3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(10):961-969. doi:10.7150/ijms.20121 This issue Cite
Research Paper
Genotoxicity of a Low-Dose Nitrosamine Mixture as Drinking Water Disinfection Byproducts in NIH3T3 Cells
1. Key Laboratory of Public Health Safety, Ministry of Education, Department of Environmental Health, School of Public Health, Fudan University, 130 Dongan Road Shanghai, 200032, China;
2. Department of Pharmacology, School of Pharmacy, Fudan University, 826 Zhangheng Road, Shanghai, 201203, China;
3. Department of Biostatistics, School of Public Health, Fudan University, Shanghai, 200032, China.
Received 2017-3-17; Accepted 2017-6-17; Published 2017-8-18
Abstract
N-nitrosamines (NAms), which can arise as byproducts of disinfection agents, are reportedly found in drinking water, and their potential carcinogenicity is a concern; however, little research exists regarding the genotoxicity or carcinogenicity of NAms exposure as a low-dose mixture. The three most common NAms components in China's drinking water are N-nitrosodimethylamine (NDMA), N-nitrosodiethylamine (NDEA) and N-nitrosomethylethylamine (NMEA). Thus, we measured the genotoxic and carcinogenic potential of these compounds and measured the cell cycle and gene expression. The data show that exposure to the NAms-mixture doubled the revertants in the TA98 and TA100 S. typhimurium strains and increased the DNA double-strand breaks and the micronuclear frequency in the NIH3T3 cells compared to a single exposure. After long-term NAms mixture exposure, a malignant transformation of NIH3T3 and a significantly increased G2/M distribution were observed. Furthermore, P53, CDK1, P38, CDC25A and CyclinB expressions were down-regulated in the NAms-mixture exposure group; however, P21 and GADD45A genes were up-regulated. Interestingly, the CHK1/CHK2 and CDC25A genes had two responses, depending on the NAms concentrations. Thus, we observed mutagenic, genotoxic and carcinogenic effects after a low-dose NAms-mixture exposure in drinking water, and DNA repair and apoptosis pathways may contribute to these adverse effects.
Keywords: Nitrosamines, disinfection byproducts, mixed exposure, genotoxicity, mutagenicity, transformation, DNA repair.
Introduction
N-nitrosamines (NAms) are disinfection byproducts (DBPs) [1, 2] found in drinking water, and their potential carcinogenicity is a concern [3]. Nine NAms compounds have been identified, as well as their molecular structures, physical traits, classifications and risks, and these data appear in Supplemental Table S1 and Figure S1 [4, 5]. The International Agency for Research on Cancer (IARC) has classified eight NAms compounds as potential (Group 2A) or possible carcinogens (Group 2B) to humans [6]. The US Environmental Protection Agency (EPA), the Agency for Toxic Substances and Disease Registry and the Department of Human Health Services also suggest that this group of NAms may be considered a human carcinogen that is hazardous in low concentrations. In addition, some developed countries have exposure limits for NAms. Ontario Canada has set a drinking water standard for N-nitrosodimethylamine (NDMA) of 9 ppt [7]. The California Department of Health Services has set notification levels for NDMA, N-nitrosodiethylamine (NDEA) and N-nitrosodipropylamine (NDPA) at 10 ppt and seeks to decrease this to 3 ppt [8]. The National Institute for Public Health and the Environment (RIVM, Netherlands) proposed a provisional guideline value for NDMA in drinking water of 12 ppt. These actions are precursors to formal regulations; however, most countries have not developed guidelines for NAms exposure due to a lack of sufficient risk-assessment data.
Exposure to NAms has been shown to be associated with tumors in epidemiological studies of human and laboratory animals [9]. NDMA, NDEA and N-nitrosomethylethylamine (NMEA) are highly mutagenic compounds that are suspected human carcinogens, and it is estimated that NDEA as low as 0.2 ppt in drinking water is associated with a 10-6 increased lifetime cancer risk [10]. NDMA is carcinogenic in experimental animals through several exposure routes, including the ingestion of drinking water. Most studies have focused on a single substance at a high concentration; however, many NAms coexist in drinking water, and individual toxicity may differ from a mixture exposure, which could be toxic at low doses [11, 12]. Presently, humans and environmental species are exposed to an almost infinite number of possible chemical combinations; thus, evidence of low-dose exposures to mixtures of environmental chemicals is of interest. Therefore, we investigated NAms mixtures at low doses to understand the health risk of pollutants in drinking water.
For this study, we selected a dose-addition approach and the most sensitive transformation cell line (NIH3T3) and common NAms compounds (NDMA, NDEA and NMEA) to assess the genotoxic and mutagenic potential and the possible molecular mechanism underlying low-dose exposure to a NAms mixture in drinking water.
