | 83 (54.61) | 43 (43.75) | 0.97 | (0.56-1.66) | 0.9 | 0.98 | (0.56-1.73) | 0.94 | |
| Prednisolone | Negative | 68 (44.74) | 39 (48.75) | 1 | 1 | ||||
| Positive | 84 (55.26) | 41 (51.25) | 0.52 | (0.50-1.46) | 0.56 | 0.91 | (0.53-1.59) | 0.75 | |
| ESR | < 20b | 66 (43.42) | 41 (51.25) | 1 | 1 | ||||
| ≥ 20b | 86 (56.58) | 39 (48.75) | 0.73 | (0.42-1.26) | 0.26 | 0.78 | (0.45-1.35) | 0.37 | |
| CRP | < 8c | 93 (61.18) | 49 (61.25) | 1 | 1 | ||||
| ≥ 8c | 59 (38.82) | 31 (38.75) | 0.99 | (0.57-1.74) | 0.99 | 0.97 | (0.55-1.71) | 0.91 | |
| rs2249825 | CC (n=180) | CG+GG (n=52) | |||||||
| RF | Negative | 26 (14.44) | 8 (15.38) | 1 | 1 | ||||
| Positive | 154 (85.56) | 44 (84.62) | 0.93 | (0.39-2.20) | 0.87 | 0.9 | (0.38-2.15) | 0.81 | |
| ACPA | Negative | 41 (22.78) | 8 (15.38) | 1 | 1 | ||||
| Positive | 139 (77.22) | 44 (84.62) | 1.62 | (0.71-3.72) | 0.25 | 1.81 | (0.78-4.23) | 0.17 | |
| Anti-TNF | Negative | 109 (60.56) | 32 (61.54) | 1 | 1 | ||||
| Positive | 71 (39.44) | 20 (38.46) | 0.96 | (0.51-1.81) | 0.9 | 0.93 | (0.48-1.78) | 0.82 | |
| Methotrexate | Negative | 81 (45.00) | 25 (48.08) | 1 | 1 | ||||
| Positive | 99 (55.00) | 27 (51.92) | 0.88 | (0.48-1.64) | 0.69 | 1.01 | (0.53-1.92) | 0.99 | |
| Prednisolone | Negative | 84 (46.67) | 23 (44.23) | 1 | 1 | ||||
| Positive | 96 (53.33) | 29 (55.77) | 1.1 | (0.59-2.05) | 0.76 | 1.14 | (0.60-2.14) | 0.69 | |
| ESR | < 20b | 80 (44.44) | 27 (51.92) | 1 | 1 | ||||
| ≥ 20b | 100 (55.56) | 25 (48.08) | 0.74 | (0.40-1.37) | 0.34 | 0.77 | (0.41-1.44) | 0.42 | |
| CRP | < 8c | 113 (62.78) | 29 (55.77) | 1 | 1 | ||||
| ≥ 8c | 67 (37.22) | 23 (44.23) | 1.34 | (0.75-2.50) | 0.36 | 1.26 | (0.67-2.38) | 0.48 | |
*RF= rheumatoid factor; ACPA= anti-citrullinated protein antibodies; ESR= erythrocyte sedimentation rate; CRP= C-reactive protein;
a Logistic regression with adjustment for age and gender. b normal value of ESR C normal value of CRP
HMGB1 rs1360485/rs2249825/rs1412125 haplotype frequencies and the association with risk of RA.
