3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(1):23-31. doi:10.7150/ijms.10019 This issue Cite
Research Paper
Genome-based Proteomic Analysis of Lignosus rhinocerotis (Cooke) Ryvarden Sclerotium
1. Department of Molecular Medicine, Faculty of Medicine, University of Malaya, 50603 Kuala Lumpur, Malaysia;
2. Ligno Biotech Sdn. Bhd., 43300 Balakong Jaya, Selangor, Malaysia.
Received 2014-7-1; Accepted 2014-10-13; Published 2015-1-1
Abstract
Lignosus rhinocerotis (Cooke) Ryvarden (Polyporales, Basidiomycota), also known as the tiger milk mushroom, has received much interest in recent years owing to its wide-range ethnobotanical uses and the recent success in its domestication. The sclerotium is the part with medicinal value. Using two-dimensional gel electrophoresis coupled with mass spectrometry analysis, a total of 16 non-redundant, major proteins were identified with high confidence level in L. rhinocerotis sclerotium based on its genome as custom mapping database. Some of these proteins, such as the putative lectins, immunomodulatory proteins, superoxide dismutase, and aegerolysin may have pharmaceutical potential; while others are involved in nutrient mobilization and the protective antioxidant mechanism in the sclerotium. The findings from this study provide a molecular basis for future research on potential pharmacologically active proteins of L. rhinocerotis.
Keywords: Lignosus rhinocerotis, proteomic analysis, LC-MS, MALDI-MS, proteins.
Introduction
Lignosus rhinocerotis (Cooke) Ryvarden (Polyporales, Basidiomycota) is a white-rot fungus that is characterized by having a centrally stipitate pilei arising from the underground tuber-like sclerotium. It is mainly distributed in China, Malaysia, Sri Lanka, the Philippines, Australia, and East Africa [1]; and more commonly known as tiger milk mushroom in Malaysia. In recent years, this mushroom has received much attention owing to its wide-range ethnobotanical uses as a folk medicine. This is also made possible due to the recent success in the domestication of this once very rare and expensive mushroom [2, 3]. This mushroom has been used by the local communities to treat numerous ailments including fever, whooping cough, asthma, cancer, food poisoning, wounds, chronic hepatitis, and gastric ulcers [4, 5].
On-going scientific research has further validated some of the traditional claims on L. rhinocerotis. Its petroleum ether, chloroform, methanol, and water sclerotial extracts displayed strong antimicrobial activity against selected human pathogens including gram-positive and gram-negative bacteria and fungi in disk diffusion test [6]. It has also been reported that the aqueous extract of L. rhinocerotis sclerotium enhanced neurite outgrowth in PC-12 Adh pheochromocytoma and Neuro-2a mouse neuroblastoma cell lines [7, 8]. Several authors also demonstrated the presence of antiproliferative activity in aqueous (hot and cold) or methanol pressurized liquid extracts, and hot water-soluble polysaccharides isolated from L. rhinocerotis sclerotium against human breast carcinoma (MCF7), lung carcinoma (A549) and colorectal cancer (HCT 116) cells, as well as various types of leukemic cells including acute promyelocytic leukemia cells (HL-60), chronic myelogenous leukemia cells (K562), and human acute monocytic leukemia cells (THP-1), through apoptosis and/or cell cycle arrest [9-11]. Wong et al. demonstrated that Polyporus rhinocerus (synonym to L. rhinocerotis) sclerotial polysaccharides exhibited immunomodulatory effects by activation of innate immune cells and T-helper cells in normal and athymic BALB/c mice [12]. The non-digestible carbohydrates extracted from P. rhinocerus was also shown to stimulate the growth of Bifidobacterium longum and Lactobacillus brevis, thus suggesting its potential application as novel prebiotics for gastrointestinal health [13]. Moreover, the mushroom sclerotial extract was shown to exhibit strong superoxide anion radical scavenging activity comparable to rutin [14]. A 180-day chronic toxicity study of L. rhinocerotis cultivar (termed TM02) sclerotial powder in Sprague Dawley rats indicated that the no-observed-adverse-effect level dose is higher than 1,000 mg/kg; thus establishing its safety for human consumption [15].
The sclerotium of the mushroom is the part with medicinal value. Substantial amount of L. rhinocerotis sclerotial proteins, especially in the cultivar strain, are believed to constitute a crucial part not only for its functionality as nutritional reserves but also with pharmaceutical potential [14, 16]. Mushrooms are known to consist of large number of pharmacologically active proteins and peptides. These include lectins, fungal immunomodulatory proteins (FIP), ribosome inactivating proteins (RIP), antimicrobial proteins, ribonucleases, and laccases; all with interesting pharmacological activities and may act as natural antitumor, antiviral, antimicrobial, antioxidative, and immunomodulatory agents [17]. It is believed that the sclerotium of L. rhinocerotis also contains some of these pharmacologically active proteins with biomedical potential. However, to date, a systematic profiling of L. rhinocerotis proteins is still lacking. Although Lau et al. have previously reported the surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) profiling of low molecular-mass protein/peptides (< 20 kDa) from L. rhinocerotis cultured by liquid fermentation, none of the proteins have been identified [18]. In this study, we report the two-dimensional gel electrophoresis (2DE) separation of the sclerotial proteins and identification of the main protein spots using liquid chromatography-mass spectrometry (LC-MS), taking advantage of the recently available L. rhinocerotis genome database [19]. A number of proteins including several pharmacologically active proteins were identified with high level of confidence based on the predicted open reading frames (ORFs). The proteome obtained will facilitate future work on characterization of the pharmacologically active proteins from the mushroom.