Materials and Methods
Bacterial strain and cells
The T97, TA98, TA100 and TA102 S. typhimurium strains were obtained from Ames Laboratory in USA. The NIH3T3 cells (ATCC, CRL-1658) were cultured in DMEM (Gibco, Grand Island, NY, USA) containing 10% FBS (Gibco, USA), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a humidified 5% CO2 atmosphere.
Database of exposure
Meta-analysis was performed to summarize average NAms in drinking water. PubMed, Web of Science and Google Scholar were used to review the existing literature (from 1980 to 2015) [13-20] regarding NAms exposure in China's drinking water. The search terms were "N-nitrosamines", "Disinfection Byproducts or DBPs", "Drinking Water" and "China or Chinese" in various combinations. We chose eligible full texts and contacted the authors to confirm information when necessary.
Cell viability assay
Cytotoxicity was measured using a CCK-8 assay kit (Kumamoto, Japan) [21]. We used 1-30,000-fold concentrations of NAms to measure toxicity in NIH3T3 cells. Ten thousand cells with five replicates were plated in 96-well microplates and cultured for 24 h at 37°C. Then, the cells were incubated for an additional 72 h in media containing different NAms concentrations. Subsequently, optical density (OD) was measured at 450 nm with a Bio-Rad microplate reader. Each experiment was repeated three times. The 50% lethal concentration (LC50) was calculated using a dose-response curve.
Ames test
TA98, TA100, T97 and TA102 Salmonella typhimurium strains were cultured (1 × 109 cells/ml) overnight, and 0.5 ml S9 mix or PBS, 0.1 ml NAms and 0.1 ml bacterial suspension were mixed in tubes and cultured for 1 h at 37℃ with shaking (100 times/min). Then, 2 ml of top agarose was added to each tube and poured onto the underlying medium. The mixture was incubated for 48 h at 37°C before counting the revertant colonies. Each test was performed in triplicate with positive and negative controls, as shown in Table S2. A chemical was regarded as positive when the number of revertant colonies was at least twice the negative control [22].
Comet assay
A comet assay was performed similar to previous studies [23]. NIH3T3 cells were treated with different concentrations of NAms for 24 h, and cell viability >75% H2O2 (500 μg/ml) and DMSO (0.5%) were used as positive and negative controls, respectively. Cells were embedded in an agarose micro-gel and lysed. DNA was denatured and electrophoresed under alkaline conditions (pH=13) and stained with EB solution (20 μg/ml) for 10 min. At least 100 randomly selected cells were analyzed for each group, with triplicates, using fluorescentmicroscopy (Nikon, Japan). For quantifying DNA damage, the percentage of tail DNA was calculated using a CASP image analysis system (CaspLab, Poland) [24].
8-OHdG assay
After exposure to NAms, the NIH3T3 cells' supernatant was centrifuged at 3,000 rpm for 10 min. We added 50 μl of standard solution to standard wells, 10 μl sample and 40 μl dilution buffer to sample wells and then 100 μl of HRP-conjugate reagent to the standard well and sample well, respectively. The plate was then incubated 1 h at 37°C and was washed five times. Next, 50 μl of TMB and HRP chromogenic substrates were added to each well and incubated for 15 min in the dark at 37°C and then stopped with 50 μl stop solution. OD was measured and the 8-OHdG was calculated. Each treatment was carried out in triplicate [25].
Cytoplasm block micronucleus (CBMN) assay
A CBMN assay was performed following the Organization for Economic Co-operation and Development's method (OECD-T487) [26]. The NIH3T3 cells were exposed to different NAms levels for 40 h (1.5-2 normal cell cycles). Mitomycin C (1μM) and 0.5% DMSO were used as positive and negative controls. At least 2,000 binucleated cells were scored per group under fluorescence microscope (Nikon, Japan). Micronucleus (MNi), Nuclear Budding (NBUDs) and Nucleoplasmic Bridge (NPB) were calculated [26, 27]. The experiments were repeated three times.
Cell colony formation assay
NIH3T3 cells were used due to their wide applicability in cell malignant transformation studies. We seeded the NIH3T3 cells into a 6-well plate (100 cells/well). After culturing for 24 h at 37°C, the cells were treated with different NAms concentrations, positive control (3-methylcholanthrene, 3-MCA), solvent control (0.5 % DMSO) and negative control (distilled water) for 72 h, respectively. After washing twice with PBS, the cells were continually cultured for seven days at 37°C, and the medium was refreshed every three days. Then, the cells were fixed with methyl alcohol and stained with 10% Giemsa, and the colonies with more than 50 cells were counted [21, 28]. This was used to quantify colony-forming efficiency (CFE) and relative colony-forming efficiency (RCFE). CFE and RCFE were calculated as follows:
CFE (%) = (number of colonies induced)/(number of cells seeded) × 100%
RCFE (%) = [(CEF of treatment group)/(CEF of negative control group)] × 100%
Cell transformation assay
The NIH3T3 cells were seeded at a density of 2,000 cells/dish (10 cm), and the cells were cultured for 24 h. Cells were treated the same as the cell colony formation assay for 72 h. After rinsing with PBS, the cells were continually cultured for 14 days at 37°C, and the medium was replaced every three days. The cells were stained with 10% Giemsa, and the transformation frequency (TF) was calculated as follows [28]:
TF = [total number of transformed colonies per treatment/(total cells plated per treatment × CFE)] × 100%
Concanavalin A (Con A) agglutination
The transformed malignant cells induced by NAms were seeded (1,000 cells/dish) for the Con A agglutination assay, and the untransformed cells were labeled as negative controls. On day 14, the cells were harvested by adjusting them to 104 cells /ml with PBS. Then, 100 μl of single-cell suspensions and different concentrations of Con A were added to 24-well microplates for 10 min. Cell agglutination with Con A was observed by microscope (Nikon, Japan) [21].
