3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(10):790-796. doi:10.7150/ijms.12609 This issue Cite
Research Paper
Single Nucleotide Polymorphisms in IL-10, IL-12p40, and IL-13 Genes and Susceptibility to Glioma
1. Medical research Unit, Microbiology Department, School of Medicine, University of AL-Nahrain, Baghdad Iraq,
2. Biotechnology Department, School of Science, University of Baghdad,
3. Neuroscience Hospital, Neurosurgery, Iraq, Baghdad,
4. Center for Translational Studies, Medical Services, VA Medical Center, Department of Veteran Affairs, Washington DC, USA,
5. Department of Pathology, Microbiology and Immunology, School of Medicine, University of South Carolina, SC, USA
Received 2015-5-5; Accepted 2015-9-2; Published 2015-9-19
Abstract
Glioma is one of the most aggressive and most common tumors of the central nervous system (CNS) in humans. The exact causes of glioma are not well known, but evidence suggests the involvement of genetic factors in addition to environmental risk factors. The present study aimed to determine whether polymorphisms in IL-10-1082A/G, IL-12p40 1188C/A, and IL-13+2044G/A (rs20541) are associated with the incidence of glioma in Iraqi patients. Ninety-six patients with different grades of glioma and 40 apparently healthy individuals were recruited. A blood sample and genomic DNA were collected from all subjects. The amplification refractory mutation system and sequence-specific primer polymerase chain reaction (PCR) were used for genotyping of IL-10-1082A/G and IL-12p40 1188C/A, respectively; whereas, the IL-13+2044G/A was detected by DNA sequencing after amplification of the genes by PCR.
All SNPs were within Hardy-Weinberg equilibrium and each appeared in three genotypes in patients and controls. In IL-10-1082A/G, these genotypes frequencies were AA (75%), AG (22.93%) and GG (2.07%) in patients as compared to similar frequencies (62.5%), (27.5%) and (10%) respectively, in controls. The variant IL-12p40 1188C/A genotype was AA (72.92%), AC (23.96%), and CC (3.13%%) in patients as compared to 65%, 30%, and 5%, respectively, in controls. The frequencies of IL-13+2044G/A genotypes (GG, GA, and AA) were 89.58%, 9.37%, and 1.04% among patients versus 47.5%, 32.5% and 20%, respectively, among controls. These results suggest a protective role of mutant alleles G and A in IL-10-1082A/G and IL-13+2044G/A against gliomas. Further studies with more rigorous parameter designs will be needed to confirm the current findings.
Keywords: glioma, single nucleotide polymorphism (SNPs), IL-10-1082A/G, IL-12p40 1188C/A, IL-13+2044G/A
Introduction
Tumors of the central nervous system (CNS) represent about 2% of all cancers. Gliomas account for ≥70% of all brain tumors [1]. In Iraq, as published in the cancer registry, CNS tumors are the fifth most common tumor in adults and the second most common in children. The exact etiology of glioma remains unclear [2]. The most important established environmental risk factor for gliomas and meningiomas is ionizing radiation exposure to the head and neck [3]. Inherited syndromes such as neurofibromatosis 1 and 2, tuberous sclerosis, Li-Fraumeni syndrome, and Turcot syndrome can also increase risk of both malignant and benign brain tumors, as well as for other cancers [4]. Several studies have examined the effects of single nucleotide polymorphisms (SNPs) in inflammation-associated genes on the risk of developing glioma, focusing on their potential role in survival. Certain SNPs in cytokine genes affect the expression of corresponding cytokines and therefore may interfere with individuals' susceptibility to different type of malignancies [5].
Interleukin (IL)-10 is an important anti-inflammatory cytokine secreted by various cells of the immune system [6]. IL-10 inhibits the production of pro-inflammatory cytokines such as IL-1, IL-6, IL-12, IL-18 and TNF-α, as well as co-stimulatory molecules on antigen-presenting cells (APCs) [7]. High serum levels of IL-10 have suppressive roles in tumor-specific immunity and could reduce tumor growth [7-9]. Three single SNPs have been confirmed in the promoter region of IL-10; the most important is -1082 A>G (rs1800869). Respectively, alleles A and G of this SNP seem to be associated with low and high levels of IL-10 production [10]. Several recent studies have shown that this SNP increases the susceptibility to digestive cancer, schizophrenia, gastric cancer, type-2 diabetes mellitus, and thyroid cancer [11-15].
Interleukin-12 (IL-12) is a disulphide-linked heterodimer composed of a light chain of 35kDa (IL-12A) and a heavy chain of 40kDa, which are encoded by separated genes [16]. The anti-tumor and anti-metastatic activities of IL-12 have been extensively examined in a variety of murine tumor models and found to delay, reduce and, in some cases, prohibit tumor development [17]. It has been shown that IL-12 induces the proliferation of natural killer (NK) cells and T cells, as well as the production of IFN-γ and the activation of cytotoxic T lymphocytes (CTLs) through the STAT4 signaling pathway [18, 19]. The gene encoding IL-12 is TaqI (A/C) in the 3'-untranslated region (3'UTR) at position 1188, which has been found to be associated with several tumors such as nasopharyngeal carcinoma [20] and breast cancer [21]. This SNP may affect individuals' susceptibility to glioma.