Materials and methods
Materials
Sclerotia of cultivated L. rhinocerotis (TM02) were obtained from Ligno Biotech Sdn. Bhd. (Selangor, Malaysia). The fungus was identified by the internal transcribed spacer regions of ribosomal RNA [3]. Chemicals and reagents of electrophoresis- and LC/MS-grade were purchased from Sigma-Aldrich (Missouri, USA) unless otherwise specified. Urea, thiourea, 3-[(3-cholamidopropyl)-dimethylammonio]-propane-sulfonate (CHAPS), dithiothreitol (DTT), IPG buffer, 2-D Quant Kit, and 2-D Clean-Up Kit were purchased from GE Healthcare Life Sciences (Uppsala County, Sweden). Water used was of Millipore quality.
Total protein extraction by Tris-buffered phenol
Protein extraction from the sclerotium was performed according to Horie et al. with minor modification [20]. Freeze-dried sclerotia were ground into powder and sieved through 0.2 mm prior to protein extraction by mixing with Tris-buffered phenol (TBP, pH 8.8) and extraction media [0.9 M sucrose, 0.1 M Tris, 10 mM ethylenediaminetetraacetic acid (EDTA), and 0.4 % 2-mercaptoethanol, pH 8.8] for 30 min at room temperature, followed by centrifugation at 10,000 × g for 30 min at 4 °C, where the top phenol phase was collected into a new microcentrifuge tube and the aqueous phase was back-extracted using the same amount of TBP and extraction media. The suspension was centrifuged at 20,000 × g for 20 min at 4 °C and the resulting top phenol phase was transferred into the first extraction. Five volumes of 0.1 M ammonium acetate in 100 % methanol were added to precipitate the phenol-soluble proteins followed by vortexing and overnight incubation at -20 °C.
Precipitated proteins were pelleted at 20,000 × g for 20 min at 4 °C and the resulting pellet was washed twice with 0.1 M ammonium acetate in 100 % methanol, 80 % ice-cold acetone, and once in 70 % ethanol by centrifugation at 20,000 × g for 20 min at 4 °C. After the final wash, supernatant was decanted and the protein pellet was dried at 37 °C for not more than 15 min followed by solubilization with lysis buffer [7 M urea, 2 M thiourea, 4 % CHAPS, 18 mM Tris-HCl (pH 8.0), 14 mM Trizma base, and two EDTA-free proteinase inhibitor cocktail tablets (Roche Diagnostics GmbH, Baden-Württemberg, Germany) in a final volume of 100 ml buffer, 0.2 % Triton X-100 (R), containing 50 mM DTT]. Protein lysates were centrifuged at 20,000 × g for 20 min at 4 °C and the resulting supernatant was stored in aliquots at -80 °C. Protein concentration was determined using 2-D Quant Kit according to manufacturer's standard procedure.
Two-dimensional gel electrophoresis
Total protein of 500 µg was precipitated using 2-D Clean-Up Kit according to manufacturer's procedure and the pellet was resuspended in 250 µl rehydration solution (7 M urea, 2 M thiourea, 4 % CHAPS, 40 mM DTT, 0.5 % IPG buffer, 0.002 % Orange G) for first dimension isoelectric focusing (IEF). Immobiline DryStrip gel (IPG strip) pH 3-10, 13 cm (GE Healthcare Life Sciences, Uppsala County, Sweden) was rehydrated overnight with the prepared sample followed by IEF at 20 °C and current 50 µA/strip on a Ettan IPGphor 3 Isoelectric Focusing Unit (GE Healthcare Life Sciences, Uppsala County, Sweden) according to manufacturer's guidelines. Two-step gel equilibration was performed immediately prior to the second-dimension run with SDS equilibration buffer solution [6 M urea, 75 mM Tris-HCl (pH 8.8), 29.3 % glycerol, 2 % SDS, 0.002 % Orange G) containing DTT (100 mg/10 ml) or iodoacetamide (IAA, 250 mg/10 ml) for 15 min each. Equilibrated IPG strip was then laid on 15 % polyacrylamide gel and the electrophoretic run was carried out at 15 mA/gel for the first 15 min and 30 mA/gel until the end of the run. At least three replicates were done.