| Variables | Control (n=353) N (%) | Case (n=232) N (%) | OR | (95% CI) | P Value | AORa | (95% CI) | P Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| rs1360485 | rs2249825 | rs1412125 | ||||||||
| T | C | T | 488 (69.12) | 328 (70.69) | 1 | 1 | ||||
| T | C | C | 61 (8.64) | 49 (10.56) | 1.2 | (0.80-1.79) | 0.38 | 1.07 | (0.68-1.69) | 0.78 |
| T | G | T | 2 (0.28) | 1 (0.22) | 0.74 | (0.07-8.24) | 0.81 | 0.3 | (0.03-3.38) | 0.33 |
| T | G | C | 2 (0.28) | 1 (0.22) | 0.74 | (0.07-8.24) | 0.81 | 0.62 | (0.04-9.65) | 0.73 |
| C | C | T | 32 (4.53) | 24 (5.17) | 1.12 | (0.65-1.93) | 0.69 | 1.15 | (0.61-2.16) | 0.67 |
| C | C | C | 26 (3.68) | 9 (1.94) | 0.52 | (0.24-1.11) | 0.09 | 0.61 | (0.25-1.46) | 0.26 |
| C | G | T | 11 (1.56) | 3 (0.65) | 0.41 | (0.11-1.47) | 0.17 | 0.13 | (0.03-0.50) | 0.003 |
| C | G | C | 84 (11.9) | 49 (10.56) | 0.89 | (0.59-1.27) | 0.46 | 1.09 | (0.67-1.69) | 0.71 |
The linkage disequilibrium is expressed across the human genome and thus loci can be used as genetic markers to locate adjacent variants that participate to detection and treatment of disease. Haplotype analyses can provide data on the genetic contribution to disease susceptibility [29]. We evaluated the impacts of different haplotype combinations of three HMGB1 SNPs (rs1360485, rs2249825 and rs1412125) upon the risk of RA disease and found that the C/G/T haplotype was associated with a low risk for RA. It is possible that the HMGB1 C/G/T haplotype is in linkage disequilibrium with other functional polymorphisms that are responsible for susceptibility to RA.
In conclusion, our results demonstrate an association between HMGB1 gene variants and the risk of RA. We show that the HMGB1 rs1360485 and rs2249825 polymorphisms significantly lower the risk of RA progression Chinese Han females. This study is the first to report a correlation between HMGB1 polymorphisms and RA risk. HMGB1 may serve as a predictive marker for RA therapy.
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 106-2320-B-029-002) and China Medical University (CMU106-ASIA-03).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376:1094-108
2. Su CM, Huang CY, Tang CH. Characteristics of resistin in rheumatoid arthritis angiogenesis. Biomarkers in medicine. 2016;10:651-60
3. Suzuki A, Yamamoto K. From genetics to functional insights into rheumatoid arthritis. Clinical and experimental rheumatology. 2015;33:S40-3
4. Castro-Santos P, Diaz-Pena R. Genetics of rheumatoid arthritis: a new boost is needed in Latin American populations. Revista brasileira de reumatologia. 2016;56:171-7
5. Yau AC, Holmdahl R. Rheumatoid arthritis: identifying and characterising polymorphisms using rat models. Disease models & mechanisms. 2016;9:1111-23
6. Yamamoto K, Okada Y, Suzuki A, Kochi Y. Genetic studies of rheumatoid arthritis. Proceedings of the Japan Academy Series B, Physical and biological sciences. 2015;91:410-22
7. Damen MS, Agca R, Holewijn S, de Graaf J, Dos Santos JC, van Riel PL. et al. IL-32 promoter SNP rs4786370 predisposes to modified lipoprotein profiles in patients with rheumatoid arthritis. Scientific reports. 2017;7:41629
8. Paradowska-Gorycka A, Pawlik A, Romanowska-Prochnicka K, Haladyj E, Malinowski D, Stypinska B. et al. Relationship between VEGF Gene Polymorphisms and Serum VEGF Protein Levels in Patients with Rheumatoid Arthritis. PloS one. 2016;11:e0160769
9. Mariaselvam CM, Tamouza R, Krishnamoorthy R, Charron D, Misra DP, Jain VK. et al. Association of NKG2D gene variants with susceptibility and severity of rheumatoid arthritis. Clinical and experimental immunology. 2017;187:369-75
10. Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA. Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem. 1989;185:303-10
11. Bustin M, Lehn DA, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231-43
12. Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237-46