Soft agar assay
A 3-ml aliquot of 1.2% agar in a culture medium was plated in 60-mm dishes. Then 1,000 cells of transformed malignant or untransformed cells were mixed with 3 ml of 0.35% agar in a medium and plated on the solidified bottom agar. When the top agar solidified, the dishes were transferred to an incubator and cultured for 30 days. Two or three drops of the medium were added to each dish three times a week. After culturing for 30 days, the visible cell colonies were photographed and counted [29].
Cell cycle determination
NIH3T3 cells were seeded at a density of 3.2 × 104 and cultured for 24 h. The cells were treated with different concentrations of NAms for 72 h. After rinsing with cold PBS, the cells were fixed with cold 70% ethanol for 12-24 h. Then, the cells were rinsed twice with cold PBS and stained using 0.5 ml of a stain agent (0.25% Triton X-100, 10 μg/ml PI, 100 μg/ml RNase) for 30 min in the dark. Measurements were performed with flow cytometry (BD, USA).
RNA extraction and real-time RT-PCR
RNA samples were extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified by Nanodrop (Thermo, Wilmington, DE). Total RNA was converted to cDNA by using SYBR Green PCR Kit (Qiagen, Germany). The primers were designed using Primer Express Software v2.0 (Applied Biosystems, Carlsbad, CA, USA) and synthesized by the Beijing Genomics Institute (BGI, China). All the primers' sequences are shown in the Supplemental (Table S3). The RT-PCR reactions were performed with ABI ViiA7 Sequence Detection Real-Time PCR System (Applied Bio-systems, USA). The cycle threshold (Ct) values were used to show the relative gene expression. Eighteen ribosomal RNA genes (rRNA, Hs99999901_s1, 18S) were used as an internal control. The difference in each group's gene expression was calculated with the 2-ΔΔCt method [12, 30]. All experiments were run in triplicate.
Western blot
Western blot analysis was performed as the previous description [12]. The cells were harvested after being washed three times with cold PBS and placed in a lysis buffer (Beyotime Institute of Biotechnology, China) on ice for 15 min. The cell lysates were centrifuged at 12,000 × g at 4°C for 10 min, and the supernatants were collected. The concentration of protein was detected via the BCA method. Proteins were separated in 8% SDS polyacrylamide gel by electrophoresis and transferred onto nitrocellulose immunoblot membranes (Millipore, Bedford, MA, USA) after 4h of blocking in TBST solution containing 5% skim milk, and then they were incubated with primary antibodies (Abcam, Cambridge, UK) overnight at 4°C. The antibodies were diluted as follows: P21 (1: 1000), P53 (1: 1000), P38 (1: 1000), CDK1 (1: 1000), cyclinB1 (1: 2000), ChK1 (1: 1000), GADD45A (1: 1000), CDC25B (1: 1000), CHK2 (1: 1000), CDC25A (1: 1000) and GAPDH (1: 2000). The membranes were washed and then incubated with horseradish peroxidase-conjugated (HRP) secondary antibodies for 1 h at room temperature. Proteins were detected by enhanced chemiluminescence using ECL reagent (Beyotime Institute of Biotechnology, China) and visualized on an image system (Image Quant LAS 4000 mini, USA). Signal densities were quantified using software Image J 1.44 (National Institutes of Health, Bethesda, MD, http://rsbweb.nih.gov/ij/).
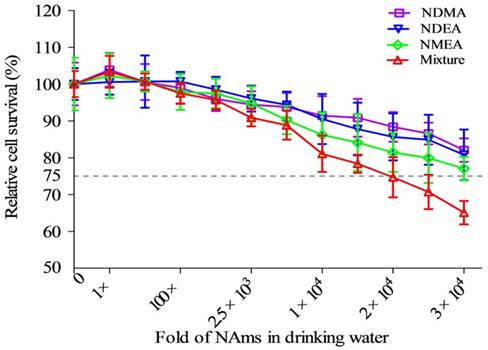
Cytotoxicity of individual NAms and a mixture in drinking water. Both exposures inhibited cell survival, and this was dose-dependent. The mixture contained NDMA, NDEA and NMEA. The exposure dose was set as a fold of actual environmental exposure level: 1× represents 10 ng/L for NDMA; 1× represents 5 ng/L for NDEA and NMEA; 1× represents (10NDMA+5NDEA+5NMEA) ng/L for the mixture.