Gel visualization and image analysis
Protein spots were visualized by Coomassie Blue R-250 staining according to Neuhoff et al. and the resulting gel image was digitized using ImageScanner III (LabScan 6.0, Swiss Institute of Bioinformatics) [21]. ImageMaster 2D Platinum 7.0 software version 7.02 (GE Healthcare Life Sciences, Uppsala County, Sweden) was used for spot detection (cut-off volume value ≥ 0.2), background subtraction, and relative quantification. Protein spot intensities were normalized based on the total detection volumes and each spot were expressed as a relative spot volume (% spot volume/total volume of all spot in the gel).
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)
Protein spots of interest from 2DE gel were manually excised using a clean razor blade and in-gel protein digestion was performed using Trypsin Gold (Promega, Massachusetts, USA) according to manufacturer's procedure. The extracted peptides were purified and concentrated using ZipTip® pipette tips (Millipore Corporation, Massachusetts, USA) following the manufacturer's instructions. Eluted peptides in 2.5 μl of 70 % acetonitrile (ACN)/0.1 % trifluoroacetic acid containing 10 mg/ml α-cyano-4-hydroxycinnamic acid were spotted directly onto MALDI plate for subsequent MALDI-TOF MS analysis by 4800 Plus MALDI TOF/TOF™ Analyzer (AB SCIEX, Massachusetts, USA). MS/MS scans were analyzed using Mascot Server (http://www.matrixscience.com) to search against the NCBInr protein database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/); choosing fungi as the taxonomic category. The following search parameters for sequence query were implemented: complete carbamidomethylation of cysteines and/or oxidation of methionines, unrestricted protein mass (monoisotopic mass values), peptide mass tolerance of ± 100 ppm, fragment mass tolerance of ± 0.2 Da, and maximum of one missed cleavage allowed. Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits.
Liquid chromatography-mass spectrometry (LC-MS)
Excised major protein spots for identification were de-stained with 200 µl of destaining buffer (100 mM ammonium bicarbonate/50 % ACN) at 37 °C prior to reduction and alkylation with 5 mM Tris(2-carboxyethyl)phosphine hydrochloride solution and 100 mM IAA solution, respectively. In-gel protein digestion was performed using Pierce™ Trypsin Protease (Thermo Scientific, Massachusetts, USA) according to manufacturer's procedure. Cleaned up peptide mixtures were further separated using Agilent 1200 HPLC-Chip/MS Interface, coupled with Agilent 6520 Accurate-Mass Q-TOF LC/MS (Agilent Technologies, California, USA).
Total of 1 μl sample in Solution A (0 .1 % formic acid in water) was injected onto the microfluidic nanospray chip containing a 160-nl enrichment column packed with C18 (300 Å) at 4 µl/min. Sequential peptides elution was accomplished over the pre-column in-line with a 75 µm x 150 mm analytical column at 0.3 µl/min in a linear gradient from Solution A to 95 % Solution B (90 % acetonitrile, 0.1 % formic acid in water) in 47 min including post-run of 8 min. For subsequent MS (rate: 8 spectra/s, time: 125 ms/spectrum) and MS/MS (rate: 4 spectra/s, time: 250 ms/spectrum) analyses, spectra were acquired in aMSMS mode with scan range from 110 to 3000 m/z and 50 to 3000 m/z, respectively. Capillary voltage was 1.9 kV with drying gas flow rate of 5.0 L/min at 325 °C.
Acquired data were searched against L. rhinocerotis genome database using Agilent Spectrum Mill MS Proteomics Workbench software packages (http://spectrummill.mit.edu/) and the following parameters and filters were implemented for protein and peptide identification: MH+ scan range from 600 to 4000 Da, complete carbamidomethylation of cysteines, protein score > 11, peptide score > 6, and % scored peak intensity > 60. Only results with “Distinct Peptide” identification of 2 or greater than 2 are considered significant. Relative protein content in terms of percentage in a protein spot was derived from the formula x/(∑x) × 100 % where x is the (Number of spectra × Mean peptide spectral intensity)/(Total number of spectra × Total mean peptide spectral intensity).
Data availability
For LC-MS analysis, the genome sequences of L. rhinocerotis cultivar TM02 were used for protein identification based on matches with the predicted ORFs and the ORFs homologs were searched in the NCBInr (Fungi) database. The Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AXZM00000000. The version used in this paper is version AXZM01000000 [19].
