3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(9):696-700. doi:10.7150/ijms.16259 This issue Cite
Research Paper
Prognostic Value of Prostaglandin-endoperoxide Synthase 2 Polymorphisms in Prostate Cancer Recurrence after Radical Prostatectomy
1. Department of Urology, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan;
2. Graduate Institute of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan;
3. Department of Pharmacy, Linsen Chinese Medicine Branch, Taipei City Hospital, Taipei, Taiwan;
4. Department of Pharmacy, China Medical University, Taichung, Taiwan;
5. Department of Urology, Faculty of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan;
6. Department of Urology, National Taiwan University Hospital, College of Medicine, National Taiwan University, Taipei, Taiwan;
7. Department of Urology, E-Da Hospital, Kaohsiung, Taiwan;
8. School of Medicine for International Students, I-Shou University, Kaohsiung, Taiwan;
9. Division of Urology, Department of Surgery, Kaohsiung Veterans General Hospital, Kaohsiung, Taiwan;
10. Department of Urology, School of Medicine, National Yang-Ming University, Taipei, Taiwan;
11. Department of Pathology, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan;
12. Department of Pathology, Faculty of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
13. Sex Hormone Research Center, China Medical University Hospital, Taichung, Taiwan;
14. Department of Nursing, Asia University, Taichung, Taiwan.
Received 2016-5-23; Accepted 2016-7-20; Published 2016-8-11
Abstract
Backgroud: Increasing evidence suggests the involvement of chronic inflammation in the progression of prostate cancer, and prostaglandin-endoperoxide synthase 2 (PTGS2), also known as cyclooxygenase-2, catalyzes the rate-limiting steps of the pathway. We hypothesized that genetic variants of PTGS2 can influence the outcome of prostate cancer patients.
Methods: We genotyped five haplotype-tagging single-nucleotide polymorphisms (SNPs) to detect common genetic variations across the PTGS2 region in 458 prostate cancer patients treated with radical prostatectomy.
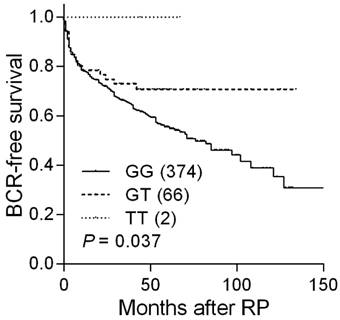
Results: One SNP, rs4648302, was associated with disease recurrence. Five-year recurrence-free survival rate increased according to the number of variant alleles inherited (55.6%, 70.7%, and 100.0% for patients with different genotypes; P = 0.037), and the effect was maintained in multivariable analysis. Public dataset analyses also suggested that PTGS2 expression was correlated with prostate cancer prognosis.
Conclusion: Our results indicated that PTGS2 could be a potential prognostic marker to improve the prediction of disease recurrence in prostate cancer patients.
Keywords: biochemical recurrence, prostate cancer, radical prostatectomy, single-nucleotide polymorphism, PTGS2, inflammation.
Introduction
Prostate cancer is the most commonly diagnosed cancer in the western world, and it has become a serious public health issue in Taiwan with incidence rate increasing over the past two decades [1]. Many men diagnosed with prostate cancer have early-stage disease, and radical prostatectomy (RP) is one of the most effective means of curing the localized cancer. Although RP provides durable cancer control with excellent quality of life, a significant rise in prostate-specific antigen (PSA) levels are observed in many patients, indicating biochemical recurrence (BCR) [2]. Understanding the molecular mechanisms underlying prostate cancer recurrence could facilitate the discovery of novel biomarkers and therapeutic strategies to control this malignancy.
Studies have suggested chronic inflammation contributes to a variety of malignancies, including that of prostate cancer [3]. Prostaglandin-endoperoxide synthase 2 (PTGS2, also known as cyclooxygenase-2, COX-2) catalyzes the rate-limiting steps in prostaglandin biosynthesis, and its expression is controlled by inflammatory signaling at both transcriptional and post-transcriptional levels [4]. Prostaglandins could interact with their receptors to transduce downstream signaling, which could promote cancer cell proliferation, survival, angiogenesis, and metastasis [5].