Statistical analysis
Data were analyzed by SPSS version 18.0 (Armonk, IBM Corp, NY, US) and presented as means ± standard deviations (SD), with p <0.05 considered statistically significant. Results were analyzed using the GraphPad Prism software (version 5.0, GraphPad Prism Inc., San Diego, CA). A Dunnett-t multiple comparison analysis of treatments versus controls was applied.
Results
Occurrence and concentration of NAms in China's drinking water
According to the literature [13-20], nine NAms compounds are found in China's drinking water. NDMA, NDEA and NEMA are the most common and are found at the greatest concentration; thus, in our study, we used 10, 5 and 5 ng/L, respectively. Upon examination of the chemicals' toxic effects, cytotoxicity and proliferation rates were determined to be confounding factors. To avoid this, we measured the viability of NIH3T3 cells exposed to these compounds, as well as a mixture of all. Table S2 shows these data and the actual concentrations for the biological experiment.
Cytotoxicity of NAms in NIH3T3 cells
Cell survival data appears in Figure 1. Survival decreased with an increased concentration of NAms in both single and mixture exposure groups. Treatment with mixed NAms reduced viability the most. The data show that a 1,000-fold concentration of mixture exposure was chosen as an optimal concentration, according to the OECD's proposed genotoxicity dose requirements.
NAms mixture exposure increased Ames assay colonies
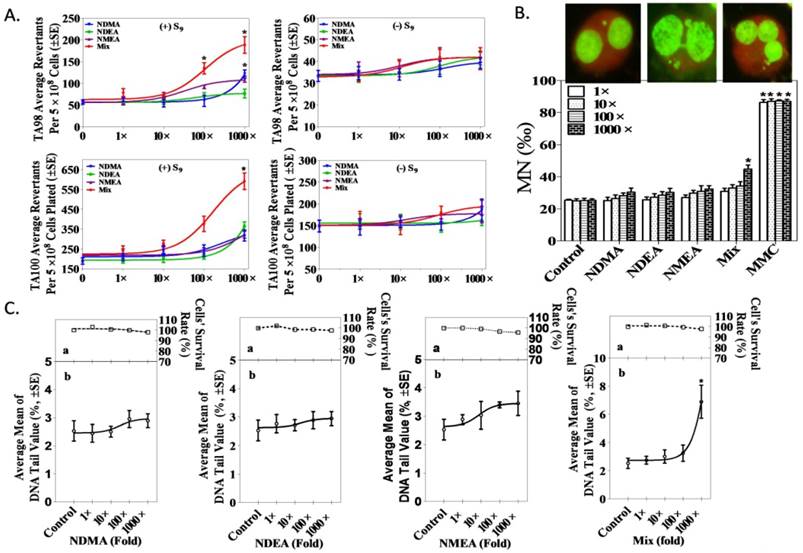
An Ames test was performed in TA97, TA98, TA100 and TA102 S. typhimurium strains, and the data demonstrate that 100-fold (20 × 102 ng/L) and 1,000-fold (20 × 103 ng/L) NAms mixtures increase colonies, as shown in Figure 2A. The mutagenic index (MI) data appear in Supplemental Table S2. No differences were observed for S. typhimurium revertant colonies among the three NAms alone or in the mixture of TA97 and TA102 strains (Supplemental Figure S2).
NAms mixture exposure caused chromosomal damage according to a CBMN test
A CBMN test was used to measure chromosomal damage after exposure of an NIH3T3 cell line to NAms alone or a mixture, and the data show that the mixture increased MNi in NIH3T3 cells but that a single exposure has no influence (Figure 2B). NBUDs and NPB were similar (Supplemental Figure S4).