IL-13 is a Th-2-derived proinflammatory cytokine known to induce apoptosis in many cell types, including glioblastoma multiforme (GBM) [22]. The IL-13 gene has many SNPs, which mainly associate with allergy and atopic diseases, as well as effector functions that induce airway hyper-responsiveness, allergic inflammation, immunoglobulin E production, and tumor cell growth [23]. Further, high systemic levels of IL-13 are associated with increases in the occurrence in different cancers, including colorectal and ovarian cancer [24]. This association is thought to enhance the extracellular-signal-regulated kinase (ERK) and increase transcription factor Ap-1 and matrix metalloproteinase (MMP) activities. Hsi et al. [25] found a reduction in glioblastoma multiforme growth in mice after knockdown of IL-13 Rα2 by siRNA, which suggests that IL-13 is a risk factor for this malignancy. Recently, the SNPs for GBM on the polymorphism in exon 4(+2044G/A) have been studied in detail [26]. The impact of IL13+2044GA on the functional properties of IL-13 shows that this polymorphism is more active than normal IL-13 in inducing STAT6 phosphorylation and CD23 expression in monocytes and hydrocortisone-dependent IgE switching in B cells [27]. Thus, decreased neutralization with carriers of IL13+ 2044A eventually contributes to increased allergic inflammation as a consequence of increased IL-13-mediated Th2 effector function.
Based on current evidence, glioma is a major concern in Iraq. Very little information is available to demonstrate an association between polymorphisms in IL-10-1082A/G, IL-12p40 1188C/A, and IL-13+2044G/A (rs20541) and glioma development and progression. Studying these SNPs may allow us to move forward in developing possible diagnostic and interventional strategies. Accordingly, the present study was designed to investigate the association of SNPs in IL-10, IL-12p40 and IL-13 with the incidence of glioma in Iraqi patients.
Materials and Methods
Study Population
This case-control study involved 96 glioma patients (44 male and 52 female, mean age ± 39.18 years) who attended Neuroscience Hospital between January 2013 and March 2014 and 40 (15 male and 25 female, mean age ±37.06 years) apparently healthy individuals. Selection of patients was done with the assistance of surgeons in the hospital. All patients had glioma brain tumors. Data were collected through direct interviews with patients and examination of hospital records and previous medical reports. Patient's claims were followed as an alternative source of information when their previous medical reports were not available. The data collected included subjects' age, gender, smoking habits (never, ex-smoker and current), family history of brain cancer, body mass index, and area of residence. Patients were excluded from the study if they had undergone prior treatment with radiotherapy or chemotherapy or had neuropathology that had not been confirmed as glioma.
Blood Samples
Two milliliters (ml) of blood were taken from controls and patients before the initiation of chemotherapy or radiation therapy.
DNA Extraction and Genotyping
DNA was extracted from whole blood samples using a kit (g SYNCTM DNA Mini Kit Whole Blood Protocol, Geneaid, Korea) according to the manufacturer's instructions. An amplification- refractory mutation system PCR (ARMS-PCR) was used for amplification and genotyping of IL-10 (-1082 G/A). Allele A was amplified with forward sense primer A (5'-AACACTAAGGCTTCTTTGGGTA-3'); allele G was amplified with the primer G (5'-AACACTACTAAGGCTTCTTTGGGTG-3'). The common antisense primer (5'-GTAAGCTTCTGTCGCTGGTGAGTC-3') was added in both reactions, which amplify allele- specific sequences of 161 bp of the promoter region of IL-10. The cycling conditions involved an initial denaturation at 95ºC for 5 min, followed by 35 cycles at 95ºC for 30 sec, 63ºC for 30 sec, and 72ºC for 30 sec. The final extension step was at 72ºC for 5 min.
Amplification and genotyping of IL-12p40 gene was done using two tubes of sequence- specific primer-PCR (SSP-PCR) with sequence-specific reverse primers 5'-GTTTCAATGAGCATTTAGCATCT-3' and 5'-GTTTCAATGAGCATTTAGCATCTCG-3' in combination with the consensus forward primer 5'-ATCTTGGAGCGAATGGGCAT-3'. Thermo-cycling was done at 95ºC for 2 min, then 35 cycles at 95ºC for 20 sec, 60ºC for 20 sec, and 72ºC for 30 sec, followed by a 5-min final extension at 72ºC. The expected product size was 780 bp. The amplification products were separated by electrophoresis on a 1.5% agarose gel stained with ethidium bromide. The gel was visualized under a UV transluminator with a 100-base-pair ladder. The primer pairs used for amplification of IL-13 were gene-forward primer 5'-CTTCCGTGAGGACTGAATGAACGGTC-3' and reverse primer 5'-GCAAATAATGATGCTTTCGAAGTTTCAGTGGA-3'. Cycling conditions were 94ºC for 2 min, followed by 30 cycles at 94ºC for 30 sec, 65ºC for 30 sec, 72ºC for 45 sec, and a final extension step of 72ºC for 10 min. This amplification resulted in 236 bp of PCR product.