Results
Protein extraction and 2DE gel profile of L. rhinocerotis sclerotia
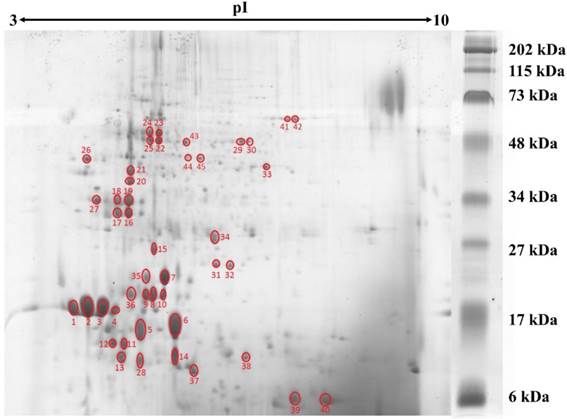
Protein concentration in L. rhinocerotis sclerotial extract was quantified by 2-D Quant Kit based on the specific binding of copper ions to the precipitated protein while leaving interfering contaminants in solution. Using phenolic extraction method adapted from Horie et al. [20], L. rhinocerotis sclerotial extract had a protein content of 2.48 ± 0.02 g/100 g dry weight. The proteins were resolved by 2DE using IEF with a linear pH 3-10 gradient prior to 15 % SDS-polyacrylamide gel electrophoresis. Fig. 1 shows a representative separation of the proteins by 2DE according to their molecular mass. A total of 110 protein spots were identified by ImageMaster 2D Platinum 7.0 with cut-off volume value of 0.2 (Smooth: 2; Saliency: 1; Min area: 5). The majority of the protein spots were concentrated in between 10 to 75 kDa with pI range from 4 to 6.
Protein identification by MALDI-MS
A total of 45 major, well-defined, well-separated, and reproducible protein spots were subjected to MALDI-MS analysis and the resulting MS/MS scans were searched against the NCBInr (Fungi) database using Mascot Server; but only eight of them were detected with significant protein scores of p less than 0.05. Protein identification data for these eight protein spots are shown in Table 1. Only five different putative proteins were identified including manganese superoxide dismutases (Mn-SOD), catalases (CAT), NAD-dependent formate dehydrogenase, enolase, and 70 kDa heat shock proteins.
2DE gel profile for the proteome of L. rhinocerotis sclerotial extract. The proteins (500 µg) were resolved by 2DE using IEF along a linear pH 3-10 gradient (13 cm) prior to 15 % SDS-polyacrylamide gel electrophoresis. Molecular weight markers are indicated on the right (30 μL/gel; Bio-Rad, California, USA). Protein spots were visualized by Coomassie Blue R-250 staining and gel image presented is representative from at least three triplicate analyses. Red circles indicate protein spots that are selected for peptide sequencing by mass spectrometry.
L. rhinocerotis sclerotial proteins identified by MALDI-MS.
| Spot | Spot volume (%) | MW (kDa) | pI | Score | Accession | Description | Matching peptide (#) | AA coverage (%) |
|---|---|---|---|---|---|---|---|---|
| 31 | 0.43 | 20 | 6.1 | 163 | gi|2500828 | Manganese superoxide dismutase | 5 | 44 |
| 32 | 0.41 | 20 | 6.3 | 268 | gi|1519016 | Manganese superoxide dismutase | 4 | 36 |
| 33 | 0.34 | 45 | 7.0 | 92 | gi|164564766 | NAD-dependent formate dehydrogenase | 4 | 13 |
| 41 | 0.22 | 70 | 7.3 | 93 | gi|30172926 | Peroxisomal catalase | 3 | 9 |
| 42 | 0.38 | 70 | 7.5 | 101 | gi|28558774 | Catalase | 4 | 10 |
| 43 | 0.30 | 59 | 5.6 | 84 | gi|169845435 | Enolase | 5 | 8 |
| 44 | 0.21 | 50 | 5.6 | 147 | gi|172713 | 70kDa heat shock protein | 6 | 16 |
| 45 | 0.26 | 50 | 5.9 | 461 | gi|172713 | 70kDa heat shock protein | 8 | 22 |
NCBInr (Fungi) database was employed for the identifications. The molecular weight and pI of each spot were estimated from the 2DE gel. Sequences of the matching peptides and functional classification of the identified proteins are available at Supplementary Material: Table S1. Abbreviations: MW, molecular weight; AA, amino acid.
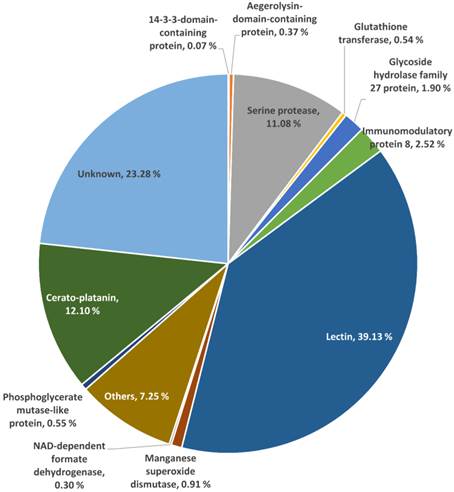
The proteome of L. rhinocerotis sclerotial extract. Overview percentage distributions of identified proteins based on the predicted open reading frames of L. rhinocerotis genome are shown. About 76.72 % of total spot volumes were subjected to LC-MS analysis. The three main identified protein families are lectins, cerato-platanin, and serine proteases. Identities of the remaining 23.28 % which were not analyzed are grouped as unknown.