NAms mixture exposure can induce DNA double-strand breaks, as measured with a comet assay
A comet assay confirmed that a mixture of 1,000-fold (20 × 103ng/L) NAms exposure significantly increased the percentage of tail DNA (p <0.05) in NIH3T3 cells compared to the controls; however, no such changes were observed in NAms alone (1 to 1,000-fold) or with low-dose mixed exposure (1 to 100-fold; Figure 2C). The 8-OHdG data agree with the comet assay (Supplemental Figure S3)
Colony formation assay
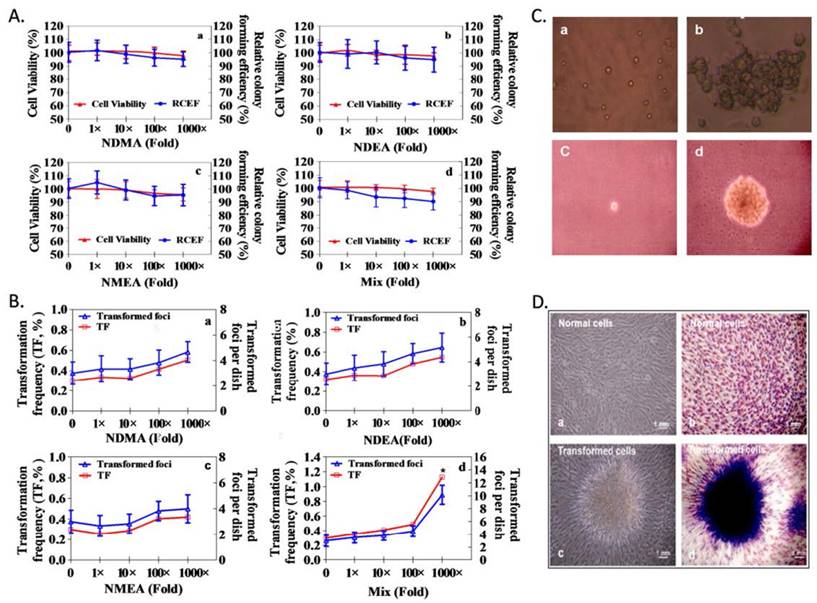
Cell colonies were counted as depicted in the Methods section, and CFE (%) and RCFE (%) data appear in Figure 3A.Compared with the controls, RCFE (%) was not significantly different among NDMA, NDEA, NMEA and mixture exposures (p >0.05). RCFE for each treatment was expressed as a percentage of CFE from the negative controls.
NAms mixture exposure transformed NIH3T3 cells
Individual NAms do not induce NIH3T3 cell transformation; however, the mixture increased TF in the NIH3T3 cell (Figure 3B). Con A and the soft agar assay confirmed phenotypic changes in the transformed cells (Figure 3C and Figure 3D). Furthermore, the untransformed NIH3T3 cells were agglutinated with only a high (100µg/ml) concentration of Con A, whereas NAms-transformed cells were agglutinated with 25µg/ml Con A (Supplemental Table S4). Negative controls failed to grow when suspended in soft agar; however, NAms-transformed cells grew in soft agar (Supplemental Table S5).
Genotoxic potential of low-dose exposure to single or NAms mixture in NIH3T3 cells. A. Mutagenicity concentration-response curves for single and mixed NAms with TA98 and TA100 S. typhimurium. B. Determination of micronucleus formation induced by NAms in NIH3T3 cells. Binucleated cells with micronuclei after exposure to NAms for 40 h. The upper image shows the MNi, NPBs and NBUDs, respectively. MNi were fewer in the lower panel. Compared to the control, a mixture of 1,000-fold (20 × 103 ng/L) NAms can cause an increased MNi in NIH3T3 cells (p <0.05). C. The percentage of tail DNA induced by NAms was measured using a comet assay. The upper panel (a) represented the cytotoxicity at the same concentration of NAms, and the lower panel (b) represented DNA damage. Compared to the control, a mixture of 1,000-fold (20 × 103 ng/L) NAms significantly increased the percentage of tail DNA (p <0.05) in NIH3T3 cells.
NAms-induced transformation of NIH/3T3 cells. A. Colony-forming assay results showed that relative colony-forming efficiency (RCFE) was not significantly different among NDMA, NDEA, NMEA and mixture exposures. B. Transformation assay results for the combined exposure of three NAms at 1,000-fold (20 × 103 ng/L) concentrations can increase transformation frequency (TF) in NIH3T3 cells. C. The results of Con A agglutination (a, b) and the results of soft agar assay (c, d); a: dispersive cells; b: agglutinated cells; c: non-survival cells; d: NAms-transformed cells. D. Transformed cell images: normal cells (a, b) and NAms-transformed cells (c, d).