PCR Product Purification
For IL-12p40 PCR product, purification was done using a gSYNCTM PCR Product Purification Mini Kit (Geneaid/Korea) according to the manufacturer's instructions.
Measurement of DNA Concentration
The DNA concentration of purified PCR products was measured using Nanodrop/UVS-99 (ACTGene; Piscataway, NJ USA). All products had a concentration of more than 100 ng/mL and sequenced DNA.
DNA Sequencing
Polymerase chain reaction products of the IL-13 gene were sent to Bioneer Company (Korea) for DNA sequencing. The obtained sequences, aligned using ClustalW software (Accession Number NC_000005-10) with a normal sequence from GenBank (National Center of Biotechnology Information), were examined for the presence of polymorphisms.
Statistical Analysis
The statistical package for social sciences, version 16.0 (SPSS, Chicago) was used for statistical analysis. Chi square test was used to test the deviation from Hardy-Weinberg Equilibrium (HWE) of SNPs by comparing the observed and expected frequencies. The association of different genotypes with the risk of glioma was estimated by calculation of odds ratios (OR) with 95% confidence intervals (CI) using logistic regression. Statistical significance was set at p <0.05.
Results
Gliomas' Risk Factors
The characteristics of glioma patients as compared to normal healthy controls are shown in Table 1. Among the studied risk factors, residence showed a significant association with the incidence of glioma, with 60.42% of patients and 40% of controls coming from urban areas (OR= 0.437, 95% CI= 0.206-0.928). Thus, environmental factors may increase individuals' susceptibility to glioma. No other risk factors had any association with glioma.
Association of risk factors with the incidence of glioma
| Risk Factors | Cases N=96 | Control N=40 | P-value | OR (95%CI) |
|---|---|---|---|---|
| Mean age ± SD (years) | 39.18±8.74 | 37.06±9.11 | 0.229 | 1.038 (0.977- 1.104) |
| Gender | 0.372 | |||
| Male | 44 (45.83%) | 15 (37.5%) | 1.0 | |
| Female | 52 (54.17%) | 25 (62.5%) | 0.709 (0.333-1.510) | |
| Family history | 0.396 | |||
| No | 84 (87.5%) | 37 (92.5%) | 1.0 | |
| Yes | 12 (12.5%) | 3(7.5%) | 1.762 (0.469-6.615) | |
| Mean BMI ±SD | 26.44±4.27 | 24.71±5.02 | 0.077 | 1.108 (0.989- 1.243) |
| Smoking | ||||
| Never | 63 (65.63%) | 18 (45%) | 0.123 | 1.0 |
| Ex-smoker | 18 (18.75%) | 13 (32.5%) | 0.108 | 3.342 (0.766-14.57) |
| Current | 15(15.62%) | 9 (22.5%) | 0.886 | 1.116(0.247-5.048) |
| Residency | 0.029 | |||
| Urban | 58 (60.42%) | 16 (40%) | 1.0 | |
| Rural | 38 (39.58%) | 24 (60%) | 0.437 (0.206- 0.928) |
(BMI, body mass index; CI, confidence interval; N, number; OR, odds ratio; SD: standard deviation)
Association of IL-10 (-1082A/G) with Gliomas
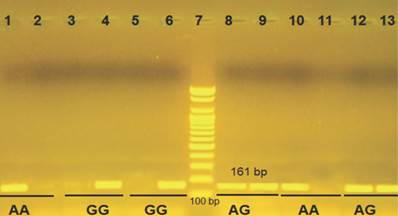
Genotyping of IL-10 -1082 A/G revealed three genotypes: AA, AG, and GG, which, respectively, had frequencies of 75%, 22.93% and 2.07% among patients and 62.5%, 27.5% and 10% respectively among controls. Despite these variations in percentages, the differences were not significant. However, at the allele level, a chi-square test demonstrated that the mutant allele (G) had a significant protective effect against glioma, with 13.54% of patients and 23.75% of controls having this allele (OR = 0.503, 95% CI = 0.260-0.973) (Fig. 1, Table 2).