Protein identification by LC-MS
A total of 40 selected protein spots which cover 76.72 % of total spot volume were subjected to LC-MS analysis. These 40 protein spots are from the same cohort examined by MALDI-MS, excluding the five spots that have already been identified by MALDI-MS as described earlier. The resulting data were searched against the predicted ORFs of L. rhinocerotis genome. Each spot consists of one major protein (> 50 % of total spot volume) and four to five other proteins of lower percentage (Supplementary Material: Table S2). Identification with the highest number of “Distinct Peptide” for each protein spot is tabulated in Table 2. Fig. 2 shows the overview percentage distribution of the identified proteins, depicted as a pie chart. Some of the identified proteins of interest are discussed and their complete coding sequences (by Gene ID) are available at Supplementary Material: Table S3.
Thirty percent of the identified L. rhinocerotis sclerotial proteins in Table 2 are involved in the following five functional categories: posttranslational modification, protein turnover, chaperones (15 %); cell wall/membrane/envelope biogenesis (5 %); inorganic ion transport and metabolism (5 %); signal transduction mechanisms (2.5 %); and energy production and conversion and coenzyme transport and metabolism (2.5 %).
Of the 45 spots, 16 are putative lectins from three isoforms encoded by GME270_g (184 amino acids), GME272_g (173 amino acids), and GME273_g (598 amino acids). They appear to be the major protein constituents of L. rhinocerotis sclerotium and account for up to 39.13 % of the total volume. The putative lectins of L. rhinocerotis are mostly concentrated in the lower left quadrant of the 2DE gel with high degree of post-translational modifications, especially for GME273_g isoforms. Serine proteases (spots 16, 18, 20, 21, and 27) are another group of proteins with relatively high abundance in the sclerotial extract and it accounts for 11.08 % of the total volume. The glycoside hydrolase family 27 (GH27) which was identified from spots 22, 23, 24, and 25 is a family of glycoside hydrolases which is involved in the hydrolysis of glycosidic bonds in complex sugars. Two Mn-SOD isoforms (spots 31 and 32) and a glutathione transferase (GST) which covers 0.54 % of the total volume was identified from spot 34 (25 kDa, pI 6.1) by LC-MS analysis. Mn-SOD, GST, and CAT (spots 41 and 42, identified from MALDI-MS) together form the antioxidants defense system against oxidative stress in the mushroom sclerotium. The highly conserved 14-3-3-domain-containing protein was identified from spot 17 (31 kDa, pI 4.5). A protein with amino acid sequence homolog to ling zhi-8, an immunomodulatory protein isolated from Ganoderma lucidum was identified from spot 37 (8 kDa, pI 5.8). This protein covers 2.52 % from the total volume of L. rhinocerotis sclerotial proteins and is encoded by GME10641_g (141 amino acids), with a Fve domain (a major fruiting body protein from Flammulina velutipes which possessed immunomodulatory activity) [22]. Two isoforms of phosphoglycerate mutase-like protein (59 kDa) in different phosphorylation states were identified from spot 29 and 30 with pI values of 6.5 and 6.6, respectively and two cerato-platanin (CP) isoforms which cover 12.10 % of total volume were identified from spot 6 and 14. As the molecular weights of these CP isoforms are different, it is possible that these proteins are glycosylated; however, more studies are needed to confirm these modifications. An aegerolysin-domain-containing protein was identified from spot 5 (11 kDa, pI 4.9). This putative protein covers 0.37 % of the total volume.
L. rhinocerotis sclerotial proteins identified by LC-MS.
| Spot | Spectra (#) | Distinct peptides (#) | MPSI | AA coverage (%) | Volume (%) | MW (Kda) | pI | Gene ID | Protein name | Functional category |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 | 4 | 9.91e+05 | 9 | 3.90 | 13 | 3.8 | GME273_g | Lectin | Unclassified |
| 2 | 134 | 9 | 2.12e+06 | 11 | 8.98 | 13 | 4.0 | GME273_g | Lectin | Unclassified |
| 3 | 137 | 8 | 1.29e+06 | 11 | 7.23 | 13 | 4.3 | GME273_g | Lectin | Unclassified |
| 4 | 59 | 6 | 5.87e+05 | 14 | 1.22 | 13 | 4.5 | GME273_g | Lectin | Unclassified |
| 5 | 8 | 8 | 1.45e+05 | 56 | 0.34 | 11 | 4.9 | GME7309_g | Aegerolysin-domain-containing protein | Unclassified |