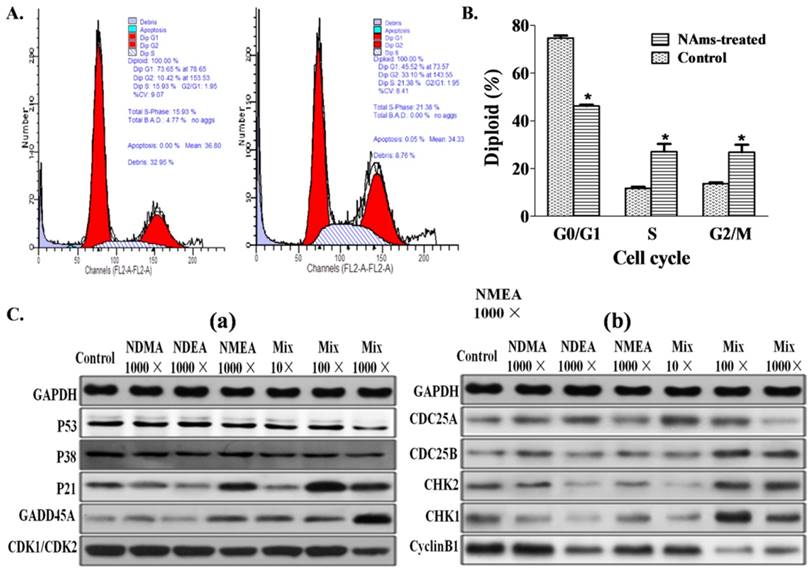
NAms mixture exposure leads to G2/M arrest by multiple gene regulation
Cell-cycle analysis showed significant G2/M arrest after 1,000-fold (20 × 103 ng/L) NAms mixture exposure (Figure 4A and 4B). Because this concentration can induce cell transformation and G2/M arrest, we speculated that some genes may regulate the effects of the mixture exposure. Gene expression data show that genes were altered, and compared to the single exposure, the mRNA and protein for P53/CDK1/CDC25A/P38/CyclinB1 were down-regulated with the 1,000-fold (20 × 103 ng/L) NAms mixture exposure group, and the P21 and GADD45A genes were up-regulated (Figure 4C and Supplemental Figure S5). After 1,000-fold NAms mixture exposure, the CHK1/CHK2 genes were up-regulated, and the CDC25A genes were down-regulated (Figure S5).
Discussion
NAms are nitrogen non-halogen DBPs mainly generated during chloramine disinfection [31, 32], and they may be carcinogenic. Thus, some countries have guidelines for specific NAms components; however, these guidelines are based on single NAms exposures. Better data about NAms mixtures may help establish safety limits for these types of exposures in drinking water, yet no reports for mixtures are available [33, 34].
Furthermore, the genotoxic assessments of NAms are usually assessed as mg/L, which is 40-50 million times the concentrations present in drinking water. For example, NDMA has been studied using an Ames assay at 20 mM [35] and with a comet assay at 2.39 mM [36]. Previous genotoxicity studies with NAms-DBPs did not assess the genotoxicity thoroughly with different end points [35, 36]. Research may also ignore potent adverse effects when investigating the predicted effects (or risk assessment) for mixed chemicals at a low dose. Thus, we must better characterize the genotoxicity and mutagenicity of NAms [37]. Therefore, we used a genotoxicity test proposed by the International Council for Harmonization (ICH) [38], including an Ames, comet, 8-OHdG and CBMN assays, to evaluate the genotoxicity of NAms individually and as mixtures in vitro. We found increased mutagenicity in double revertants, chromosome abbreviation and DNA double-strand breaks after exposure to a three-compound mixture at 1,000-fold (20 × 103 ng/L) of actual NAms in drinking water, and at this concentration, genotoxic and carcinogenic effects were noted. The mutagenic potential of NAms was measured using a cell transformation assay in vitro [39], and NAms in a 1,000-fold (20 × 103 ng/L) mixture induced a malignant transformation in NIH3T3 cells. Thus, the mixture had greater genotoxicity and mutagenicity than the individual constituents.
To evaluate genotoxicity and mutagenicity of a NAms mixture exposure, we conducted cell-cycle and gene-expression evaluations in the transformed cells. The data show that 100-fold and 1,000-fold concentrations of NAms exposure could lead to G2/M arrest; however, only the 1,000-fold concentrations induced cell transformation. RT-PCR and western blot showed that p53, p21, CDC25A/B1, CHK1/2 and CDK1/2 expressions were up-regulated in the 100-fold (20 × 102 ng/L) exposure but down-regulated after a 1,000-fold exposure. DNA damage from NAms included DNA adducts and an activated DNA damage checkpoint. The NIH3T3 cells' exposure to NAms caused DNA damage and initiated DNA repair pathways, triggering cell-cycle arrest to repair the damage. However, the activated repair response was not effective when the exposure exceeded a threshold that produced a transformed malignant cell. Our study had some limitations in that we did not elucidate how each NAms component contributed to toxicity independently as a carcinogen. Thus, more study is needed. Furthermore, the gene-expression data were uncertain, as activity varied with increasing concentrations. We require RNA and/or protein data to better understand these preliminary observations.
Effect of low-dose NAms mixture exposure and cell cycle and gene expression. (A) Representative flow cytometry image of cell-cycle results. (B) NIH3T3 cells were in G2/M arrest after 1,000-fold (20 × 103 ng/L) mixed NAms exposure compared to control cells (p <0.05). C. Expression of protein for cell-cycle regulation genes; (a) expression of P53, P38, p21, GADD45A and CDK1 (CDK2); (b) expression of CDC25A, CDC25B, CHK2, CHK1 and CyclinB1; * indicates p <0.05 vs control.
This is the first genotoxicity and carcinogenicity study of low-dose NAms mixture exposure, and the data suggest NAms are hazardous to public health; however, their toxicity and mechanism must be clarified. Human epidemiological studies must be performed to understand NAms' adverse effects on public health and our environment.