More evidence indicates that ethnic background and family history are associated with the risk of prostate cancer, suggesting genetic susceptibility in prostate cancer predisposition [6]. Several single-nucleotide polymorphisms (SNPs) of PTGS2 are associated with susceptibility to prostate cancer [7-9], but no study to date has examined their ability to predict disease progression. Therefore, the aim of this study was to systematically evaluate the prognostic significance of PTGS2 SNPs on BCR in localized prostate cancer patients after RP.
Materials and Methods
Patient recruitment and data collection
We recruited 458 localized prostate cancer patients who underwent RP as initial therapy at the National Taiwan University, Kaohsiung Medical University, E-Da, and Kaohsiung Veterans General hospitals, as described previously [10-13]. Demographic, clinical, and follow-up data were obtained from the medical records. BCR was defined as two consecutive PSA values of at least 0.2 ng/mL [14, 15]. The present study was approved by the Institutional Review Board of Kaohsiung Medical University Hospital. Written informed consent was obtained from each patient, and the study was carried out in accordance with the approved guidelines.
Basic characteristics of 458 localized prostate cancer patients who received RP are described in Table S1. The median age of the patients was 66 years, and the 5-year BCR-free survival rate was 56.5%. Disease recurred in 184 (40.2%) patients during the median follow-up of 54 months. PSA at diagnosis, pathologic Gleason score, and pathologic stage were significantly associated with BCR (P ≤ 0.001).
SNP selection and genotyping
Genomic DNA was extracted from the peripheral blood with QIAamp DNA Blood Maxi Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol, and was stored until use. We utilized a haplotype-tagging SNP method to cover most of the haplotype diversity in the PTGS2 gene region. Haplotype-tagging SNPs were selected using the Haploview Tagger with pairwise tagging [16], a minimum r2 of 0.8, and a minor-allele frequency ≥ 2.5% on the basis of the data on 1000 genomes from Han Chinese in Beijing and Southern Han Chinese [17]. We identified six SNPs, which were genotyped at the National Center for Genome Medicine, Taiwan, using the Agena Bioscience MassARRAY iPLEX technology. For quality control, we randomly selected 10 samples as duplicates, and the concordance rate was 100% for all SNPs assayed. The average call rate was 99.2%, but rs5275 with a Hardy-Weinberg equilibrium < 0.05 was removed. Thus, five SNPs were included for further statistical analyses.
Statistical analysis
Patient characteristics were summarized as either the numbers and percentages of patients, or the median and interquartile range of values. Individual SNPs were initially assessed using the Kaplan-Meier analysis with log-rank test for the three genetic models of inheritance: dominant (common homozygotes versus variant allele carrying genotypes), recessive (common allele carrying genotypes versus variant homozygotes), and additive (P for trend). Only dominant and additive models were considered if the variant homozygotes were observed in < 5% of the study population. Cox proportional hazards regression analyses were used to assess the effect of each SNP on BCR, with or without adjusting for known prognostic factors, including age, PSA at diagnosis, pathologic Gleason score, and stage, as previously described [11]. The Statistical Package for the Social Sciences software, version 22.0.0 (IBM, Armonk, NY, USA), was used for other statistical analyses. A two-sided P value of < 0.05 was considered statistically significant.
Bioinformatics analysis
SNPinfo [18] was used to identify the regulatory potential of the SNP. Publicly available datasets [19, 20] were used to analyze PTGS2 expression and prostate cancer progression.
Results
Analysis of five PTGS2 SNPs showed a significant correlation between rs4648302 and BCR in localized prostate cancer patients receiving RP (Table 1). Five-year BCR-free survival rates were 55.6, 70.7, and 100.0% for patients with rs4648302 GG, GT, and TT genotypes (Table 2 and Figure 1). The BCR risk decreased according to the number of variant alleles inherited at PTGS2 rs4648302 [hazard ratio (HR) 0.61, 95% confidence interval (CI) 0.38-0.98, P = 0.040; Table 2], and the impact persisted upon multivariable analysis (HR 0.61, 95% CI 0.37-0.99, P = 0.046).