| 6 | 14 | 3 | 1.29e+06 | 34 | 6.19 | 12 | 5.5 | GME3505_g | Cerato-platanin | Unclassified |
| 7 | 20 | 11 | 3.60e+06 | 76 | 4.39 | 18 | 5.3 | GME270_g | Lectin | Unclassified |
| 8 | 24 | 9 | 1.66e+06 | 20 | 2.18 | 15 | 5.1 | GME273_g | Lectin | Unclassified |
| 9 | 16 | 11 | 9.52e+05 | 56 | 1.21 | 15 | 5.0 | GME272_g | Lectin | Unclassified |
| 10 | 17 | 10 | 8.64e+05 | 56 | 1.08 | 15 | 5.3 | GME272_g | Lectin | Unclassified |
| 11 | 19 | 8 | 2.10e+06 | 72 | 1.56 | 10 | 4.5 | GME4537_g | TPA: conserved hypothetical protein | Unclassified |
| 12 | 7 | 5 | 2.95e+05 | 39 | 0.28 | 10 | 4.3 | GME4537_g | TPA: conserved hypothetical protein | Unclassified |
| 13 | 7 | 3 | 9.10e+05 | 8 | 1.62 | 9 | 4.5 | GME273_g | Lectin | Unclassified |
| 14 | 6 | 3 | 7.68e+05 | 34 | 3.49 | 9 | 5.5 | GME3505_g | Cerato-platanin | Unclassified |
| 15 | 23 | 8 | 2.02e+06 | 18 | 0.91 | 23 | 5.1 | GME273_g | Lectin | Unclassified |
| 16 | 13 | 7 | 1.33e+06 | 19 | 1.45 | 31 | 4.8 | GME4347_g | Serine protease | Posttranslational modification, protein turnover, chaperones |
| 17 | 9 | 9 | 1.05e+05 | 40 | 0.06 | 31 | 4.5 | GME1701_g | 14-3-3-domain-containing protein | Signal transduction mechanisms |
| 18 | 10 | 7 | 1.16e+06 | 19 | 1.35 | 35 | 4.5 | GME4347_g | Serine protease | Posttranslational modification, protein turnover, chaperones |
| 19 | 6 | 5 | 3.72e+05 | 17 | 0.19 | 35 | 4.8 | GME273_g | Lectin | Unclassified |
| 20 | 11 | 7 | 1.21e+06 | 19 | 1.05 | 42 | 4.8 | GME4347_g | Serine protease | Posttranslational modification, protein turnover, chaperones |
| 21 | 12 | 7 | 1.20e+06 | 19 | 1.20 | 45 | 4.8 | GME4347_g | Serine protease | Posttranslational modification, protein turnover, chaperones |
| 22 | 15 | 10 | 7.33e+05 | 48 | 0.48 | 59 | 5.1 | GME9376_g | Glycoside hydrolase family 27 protein | Unclassified |
| 23 | 13 | 8 | 1.40e+06 | 48 | 0.63 | 64 | 5.1 | GME9376_g | Glycoside hydrolase family 27 protein | Unclassified |
| 24 | 11 | 8 | 7.23e+05 | 48 | 0.39 | 64 | 5.0 | GME9376_g | Glycoside hydrolase family 27 protein | Unclassified |
| 25 | 9 | 7 | 4.42e+05 | 43 | 0.31 | 59 | 5.0 | GME9376_g | Glycoside hydrolase family 27 protein | Unclassified |
| 26 | 26 | 5 | 5.35e+05 | 13 | 0.54 | 50 | 4.0 | GME273_g | Lectin | Unclassified |
| 27 | 11 | 7 | 7.56e+05 | 18 | 0.55 | 35 | 4.1 | GME8711_g | Serine protease | Posttranslational modification, protein turnover, chaperones |
| 28 | 4 | 2 | 1.27e+05 | 6 | 1.01 | 8 | 4.9 | GME273_g | Lectin | Unclassified |
| 29 | 22 | 13 | 8.60e+05 | 30 | 0.32 | 59 | 6.5 | GME590_g | Phosphoglycerate mutase-like protein | Cell wall/membrane/envelope biogenesis |
| 30 | 18 | 12 | 5.31e+05 | 26 | 0.17 | 59 | 6.6 | GME590_g | Phosphoglycerate mutase-like protein | Cell wall/membrane/envelope biogenesis |
| 31 | 12 | 9 | 1.77e+006 | 37 | 0.38 | 20 | 6.1 | GME441_g | Manganese superoxide dismutase | Inorganic ion transport and metabolism |
| 32 | 16 | 10 | 1.10e+006 | 34 | 0.41 | 20 | 6.3 | GME441_g | Manganese superoxide dismutase | Inorganic ion transport and metabolism |
| 33 | 43 | 23 | 1.76e+006 | 63 | 0.29 | 45 | 7.0 | GME5414_g | NAD-dependent formate dehydrogenase | Energy production and conversion; Coenzyme transport and metabolism |
| 34 | 6 | 6 | 6.42e+004 | 37 | 0.52 | 25 | 6.1 | GME7546_g | Glutathione transferase | Posttranslational modification, protein turnover, chaperones |
| 35 | 20 | 9 | 4.06e+006 | 58 | 0.73 | 18 | 5.0 | GME270_g | Lectin | Unclassified |
| 36 | 13 | 9 | 1.02e+006 | 56 | 0.42 | 15 | 4.8 | GME272_g | Lectin | Unclassified |
| 37 | 31 | 13 | 2.88e+006 | 67 | 0.96 | 8 | 5.8 | GME10641_g | Immunomodulatory protein 8 | Unclassified |
| 38 | 8 | 5 | 8.65e+005 | 11 | 0.50 | 9 | 6.6 | GME273_g | Lectin | Unclassified |
| 39 | 7 | 4 | 9.40e+005 | 40 | 1.10 | 6 | 7.5 | GME1771_g | Hypothetical protein DICSQDRAFT_165309 | Unclassified |
| 40 | 43 | 7 | 2.52e+006 | 41 | 1.14 | 6 | 8.0 | GME1771_g | Hypothetical protein DICSQDRAFT_165309 | Unclassified |
L. rhinocerotis genome database was employed for the identifications. The molecular weight and pI of each spot were estimated from the 2DE gel. Coding sequences of some selected identified proteins (by Gene ID) are available at Supplementary Material: Table S3. Abbreviations: MPSI, mean peptide spectral intensity; AA, amino acid MW, molecular weight.