Association of IL-12p40 1188A/C with Glioma
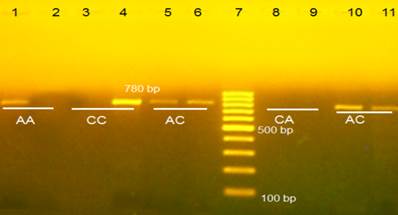
SNP IL-12p40 1188A/C also had three genotypes: AA, AC, and CC, which, respectively, accounted for 72.92%, 23.96%, and 3.13% of alleles among patients and 65%, 30% and 5% among controls (Fig. 2, Table 2). The mutant allele (allele C) frequency was 15.1% among patients and 20% among controls. Neither genotypes nor allele frequencies differed significantly in patients as compared to control subjects. Therefore, the variant IL-12p40 1188A/C may be not considered a risk factor for glioma.
Association of IL-13 +2044G/A with Glioma
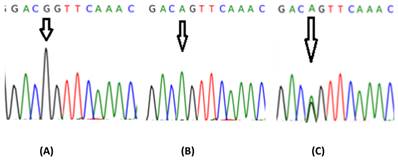
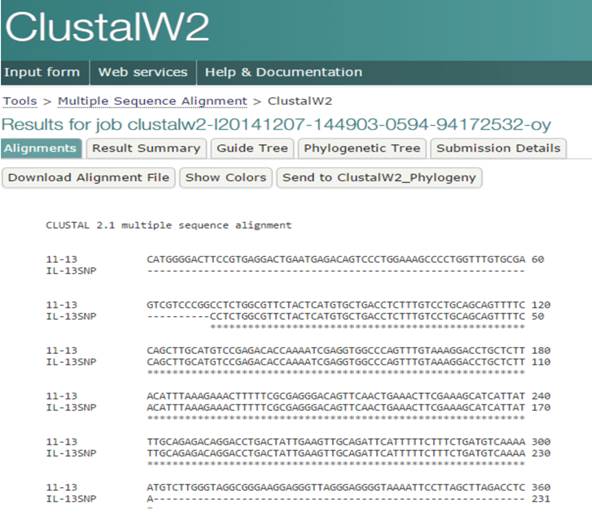
IL-13 +2044G/A showed a highly significant association with glioma at both genotype and allele levels. Specifically, 9.37% of glioma patients had the GA genotype as compared to 32.5% of controls (OR = 0.028, 95% CI = 0.003-0.234), while only 1.04% of patients had the AA genotype as compared to 20% of control subjects (OR = 0.181, 95% CI = 0.019-1.706) (Fig. 3, Table 2). Analysis of allele frequency confirmed this result, with allele A occurring in 5.73% of patients and 36.25% of controls (OR = 0.107, 95% CI = 0.050-0.229). Using ClustalW software for multiple sequence alignment showed that the sequence region of the IL-13 gene was identical to that of NCBI (Accession Number NC_000005-10) except in samples having the G nucleotide instead of A at the position 132660272(5q31) on chromosome 5 (Fig. 4).
Gel electrophoresis visualized under UV light of IL-10 (-1082 G/A) using ARMS-PCR. Lanes 1-6 control; lane 7, 100 bp DNA marker; lanes 8-13 from glioma patients. In this we used two tubes for each sample and each tube contain one allele. Thus each two wells in the agarose refer to one sample.
Gel electrophoresis visualized under UV light of IL-12p401188A/C after SSP-PCR. Lanes 1-6 control; lane 7, 100 bp DNA marker; lanes 8-11 from glioma patients. In this we used two tubes for each sample and each tube contain one allele. Thus each two wells in the agarose refer to one sample.
DNA sequencing for part of the fourth exon of IL-13. The arrow indicates the position of the SNP IL-13+2044.A: represents the genotype GG; B: AA and C: AG.
Distribution of genotypes and allele frequencies of IL-10 (-1082 A/G), IL-12p40 1188A/C, and IL-13+2044G/A in glioma patients and control subjects.
| Variables | Cases (N=96) | Control (N=40) | P-value | OR (95%CI) |
|---|---|---|---|---|
| IL-10 (-1082 AlG) | ||||
| AA | 72 (75%) | 25 (62.5%) | 0.126 | 1.0 |
| AG | 22 (22.93%) | 11 (27.5%) | 0.051 | 0.174 (0.030-1.006) |
| GG | 2 (2.07%) | 4 (10%) | 0.141 | 0.250 (0.039-1.582) |
| Alleles | ||||
| A | 166 (86.46%) | 61 (76.25%) | 1.0 | |
| G | 26 (13.54%) | 22 (23.75%) | 0.039 | 0.503 (0.260-0.973) |
| IL-12p40 1188A/C) | ||||
| AA | 70(72.92%) | 26 (65%) | 0.633 | 1.0 |
| AC | 23 (23.96%) | 12 (30%) | 0.534 | 0.557 (0.088-3.525) |
| CC | 3 (3.13%) | 2 (5%) | 0.802 | 0.783 (0.115-5.341) |
| Alleles | ||||
| A | 163 (84.9%) | 64 (80%) | 1.0 | |
| C | 29 (15.1%) | 16(20%) | 0.322 | 0.712(0.362-1.398) |
| IL-13 +2044G/A | ||||
| GG | 86(89.58%) | 19 (47.5%) | 0.001 | 1.0 |
| GA | 9 (9.37%) | 13 (32.5%) | 0.001 | 0.028(0.003-0.234) |
| AA | 1 (1.04%) | 8 (20%) | 0.135 | 0.181(0.019-1.706) |
| Allele | ||||
| G | 181 (94.27%) | 51 (63.75%) | 1.0 | |
| A | 11 (5.73%) | 29 (36.25%) | 0.001 | 0.107(0.050-0.229) |
Multiple sequence alignment of IL-13+2044 A/G. The obtained sequence was compared with that of National Center of Biotechnology Information (NCBI).