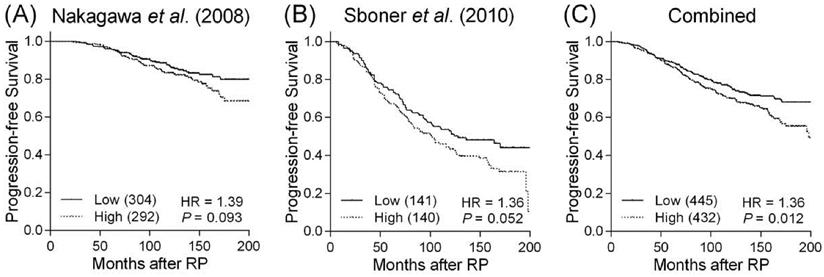
Bioinformatic analysis revealed the rs4648302 G to T transition gained a new microRNA binding site and thus could decrease PTGS2 expression. We performed an in silico evaluation of PTGS2 gene expression in relation to prostate cancer progression using publicly available datasets. When the patients were grouped based on the values higher or lower than the median value of gene expression, we observed a trend toward more favorable outcome for cancers with lower PTGS2 expression in two independent prostate cancer microarray datasets (Figures 2A and B). In a combined analysis, lower PTGS2 expression was significantly associated with a reduced risk of disease progression (P = 0.012, Figure 2C).
Kaplan-Meier analysis of BCR-free survival based on PTGS2 rs4648302 genotypes. Numbers in parentheses indicate the number of patients.
Association between haplotype tagging SNPs in PTGS2 and BCR in localized prostate cancer patients treated with RP.
| SNP ID | Location | Chromosome | Position | Alleles | MAF | P | ||
|---|---|---|---|---|---|---|---|---|
| Additive | Dominant | Recessive | ||||||
| rs4648302 | 3'-UTR | 1 | 186641218 | G:T | 0.079 | 0.037 | 0.046 | - |
| rs2066826 | Intron 6 | 1 | 186645927 | G:A | 0.035 | 0.797 | 0.989 | - |
| rs5277 | Exon 3 | 1 | 186648197 | G:C | 0.039 | 0.356 | 0.244 | - |
| rs20427 | Intron 2 | 1 | 186648355 | C:G | 0.060 | 0.053 | 0.060 | - |
| rs2745557 | Intron 1 | 1 | 186649221 | G:A | 0.045 | 0.139 | 0.139 | - |
Abbreviations: SNP, single-nucleotide polymorphism; BCR, biochemical recurrence; RP, radical prostatectomy; MAF, minor allele frequency; UTR, untranslated region.
P values for log-rank test.
P < 0.05 is in boldface.
Kaplan-Meier analysis of progression-free survival based on PTGS2 expression. Expression of PTGS2 mRNA is compared with progression-free survival in datasets from (A) Nakagawa et al., (B) Sboner et al., and (C) in combined analysis. Patients were divided into high and low groups according to the median mRNA expression values of PTGS2. Numbers in parentheses indicate the number of patients.
Univariate and multivariate analyses of PTGS2 rs4648302 and BCR after RP.
| SNP Genotype | No BCR, n (%) | BCR, n (%) | 5-year BCR-free survival, % | HR (95% CI) | P | HR (95% CI)* | P* |
|---|---|---|---|---|---|---|---|
| rs4648302 | |||||||
| GG | 220 (58.8) | 154 (41.2) | 55.6 | 1.00 | 1.00 | ||
| GT | 48 (72.7) | 18 (27.3) | 70.7 | 0.64 (0.39-1.04) | 0.071 | 0.63 (0.38-1.05) | 0.074 |
| TT | 2 (100.0) | 0 (0.0) | 100.0 | - | - | ||
| GT/TT vs GG | 0.61 (0.38-1.00) | 0.050 | 0.61 (0.37-1.01) | 0.055 | |||
| Trend | 0.61 (0.38-0.98) | 0.040 | 0.61 (0.37-0.99) | 0.046 |
Abbreviations: BCR, biochemical recurrence; RP, radical prostatectomy; SNP, single-nucleotide polymorphism; HR, hazard ratio; CI, confidence interval; PSA, prostate-specific antigen.