13. Martinotti S, Patrone M, Ranzato E. Emerging roles for HMGB1 protein in immunity, inflammation, and cancer. ImmunoTargets and therapy. 2015;4:101-9
14. Wang B, Yeh CB, Lein MY, Su CM, Yang SF, Liu YF. et al. Effects of HMGB1 Polymorphisms on the Susceptibility and Progression of Hepatocellular Carcinoma. International journal of medical sciences. 2016;13:304-9
15. Wu HH, Liu YF, Yang SF, Lin WL, Chen SC, Han CP. et al. Association of single-nucleotide polymorphisms of high-mobility group box 1 with susceptibility and clinicopathological characteristics of uterine cervical neoplasia in Taiwanese women. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016
16. Yang MD, Lin KC, Lu MC, Jeng LB, Hsiao CL, Yueh TC. et al. Contribution of matrix metalloproteinases-1 genotypes to gastric cancer susceptibility in Taiwan. Biomedicine (Taipei). 2017;7:10
17. Chen SY, Chen CH, Huang YC, Chan CJ, Chen DC, Tsai FJ. Genetic susceptibility to idiopathic membranous nephropathy in high-prevalence Area, Taiwan. Biomedicine (Taipei). 2014;4:9
18. Wang Y, Li XP, Yin JY, Zhang Y, He H, Qian CY. et al. Association of HMGB1 and HMGB2 genetic polymorphisms with lung cancer chemotherapy response. Clinical and experimental pharmacology & physiology. 2014;41:408-15
19. Lin YJ, Ho TJ, Lin TH, Hsu WY, Huang SM, Liao CC. et al. P-coumaric acid regulates exon 12 splicing of the ATP7B gene by modulating hnRNP A1 protein expressions. Biomedicine (Taipei). 2015;5:10
20. Li TC, Li CI, Liao LN, Liu CS, Yang CW, Lin CH. et al. Associations of EDNRA and EDN1 polymorphisms with carotid intima media thickness through interactions with gender, regular exercise, and obesity in subjects in Taiwan: Taichung Community Health Study (TCHS). Biomedicine (Taipei). 2015;5:8
21. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
22. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-5
23. Aletaha D, Bingham CO 3rd, Tanaka Y, Agarwal P, Kurrasch R, Tak PP. et al. Efficacy and safety of sirukumab in patients with active rheumatoid arthritis refractory to anti-TNF therapy (SIRROUND-T): a randomised, double-blind, placebo-controlled, parallel-group, multinational, phase 3 study. Lancet. 2017;389:1206-17
24. Chou PY, Su CM, Huang CY, Tang CH. The characteristics of thrombin in osteoarthritic pathogenesis and treatment. BioMed research international. 2014;2014:407518
25. Kuo SJ, Yang WH, Liu SC, Tsai CH, Hsu HC, Tang CH. Transforming growth factor beta1 enhances heme oxygenase 1 expression in human synovial fibroblasts by inhibiting microRNA 519b synthesis. PloS one. 2017;12:e0176052
26. Li YB, Xu P, Xu K, Cai YS, Sun MY, Yang L. et al. Methotrexate affects HMGB1 expression in rheumatoid arthritis, and the downregulation of HMGB1 prevents rheumatoid arthritis progression. Molecular and cellular biochemistry. 2016;420:161-70
27. Sun XH, Liu Y, Han Y, Wang J. Expression and Significance of High-Mobility Group Protein B1 (HMGB1) and the Receptor for Advanced Glycation End-Product (RAGE) in Knee Osteoarthritis. Medical science monitor: international medical journal of experimental and clinical research. 2016;22:2105-12
28. Duan ZX, Zhu PF, Dong H, Gu W, Yang C, Liu Q. et al. Functional significance of the TLR4/11367 polymorphism identified in Chinese Han population. Shock. 2007;28:160-4
29. Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A. et al. A highly significant association between a COMT haplotype and schizophrenia. American journal of human genetics. 2002;71:1296-302
Author contact
![]() Corresponding author: Chih-Hsin Tang, PhD, Graduate Institute of Biomedical Science, China Medical University E-mail: chtangcmu.edu.tw
Corresponding author: Chih-Hsin Tang, PhD, Graduate Institute of Biomedical Science, China Medical University E-mail: chtangcmu.edu.tw

 Global reach, higher impact
Global reach, higher impact