Discussion
Molecular studies suggest that genetic susceptibility may be pivotal in modifying the risk of glioma. Previous studies clearly indicate a candidate gene that encodes for cytokines that regulate immune responses. Genes for DNA repair proteins have also been extensively studied [28]. Our study showed significant protective associations against glioma between both allele A of the variant IL-13+2044G/A and allele G of the variant IL-10 (-1082A/G). IL-12p40 1188A/C did not show any association with glioma.
IL-10 is an important cytokine that favors Th2 activity and the humeral arm of adaptive the immune response. It has been shown that the IL-10-1082GG genotype causes an increase in IL-10 production [29]. Our results indicate that allele G has a protective role against glioma. In the present study, our result with IL-10 (-1082A/G) corresponds with that from other studies regarding the association of this variant with other cancers. It has been shown that the IL-10 1082 AA genotype is associated with significantly increased risk of lymph-node involvement in Chinese women with breast cancer [30]. Increased rates of gastric cancer have also been shown in Chinese people who carry this variant [11]. In contrast, however, a previous study regarding the association of SNP with various cancers did not find any effect of IL-10 (-1082A/G) on susceptibility to cervical cancer among Tunisian women [31]. It is possible that this discrepancy is be related to allergy and IgE production by these patients.
Recent studies from Brazil and Saudi Arabia have shown an association between allele G (either heterozygous or homozygous) and increased serum levels of IgE [32, 33]. This finding is corroborated by the results of a meta-analysis involving 2,215 asthma patients and 2,170 controls, which confirmed the association of the G allele with asthma [34]. Systemic administration of IL-10 induced an effective, specific, and prolonged immune response against established tumors in mice [35], indicating the immune-regulatory rather than immune-suppressive effect of this cytokine. In addition, IL-10 was shown to have an anti-inflammatory effect by suppressing monocyte or macrophage function. Thus, it is one of the main predisposing factors for cancer of chronic inflammation [36].
Un-translated regions of genes are involved in the posttranscriptional regulation of gene expression by the modulation of mRNA stability, nucleo‐cytoplasm transport, subcellular localization, and translation efficiency. Thus, these un-translated regions consent the fine control of protein product [37]. SNPs in these regions may therefore affect the quantity and quality of the protein encoded by the gene involving these regions. The SNP in IL-12p40 1188A/C may affect IL-12 expression because of its location in the 3'UTR of IL-12 gene. A contradictory view on correlation between this SNP and increased IL-12 secretion [38, 39] has been shown in the past. In the present study, neither mutant genotypes (hetero or homozygous) nor mutant allele of the variant IL-12p40 1188A/C appeared to be associated with gliomas. This result is in accordance with a meta-analysis study [40]. Although these authors found significant association of the variant with overall cancer, the stratification by cancer type revealed that the variant represents risk factor for cervical and nasopharyngeal cancer only. Taken together, increase in IL-12 production seems to have no effect on the susceptibility to gliomas may be due to the presence of other factors that can interfere with this overproduction.
The murine IL-13 receptor α 1 and 2 has been cloned and characterized in detail by [41]. The result from this study clearly suggests that IL-13Rα2 and -α1 are functionally not equal and play a distinct role for each polypeptide in IL-13R complex formation and ultimately modify IL-13 signal transduction. It has been shown that substitution of arginine with neutral glutamine renders IL-13 binds less tightly to the decoy receptor and escapes the dampening mechanisms that normally restrain the activity of wild type IL-13 in-vivo [27]. This evasion increases the availability of IL-13, which can binds with IL-13Rα2 and IL-4Rα1/IL-13Rα1 heterodimer (the other receptor for IL-13) on APCs and other body cells. Further, it has been shown in same study that IL-13 R130Q is more active than wild type IL-13 in inducing STAT6 phosphorylation and in up-regulation of CD23 in monocytes, which implies an increase in IgE class switching [27]. In correspondence with this many previous study confirms the association of this SNP with high serum levels of IgE [42], asthma [43], atopy and atopic dermatitis [44] but not with glioblastoma [45]. To this end highly protective role for the variant IL-13+2044 G/A against glioma was recorded in the present study. Similarly, anti-proliferative effect of IL-13 on human solid tumors that inhibit their clonal growth and DNA synthesis is shown [46]. In contrast to this most previous studies involving this variants recorded null result for its association with different cancers such as head and neck carcinoma [47] and lung cancer [48]. Nevertheless a recent meta-analysis study suggests the possibility of IL-13+rs1800925 genotype as a risk factor for gliomas and IL-13+rs20541 contributes towards cancer [49]. Further, another recent study on Iranian patient shows that SNPs in IL-13 linked with multiple sclerosis [50].