Discussion
The protein content of L. rhinocerotis sclerotial extract (2.48 ± 0.02 g/100 g dry weight) determined from this study was only 18 % of the previously reported value of 13.80 ± 0.20 g/100 g dry weight using Kjeldahl digestion with conversion factor of 6.25 [14]. Although the universal conversion factor of 6.25 (equivalent to 0.16 g nitrogen/g of protein) is widely used for the calculation of all proteins by Kjeldahl method, Barros et al. recommended the use of factor 4.38 for mushroom protein analysis due to the high proportion of non-protein nitrogen compounds, mainly the indigestible chitin [23]. Thus, the crude protein content in L. rhinocerotis sclerotium, as quantified by Kjeldahl method, may be overestimated. Nonetheless, a large proportion of the sclerotial proteins are not extractable and they probably represent mainly storage proteins.
The majority of the protein spots did not yield identified proteins when searched against the NCBInr (Fungi) database during MALDI-MS analysis, indicating that the L. rhinocerotis sclerotial proteins are structurally quite different from other fungal proteins in the public databases. To improve the identification of the proteins, we decided to re-investigate the identities of the protein spots using the recent L. rhinocerotis genome database coupled with LC-MS. Mapping of the distinct peptides to the L. rhinocerotis genome gained significant information for all 40 spots and the approach significantly improved the accuracy of protein identification.
Accumulation of lectins in the sclerotium suggests that they may play a role as passive-defense, reserve storage proteins [24]. Lectins are non-immune, multivalent carbohydrate binding proteins that do not possess enzymatic activity and are generally thermo-stable [25]. Interestingly, lectins have been shown to possess potential pharmacological properties such as mitogenic, immunoenhancing, antiproliferative, antitumour, vasorelaxing, and hypotensive activities [26, 27]. Based on the sequence variations, at least three forms of lectins are known, encoded by GME270_g (184 amino acids), GME272_g (173 amino acids), and GME273_g (598 amino acids); each carrying a Jacalin-like plant lectin domain which occurs in various oligomerization states [28, 29]. Proteins containing this domain often bind to mono- or oligosaccharides with high specificity. Jacalin, an abundant protein in the jackfruit seed, specifically binds to the α-O-glycoside of the disaccharide Gal-β1-3-GalNAc [28, 29]. Lectins with comparable molecular weights but different pI values have probably undergo a series of heterogeneous phosphorylations, including gel spots 1, 2, 3, and 4 from GME273_g; spots 9, 10, and 36 from GME272_g; and spots 7 and 35 from GME270_g. On the other hand, probable heterogeneous glycosylation of GME273_g forms a series of spots with different molecular weights and pI values due to the nature of glycan structure. For example, gel spots 8, 15, and 26; are all GME273_g isoforms. The presence of three lower molecular weight isoforms (< 9 kDa) of GME273_g (spots 13, 28, and 38) suggests the plausible degradation of GME273_g by the relatively large quantity of serine proteases in the initiation of the storage proteins mobilization [30, 31].
Serine proteases cleave peptide bonds in proteins and are related to post-translational modification, protein turnover, and act as chaperones. Interestingly, a fungal serine protease isolated from Fusarium acuminatum has been found to act as a detergent enzyme for treating fibers, wool, hair, leather, food/feed and/or for any applications involving modification, degradation, or removal of proteinaceous material [32]. The L. rhinocerotis serine protease may have similar industrial application and thus warrants further investigation. On the other hand, GH27 is encoded by gene GME9376_g and is likely to be involved in starch utilization in L. rhinocerotis sclerotium as they share the same structural topology and catalytic mechanism with glycoside hydrolase family 31 [33]. The product of gene GME9376_g is 215 amino acids in length and carries a PLN02808 superfamily putative conserved domain of α-galactosidases.