*Adjusted by age, PSA at diagnosis, pathologic Gleason score, and pathologic stage.
P < 0.05 are in boldface.
Discussion
It has been challenging to predict BCR after RP for patients with high-risk prostate cancer. Our study was designed specifically to assess the possibility that genetic variants in PTGS2 could predict disease progression. We found that PTGS2 rs4648302 confers an independent risk of prostate cancer recurrence compared with commonly used clinicopathological factors. Moreover, patients with higher PTGS2 expression levels showed a poorer survival, further confirmed the functional relevance of PTGS2 in prostate cancer progression.
rs4648302 locates in the 3'-untranslated region of PTGS2. According to the functional prediction of SNPinfo [18], the favorable allele T, creates a putative microRNA binding site for miR-154, which might result in lower PTGS2 expression. Since PTGS2 is probably oncogenic (Figure 2), the rs4648302 G to T transition might attenuate prostate cancer progression, which is consistent with the results of our association study that individuals carrying the rs4648302 T allele had a decreased risk of BCR (Table 2 and Figure 1). It has also been demonstrated that miR-154 expression level is downregulated in prostate cancer, and overexpression of miR-154 can significantly reduce prostate cancer cell migration and invasion [21, 22]. However, future functional studies are required to gain more insights into the mechanism underlying the observed association with prostate cancer recurrence.
This present study has several strengths. First, we used a haplotype-tagging approach and systematically evaluated common SNPs within the PTGS2. Second, this is the first study to date to identify the effects of PTGS2 SNPs on treatment outcomes for prostate cancer patients after RP. In addition, the available clinical information enables us to adjust for potential confounding factors. However, our findings in Taiwanese might not be generalizable to other ethnic populations. Given the multiple comparison, there is a possibility that the significant results might be false discoveries. Additional validations with larger sample sizes are needed to confirm our findings.
In conclusion, we have identified PTGS2 rs4648302 as a prognostic predictor for prostate cancer. This variant might alter microRNA binding, and thus influence PTGS2 expression and prostate cancer progression. Clinical studies have suggested that selective PTGS2 inhibitors could reduce cancer risk [23, 24]. If validated, such knowledge could ultimately lead to targeted therapies for a subset of high-risk patients harboring this inherited PTGS2 variant.
Abbreviations
PTGS2, prostaglandin-endoperoxide synthase 2; SNP, single-nucleotide polymorphism; RP, radical prostatectomy; PSA, prostate-specific antigen; BCR, biochemical recurrence; HR, hazard ratio; CI, confidence interval.
Supplementary Material
Table S1.