The inverse correlation between different kinds of allergic conditions and the incidence of gliomas was well established in many previous works. For example, high levels of IgE were found among control than glioma patients [51], and a strong relationship between these levels and survival rate of glioma patients [52]. However, the underlined relationship in this protection role and whether IgE itself has anti-tumor activity on glioma cells or on other cells adjacent to the tumor is not clear. Nevertheless, a recent study reported that stimulation of CD23 expression on astrocytes (through binding with IgE) could induce production of nitric oxide and IL-1β to facilitate cells death by apoptosis [53].
To our knowledge, this is the first study in Iraq, which investigated the association of IL-13, IL-10 and IL-12p40 polymorphism and glioma. The results from the present study suggest that SNPs in IL-13 and IL-10 have a protective role against glioma through induction of an allergic status. However, more detailed studies in relation to allergic status are required for this conclusion and involving more SNPs in different cytokine genes are warranted for prudent conclusion.
Acknowledgements
We thank Dr. Haidar/Neuropathology in the Neuroscience Hospital for help in collection of patient samples and Dr. Qasim Sharhan for critical comments and technical assistance. We are grateful to Dr. Haider Sabah for critical review of this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492-507
2. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathologica. 2005;109(1):93-108
3. Hocking B. Occupational exposure to ionizing and non-ionizing radiation and risk of glioma. Occupational Medicine. 2008;58(2):148-149 author reply 149
4. Malmer B, Henriksson R, Gronberg H. Familial brain tumours-genetics or environment? A nationwide cohort study of cancer risk in spouses and first-degree relatives of brain tumour patients. Int J Cancer. 2003;106(2):260-263
5. Amirian E, Liu Y, Scheurer ME, El-Zein R, Gilbert MR, Bondy ML. Genetic variants in inflammation pathway genes and asthma in glioma susceptibility. Neuro Oncol. 2010;12(5):444-452
6. Cyktor JC, Turner J. Interleukin-10 and immunity against prokaryotic and eukaryotic intracellular pathogens. Infection and Immunity. 2011;79(8):2964-2973
7. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annual Review of Immunology. 2001;19:683-765
8. Tanikawa T, Wilke CM, Kryczek I, Chen GY, Kao J, Nunez G, Zou W. Interleukin-10 ablation promotes tumor development, growth, and metastasis. Cancer Res. 2012;72(2):420-429
9. Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170(6):2081-2095
10. Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. European Journal of Immunogenetics: Official Journal of the British Society for Histocompatibility and Immunogenetics. 1997;24(1):1-8
11. Li C, Tong W, Liu B, Zhang A, Li F. The -1082A>G polymorphism in promoter region of interleukin-10 and risk of digestive cancer: a meta-analysis. Scientific Reports. 2014;4:5335
12. Gao L, Li Z, Chang S, Wang J. Association of interleukin-10 polymorphisms with schizophrenia: a meta-analysis. PLoS One. 2014;9(3):e90407
13. Kuo WH, Huang CY, Fu CK, Hsieh YH, Liao CH, Hsu CM, Huang YK, Tsai CW, Chang WS, Bau DT. Effects of interleukin-10 polymorphisms and smoking on the risk of gastric cancer in Taiwan. In vivo. 2014;28(5):967-971
14. Bai H, Jing D, Guo A, Yin S. Association between interleukin 10 gene polymorphisms and risk of type 2 diabetes mellitus in a Chinese population. The Journal of International Medical Research. 2014;42(3):702-710
15. Cil E, Kumral A, Kanmaz-Ozer M, Vural P, Dogru-Abbasoglu S, Altuntas Y, Uysal M. Interleukin-10-1082 gene polymorphism is associated with papillary thyroid cancer. Molecular Biology Reports. 2014;41(5):3091-3097
16. Huang D, Cancilla MR, Morahan G. Complete primary structure, chromosomal localisation, and definition of polymorphisms of the gene encoding the human interleukin-12 p40 subunit. Genes and Immunity. 2000;1(8):515-520
17. Xu M, Mizoguchi I, Morishima N, Chiba Y, Mizuguchi J, Yoshimoto T. Regulation of antitumor immune responses by the IL-12 family cytokines, IL-12, IL-23, and IL-27. Clinical & Developmental Immunology 2010. 2010