Abbreviations
CBMN: Cytokinesis-block Micronuclei Test;
CFE: colony-forming efficiency;
Con A: Concanavalin;
DBPs: Disinfection Byproducts;
3-MCA: 3-Methylcholanthrene;
MNi: Micronucleus;
NAms: Nitrosamines;
NBUDs: Nuclear Buddings;
NDEA: N-nitrosodiethylamine;
NDMA: N-nitrosodimethylamine;
NDPA: N-nitrosodipropylamine;
NMEA: N-nitrosomethylethylamine;
NPB: nucleoplasmic bridge;
TF: Transformation frequency;
RCFE: relative colony forming efficiency.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank all researcher coordinators who contributed to this study.
Author contributions
HY, XW with the assistance of MQ, JY and TL, performed the study design, lab work and data analysis. All authors discussed the results and commented on the manuscript at every stage. HY and LD contributed to the manuscript's writing and revision.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Richardson SD, Plewa MJ, Wagner ED, Schoeny R, Demarini DM. Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: a review and roadmap for research. Mutat Res. 2007;636:178-242
2. Richardson SD, Ternes TA. Water analysis: emerging contaminants and current issues. Anal Chem. 2014;86:2813-48
3. Moudgal CJ, Lipscomb JC, Bruce RM. Potential health effects of drinking water disinfection by-products using quantitative structure toxicity relationship. Toxicology. 2000;147:109-31
4. Mitch WA, Sedlak DL. Formation of N-nitrosodimethylamine (NDMA) from dimethylamine during chlorination. Environ Sci Technol. 2002;36:588-95
5. Schreiber IM, Mitch WA. Occurrence and fate of nitrosamines and nitrosamine precursors in wastewater-impacted surface waters using boron as a conservative tracer. Environ Sci Technol. 2006;40:3203-10
6. IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans; 1982. http://monographs.iarc.fr/ENG
7. Ontario Goverment. Technical support document for Ontario Drinking Water Standards objectives and guidelines; 2006. http://www.creditvalleyca.ca/wp-content/uploads/2011/03/std01_079707.pdf
8. California Environmental Protection Agency. NDMA and other nitrosamines-drinking water issues. 2013. http://www.waterboards.ca.gov/drinking_water/certlic/drinkingwater/NDMA.shtml
9. Eichholzer M, Gutzwiller F. Dietary nitrates, nitrites, and N-nitroso compounds and cancer risk: a review of the epidemiologic evidence. Nutr Rev. 1998;56:95-105
10. EPA US. United States Environmental Protection Agency. Integrated Risk Information System Summaries(IRIS). 2015. http://cfpub.epa.gov/ncea/iris
11. Wang S, Tian D, Zheng W, Jiang S, Wang X, Andersen ME. et al. Combined exposure to 3-chloro-4-dichloromethyl-5-hydroxy-2(5H)-furanone and microsytin-LR increases genotoxicity in Chinese hamster ovary cells through oxidative stress. Environ Sci Technol. 2013;47:1678-87
12. Wang S, Zhang H, Zheng W, Wang X, Andersen ME, Pi J. et al. Organic extract contaminants from drinking water activate Nrf2-mediated antioxidant response in a human cell line. Environ Sci Technol. 2013;47:4768-77
13. Hung HW, Lin TF, Chiu CH, Chang YC. Trace Analysis of N-Nitrosamines in Water Using Solid-Phase Microextraction Coupled with Gas Chromatograph-Tandem Mass Spectrometry. Water Air & Soil Pollut. 2010;213:459-69
14. Luo Q, Wang D, Wang Z. Occurrences of nitrosamines in chlorinated and chloraminated drinking water in three representative cities, China. Sci Total Environ. 2012;437:219-25
15. Sgro LA, Simonelli A, Pascarella L, Minutolo P, Guarnieri D, Sannolo N. et al. Toxicological properties of nanoparticles of organic compounds (NOC) from flames and vehicle exhausts. Environ Sci Technol. 2009;43:2608-13
16. Wang C, Zhang X, Wang J, Liu S, Chen C, Xie Y. Effects of organic fractions on the formation and control of N-nitrosamine precursors during conventional drinking water treatment processes. Sci Total Environ. 2013;449:295-301
17. Wang CK, Zhang XJ, Wang J, Chen C. Detecting N-nitrosamines in water treatment plants and distribution systems in China using ultra-performance liquid chromatography-tandem mass spectrometry. Front EnvironSci & Eng. 2012;6:770-7
18. Wang W, Hu J, Yu J, Yang M. Determination of N-nitrosodimethylamine in drinking water by UPLC-MS/MS. J Environ Sci (China). 2010;22:1508-12
19. Wang W, Ren S, Zhang H, Yu J, An W, Hu J. et al. Occurrence of nine nitrosamines and secondary amines in source water and drinking water: Potential of secondary amines as nitrosamine precursors. Water Res. 2011;45:4930-8