Acknowledgements
This work was supported by the Ministry of Science and Technology of Taiwan (grant number: 102-2628-B-039-005-MY3, 103-2314-B-037-060, 104-2314-B-650-006, and 104-2314-B-037-052-MY3) and the Kaohsiung Medical University Hospital (grant number: KMUH103-3R43). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Chao-Shih Chen for data analysis and the National Center for Genome Medicine, Ministry of Science and Technology of Taiwan, for technical support.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Chiang CJ, Lo WC, Yang YW. et al. Incidence and survival of adult cancer patients in Taiwan, 2002-2012. J Formos Med Assoc. 2016
2. Pound CR, Partin AW, Eisenberger MA. et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591-7
3. Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology (Williston Park). 2002;16:217-26 29; discussion 30-2
4. Hla T, Bishop-Bailey D, Liu CH. et al. Cyclooxygenase-1 and -2 isoenzymes. Int J Biochem Cell Biol. 1999;31:551-7
5. Liu XH, Yao S, Kirschenbaum A. et al. NS398, a selective cyclooxygenase-2 inhibitor, induces apoptosis and down-regulates bcl-2 expression in LNCaP cells. Cancer Res. 1998;58:4245-9
6. Steinberg GD, Carter BS, Beaty TH. et al. Family history and the risk of prostate cancer. Prostate. 1990;17:337-47
7. Yang X, Li B, Si T. et al. Association between the 8473T>C polymorphism of PTGS2 and prostate cancer risk: a metaanalysis including 24,716 subjects. Onkologie. 2013;36:182-6
8. Zhang H, Xu Y, Zhang Z. et al. Association between COX-2 rs2745557 polymorphism and prostate cancer risk: a systematic review and meta-analysis. BMC Immunol. 2012;13:14
9. Zhang HT, Xu Y, Zhang ZH. et al. Meta-analysis of epidemiological studies demonstrates significant association of PTGS2 polymorphism rs689470 and no significant association of rs20417 with prostate cancer. Genet Mol Res. 2012;11:1642-50
10. Bao BY, Pao JB, Lin VC. et al. Individual and cumulative association of prostate cancer susceptibility variants with clinicopathologic characteristics of the disease. Clin Chim Acta. 2010;411:1232-7
11. Huang SP, Huang LC, Ting WC. et al. Prognostic significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol Biomarkers Prev. 2009;18:3068-74
12. Huang SP, Lan YH, Lu TL. et al. Clinical significance of runt-related transcription factor 1 polymorphism in prostate cancer. BJU Int. 2011;107:486-92
13. Yu CC, Lin VC, Huang CY. et al. Prognostic significance of cyclin D1 polymorphisms on prostate-specific antigen recurrence after radical prostatectomy. Ann Surg Oncol. 2013;20(Suppl 3):S492-9
14. Freedland SJ, Sutter ME, Dorey F. et al. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy. Prostate-specific antigen. Urology. 2003;61:365-9
15. Huang SP, Levesque E, Guillemette C. et al. Genetic variants in microRNAs and microRNA target sites predict biochemical recurrence after radical prostatectomy in localized prostate cancer. Int J Cancer. 2014;135:2661-7
16. Barrett JC, Fry B, Maller J. et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-5
17. Genomes Project C, Abecasis GR, Auton A. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56-65
18. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600-5
19. Nakagawa T, Kollmeyer TM, Morlan BW. et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3:e2318
20. Sboner A, Demichelis F, Calza S. et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8
21. Zhu C, Li J, Cheng G. et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379:69-75
22. Zhu C, Shao P, Bao M. et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol. 2014;32:31 e9-16
23. Reddy BS, Hirose Y, Lubet R. et al. Chemoprevention of colon cancer by specific cyclooxygenase-2 inhibitor, celecoxib, administered during different stages of carcinogenesis. Cancer Res. 2000;60:293-7
24. Smith MR, Manola J, Kaufman DS. et al. Celecoxib versus placebo for men with prostate cancer and a rising serum prostate-specific antigen after radical prostatectomy and/or radiation therapy. J Clin Oncol. 2006;24:2723-8
Author contact
![]() Corresponding authors: Shu-Pin Huang, Department of Urology, Kaohsiung Medical University Hospital, 100 Shih-Chuan 1st Road, Kaohsiung 807, Taiwan. Tel: +886-7-3121101 ext. 6694; Fax: +886-7-3221033; E-mail: shpihucom.tw Or Bo-Ying Bao, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5126; Fax: +886-4-22031075; E-mail: baocmu.edu.tw.
Corresponding authors: Shu-Pin Huang, Department of Urology, Kaohsiung Medical University Hospital, 100 Shih-Chuan 1st Road, Kaohsiung 807, Taiwan. Tel: +886-7-3121101 ext. 6694; Fax: +886-7-3221033; E-mail: shpihucom.tw Or Bo-Ying Bao, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5126; Fax: +886-4-22031075; E-mail: baocmu.edu.tw.

 Global reach, higher impact
Global reach, higher impact