18. Trinchieri G. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annual Review of Immunology. 1995;13:251-276
19. Gately MK, Renzetti LM, Magram J, Stern AS, Adorini L, Gubler U, Presky DH. The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annual Review of Immunology. 1998;16:495-521
20. Ben Chaaben A, Busson M, Douik H, Boukouaci W, Mamoghli T, Chaouch L, Harzallah L, Dorra S, Fortier C, Ghanem A. et al. Association of IL-12p40 +1188 A/C polymorphism with nasopharyngeal cancer risk and tumor extension. Tissue Antigens. 2011;78(2):148-151
21. Kaarvatn MH, Vrbanec J, Kulic A, Knezevic J, Petricevic B, Balen S, Vrbanec D, Dembic Z. Single nucleotide polymorphism in the interleukin 12B gene is associated with risk for breast cancer development. Scandinavian Journal of Immunology. 2012;76(3):329-335
22. Nishimura Y, Nitto T, Inoue T, Node K. STAT6 mediates apoptosis of human coronary arterial endothelial cells by interleukin-13. Hypertens Res. 2008;31(3):535-541
23. Zhou R, Qian S, Gu X, Chen Z, Xiang J. Interleukin-13 and its receptors in colorectal cancer (Review). Biomedical Reports. 2013;1(5):687-690
24. Fujisawa T, Joshi BH, Puri RK. IL-13 regulates cancer invasion and metastasis through IL-13Ralpha2 via ERK/AP-1 pathway in mouse model of human ovarian cancer. Int J Cancer. 2012;131(2):344-356
25. Hsi LC, Kundu S, Palomo J, Xu B, Ficco R, Vogelbaum MA, Cathcart MK. Silencing IL-13Ralpha2 promotes glioblastoma cell death via endogenous signaling. Molecular Cancer Therapeutics. 2011;10(7):1149-1160
26. Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, Weiland SK, Erickson RP, von Mutius E, Martinez FD. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105(3):506-513
27. Vladich FD, Brazille SM, Stern D, Peck ML, Ghittoni R, Vercelli D. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J Clin Invest. 2005;115(3):747-754
28. Jiang L, Fang X, Bao Y, Zhou JY, Shen XY, Ding MH, Chen Y, Hu GH, Lu YC. Association between the XRCC1 polymorphisms and glioma risk: a meta-analysis of case-control studies. PLoS One. 2013;8(1):e55597
29. Schaaf BM, Boehmke F, Esnaashari H, Seitzer U, Kothe H, Maass M, Zabel P, Dalhoff K. Pneumococcal septic shock is associated with the interleukin-10-1082 gene promoter polymorphism. American Journal of Respiratory and Critical Care Medicine. 2003;168(4):476-480
30. Kong F, Liu J, Liu Y, Song B, Wang H, Liu W. Association of interleukin-10 gene polymorphisms with breast cancer in a Chinese population. Journal of Experimental & Clinical Cancer Research: CR. 2010;29:72
31. Zidi S, Verdi H, Yilmaz-Yalcin Y, Yazici A, Gazouani E, Mezlini A, Atac FB, Yacoubi-Loueslati B. Involvement of Toll-like receptors in cervical cancer susceptibility among Tunisian women. Bulletin du Cancer. 2014;101(10):31-35
32. Jacob CM, Pastorino AC, Okay TS, Castro AP, Gushken AK, Watanabe LA, Frucchi VC, Oliveira LC. Interleukin 10 (IL10) and transforming growth factor beta1 (TGFbeta1) gene polymorphisms in persistent IgE-mediated cow's milk allergy. Clinics. 2013;68(7):1004-1009
33. Hussein YM, Alzahrani SS, Alharthi AA, Ghonaim MM, Alhazmi AS, Eed EM, Shalaby SM. Association of serum cytokines levels, interleukin 10 -1082G/A and interferon-gamma +874T/A polymorphisms with atopic asthma children from Saudi Arabia. Cell Immunol. 2014;289(1-2):21-26
34. Hyun MH, Lee CH, Kang MH, Park BK, Lee YH. Interleukin-10 promoter gene polymorphisms and susceptibility to asthma: a meta-analysis. PLoS One. 2013;8(1):e53758
35. Berman RM, Suzuki T, Tahara H, Robbins PD, Narula SK, Lotze MT. Systemic administration of cellular IL-10 induces an effective, specific, and long-lived immune response against established tumors in mice. J Immunol. 1996;157(1):231-238
36. Acuner-Ozbabacan ES, Engin BH, Guven-Maiorov E, Kuzu G, Muratcioglu S, Baspinar A, Chen Z, Van Waes C, Gursoy A, Keskin O. et al. The structural network of Interleukin-10 and its implications in inflammation and cancer. BMC genomics. 2014;15(Suppl 4):S2
37. A T, F S, G P, M B. Genetic alterations in medullary thyroid cancer: diagnostic and prognostic markers. Current Genomics. 2011;12(8):618-625