20. Zhang A, Li Y, Chen L. Distribution and seasonal variation of estrogenic endocrine disrupting compounds, N-nitrosodimethylamine, and N-nitrosodimethylamine formation potential in the Huangpu River, China. J Environ Sci (China). 2014;26:1023-33
21. Wei X, Wang S, Zheng W, Wang X, Liu X, Jiang S. et al. Drinking water disinfection byproduct iodoacetic acid induces tumorigenic transformation of NIH3T3 cells. Environ Sci Technol. 2013;47:5913-20
22. Mortelmans K, Zeiger E. The Ames Salmonella/microsome mutagenicity assay. Mutat Res. 2000;455:29-60
23. Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23-9
24. Konca K, Lankoff A, Banasik A, Lisowska H, Kuszewski T, Gozdz S. et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat Res. 2003;534:15-20
25. Dong H, Su C, Xia X, Li L, Song E, Song Y. Polychlorinated biphenyl quinone-induced genotoxicity, oxidative DNA damage and gamma-H2AX formation in HepG2 cells. Chem Biol Interact. 2014;212:47-55
26. OECD guideline for the testig of chemicals, In Vitro Mammalian Cell Micronucleus Test 487. 2013. http://www.oecd.org/env/ehs/testing/TG487_Dec2013_WNT4thCR.pdf
27. Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084-104
28. Swierenga SH, Bradlaw JA, Brillinger RL, Gilman JP, Nestmann ER, San RC. Recommended protocols based on a survey of current practice in genotoxicity testing laboratories: I. Unscheduled DNA synthesis assay in rat hepatocyte cultures. Mutat Res. 1991;246:235-53
29. Booden MA, Ulka AS, Der CJ. Cellular assays of oncogene transformation. In Cell Biology: A Laboratory Handbook. In: (ed.) Celis JE CN, Simons K, Small JV, Hunter T, Shotton DM. Cell Biology. Burlington: Elsevier Academic. 2006:345-52
30. Woods CG, Fu J, Xue P, Hou Y, Pluta LJ, Yang L. et al. Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol Appl Pharmacol. 2009;238:27-36
31. Chen WH, Young TM. Influence of nitrogen source on NDMA formation during chlorination of diuron. Water Res. 2009;43:3047-56
32. Shen R, Andrews SA. Demonstration of 20 pharmaceuticals and personal care products (PPCPs) as nitrosamine precursors during chloramine disinfection. Water Res. 2011;45:944-52
33. Goodson WH 3rd, Lowe L, Carpenter DO, Gilbertson M, Manaf Ali A, Lopez de Cerain Salsamendi A. et al. Assessing the carcinogenic potential of low-dose exposures to chemical mixtures in the environment: the challenge ahead. Carcinogenesis. 2015;36(Suppl 1):S254-96
34. Kienzler A, Bopp SK, van der Linden S, Berggren E, Worth A. Regulatory assessment of chemical mixtures: Requirements, current approaches and future perspectives. Regul Toxicol Pharmacol. 2016;80:321-34
35. Mori Y, Tatematsu K, Koide A, Sugie S, Tanaka T, Mori H. Modification by curcumin of mutagenic activation of carcinogenic N-nitrosamines by extrahepatic cytochromes P-450 2B1 and 2E1 in rats. Cancer Sci. 2006;97:896-904
36. Wagner ED, Osiol J, Mitch WA, Plewa MJ. Comparative in vitro toxicity of nitrosamines and nitramines associated with amine-based carbon capture and storage. Environ Sci Technol. 2014;48:8203-11
37. Hebert A, Forestier D, Lenes D, Benanou D, Jacob S, Arfi C. et al. Innovative method for prioritizing emerging disinfection by-products (DBPs) in drinking water on the basis of their potential impact on public health. Water Res. 2010;44:3147-65
38. The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH). Guidance on genotoxicity testing and data interpretation pharmaceuticals intended for human use S2 (R1); 2011. http://www.ich.org/products/guidelines/safety/safety-single/article/guidance-on-genotoxicity-testing-and-data-interpretation-for-pharmaceuticals-intended-for-human-use.html
39. Sakai A. In BALB/c 3T3 cell transformation assays for the assessment of chemical carcinogenicity. Aatex. 2007;14:367-73
Author contact
![]() Corresponding author: Xia Wang, Key Laboratory of Public Health Safety, Ministry of Education, Department of Environmental Health, School of Public Health, Fudan University. Shanghai, 200032, China. Tel: +86-13916251491 E-mail: xwang6edu.cn
Corresponding author: Xia Wang, Key Laboratory of Public Health Safety, Ministry of Education, Department of Environmental Health, School of Public Health, Fudan University. Shanghai, 200032, China. Tel: +86-13916251491 E-mail: xwang6edu.cn

 Global reach, higher impact
Global reach, higher impact