38. Seegers D, Zwiers A, Strober W, Pena AS, Bouma G. A TaqI polymorphism in the 3'UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes and Immunity. 2002;3(7):419-423
39. Hiromatsu Y, Fukutani T, Ichimura M, Mukai T, Kaku H, Miyake I, Yamada K. Interleukin-12B gene polymorphism does not confer susceptibility to Graves' ophthalmopathy in Japanese population. Endocrine Journal. 2006;53(6):753-759
40. Chen H, Cheng S, Wang J, Cao C, Bunjhoo H, Xiong W, Xu Y. Interleukin-12B rs3212227 polymorphism and cancer risk: a meta-analysis. Molecular Biology Reports. 2012;39(12):10235-10242
41. Donaldson DD, Whitters MJ, Fitz LJ, Neben TY, Finnerty H, Henderson SL, O'Hara RM Jr, Beier DR, Turner KJ, Wood CR. et al. The murine IL-13 receptor alpha 2: molecular cloning, characterization, and comparison with murine IL-13 receptor alpha 1. J Immunol. 1998;161(5):2317-2324
42. Heinzmann A, Jerkic SP, Ganter K, Kurz T, Blattmann S, Schuchmann L, Gerhold K, Berner R, Deichmann KA. Association study of the IL13 variant Arg110Gln in atopic diseases and juvenile idiopathic arthritis. J Allergy Clin Immunol. 2003;112(4):735-739
43. Chu Y, Hua L, Liu Q, Bao Y. A common variant associated with asthma, interleukin 13 R130Q, promotes the production of IgE. International Journal of Immunogenetics. 2012;39(4):308-313
44. He JQ, Chan-Yeung M, Becker AB, Dimich-Ward H, Ferguson AC, Manfreda J, Watson WT, Sandford AJ. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes and Immunity. 2003;4(5):385-389
45. Schwartzbaum JA, Ahlbom A, Lonn S, Malmer B, Wigertz A, Auvinen A, Brookes AJ, Collatz Christensen H, Henriksson R, Johansen C. et al. An international case-control study of interleukin-4Ralpha, interleukin-13, and cyclooxygenase-2 polymorphisms and glioblastoma risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16(11):2448-2454
46. Serve H, Oelmann E, Herweg A, Oberberg D, Serve S, Reufi B, Mucke C, Minty A, Thiel E, Berdel WE. Inhibition of proliferation and clonal growth of human breast cancer cells by interleukin 13. Cancer Res. 1996;56(15):3583-3588
47. Khademi B, Hashemi SB, Ghaderi A, Shahrestani A, Mohammadianpanah M. Interleukin-13 gene polymorphisms at -1055 C/T and +2044 G/A positions in patients with squamous cell carcinoma of head and neck. Brazilian Journal of Otorhinolaryngology. 2012;78(5):64-68
48. Sameni S, Ghayumi MA, Mortazavi G, Faghih Z, Kashef MA, Ghaderi A. Lack of association between interleukin-13 gene polymorphisms (-1055 C/T and +2044 G/A) in Iranian patients with lung cancer. Molecular Biology Reports. 2009;36(5):1001-1005
49. Su T, Mi Y, Zhang L, Wang S, Lu H, Shi L, Sun H, Wu X, Zhang W, Zuo L. et al. Association between IL13 gene polymorphisms and susceptibility to cancer: a meta-analysis. Gene. 2013;515(1):56-61
50. Seyfizadeh N, Kazemi T, Farhoudi M, Aliparasti MR, Sadeghi-Bazargani H, Almasi S, Babaloo Z. Association of IL-13 single nucleotide polymorphisms in Iranian patients to multiple sclerosis. Am J Clin Exp Immunol. 2014;3(3):124-129
51. Wiemels JL, Wiencke JK, Patoka J, Moghadassi M, Chew T, McMillan A, Miike R, Barger G, Wrensch M. Reduced immunoglobulin E and allergy among adults with glioma compared with controls. Cancer Res. 2004;64(22):8468-8473
52. Wrensch M, Wiencke JK, Wiemels J, Miike R, Patoka J, Moghadassi M, McMillan A, Kelsey KT, Aldape K, Lamborn KR. et al. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006;66(8):4531-4541
53. Dugas N, Lacroix C, Kilchherr E, Delfraissy JF, Tardieu M. Role of CD23 in astrocytes inflammatory reaction during HIV-1 related encephalitis. Cytokine. 2001;15(2):96-107
Author contact
![]() Corresponding author: Udai P. Singh, Ph.D., Department of Pathology, Microbiology and Immunology, University of South Carolina, Columbia, SC 29208. Phone: (803) 216-3423; Fax: (803) 216-3413; Email: udai.singhsc.edu
Corresponding author: Udai P. Singh, Ph.D., Department of Pathology, Microbiology and Immunology, University of South Carolina, Columbia, SC 29208. Phone: (803) 216-3423; Fax: (803) 216-3413; Email: udai.singhsc.edu

 Global reach, higher impact
Global reach, higher impact