SOD and CAT work as antioxidants to reduce cytotoxic reactive oxygen species where SOD catalyzes the dismutation of toxic superoxide into oxygen and hydrogen peroxide while CAT catalyze the decomposition of hydrogen peroxide to water and oxygen [34, 35]. SOD in L. rhinocerotis is encoded by GME441_g, with 204 amino acids in length. The gene product carries two conserved domains of iron/manganese superoxide dismutases at the N- (α-hairpin domain) and C-terminals, respectively. The presence of Mn-SOD in the sclerotial extract might be partially responsible for its strong superoxide anion radical scavenging activity as reported previously [14]. GST which is coded by GME7546_g (212 amino acids) catalyzes the conjugation of reduced glutathione to a variety of substrates and is likely to involve in the detoxification of endogenous compounds such as peroxidized lipids and the degradation of steroids and xenobiotics [36, 37]. The gene product consists of two GST family (Class Phi subfamily) domains at the N- (TRX-fold domain) and C-termini (α helical domain), respectively; with an active site located in a cleft between the two domains. Phi is a class of enzymes that are highly reactive toward chloroacetanilide and thiocarbamate herbicides. Other functions of Phi include the transportation of flavonoid pigments to the vacuole; shoot regeneration, and glutathione peroxidase activity [38].
The 14-3-3-domain-containing protein is crucial for signal transduction mechanisms as this protein is able to bind a large number of signaling proteins with diverse functions including kinases, phosphatases, and transmembrane receptors. This protein is involved in numerous essential cellular processes such as signal transduction, cell cycle regulation, apoptosis, stress response, cytoskeleton organization, and malignant transformation [39]. FIP is a family of bioactive proteins isolated from mushrooms. These proteins are reported to possess immunomodulatory and antitumor effects [17]. Interestingly, a protein carrying a Fve domain was identified from spot 37. Fve is a major fruiting body protein from F. velutipes that stimulates lymphocyte mitogenesis, suppresses systemic anaphylaxis reactions and oedema, enhances transcription of interleukin 2, interferon gamma and tumor necrosis factor alpha, and haemagglutinates red blood cells [22].
Phosphoglycerate mutase converts 3-phosphoglycerate to 2-phosphoglycerate through a 2,3-bisphosphoglycerate intermediate in the eighth step of glycolysis [40]. The gene that encodes the protein is GME590_g. The protein is 482 amino acids in length and carries a histidine phosphatase superfamily (branch 2) domain. Members of CP family are known as phytotoxins. For example, CP isolated from the cell wall of Ceratocystis fimbriata, the causal agent of “canker stain disease”, elicits phytoalexin synthesis (one of the first plant defense-related events) and plant cell death [41]. Thus, the identified CP isoforms in L. rhinocerotis sclerotial extract may play an important role in its defensive mechanism against predators and parasites. Aegerolysins are reported to have interesting biological properties including antitumoral, antiproliferative, and antibacterial. Other beneficial uses of these proteins are for atherosclerosis prevention, as vaccines, to improve cultivation of some commercially important edible mushrooms, and as specific markers in cell and molecular biology [42].
Conclusion
To the best of our knowledge, this is the first systematic profiling/identification of L. rhinocerotis sclerotial proteins using 2DE coupled with MALDI-MS and LC-MS. Only a few spots were identified using the MALDI-MS with public databases. The poor success rate indicated that L. rhinocerotis proteins are indeed structurally quite different from other known fungal proteins. In the LC-MS approach, using L. rhinocerotis genome as custom database, all remaining 40 spots examined were identified. Some of the proteins identified from this study are of pharmacological interest while others depicted nutrient mobilization and defense mechanisms in the L. rhinocerotis sclerotium. Putative lectins, immunomodulatory protein, aegerolysin, and antioxidant proteins such as Mn-SOD, CAT, and GST show pharmaceutical potential. The findings from this study may assist future work for the characterization of pharmacologically active sclerotial proteins of L. rhinocerotis.
Supplementary Material
Tables S1 - S3.
Acknowledgements
This study was supported by Fundamental Research Grant Scheme (FRGS) FP029-2014A from the Government of Malaysia and Postgraduate Research Fund (PPP) PV024/2012A from University of Malaya, Malaysia.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Huang NL. Identification of the Scientific Name of Hurulingzhi. Acta Edulis Fungi. 1999;6:32-4
2. Abdullah N, Haimi MZD, Lau BF, Annuar MSM. Domestication of a wild medicinal sclerotial mushroom, Lignosus rhinocerotis (Cooke) Ryvarden. Ind Crop Prod. 2013;47:256-61 doi:10.1016/j.indcrop.2013.03.012
3. Tan CS, Ng ST, Vikineswary S, Lo FP, Tee CS. Genetic Markers for Identification of a Malaysian Medicinal Mushroom, Lignosus rhinocerus (Cendawan Susu Rimau). ISHS Acta Hortic. 2010;859:161-8
4. Jones EBG, Hyde KD, Sabaratnam V. Malaysian Fungal Diversity. Kuala Lumpur, Malaysia: Mushroom Research Centre, University of Malaya. 2007
5. Wong KH, Cheung PCK. Sclerotia: Emerging Functional Food Derived from Mushrooms. In: (ed.) Cheung PCK. Mushrooms as Functional Foods. New York: John Wiley & Sons, Inc. 2009:111-46
6. Mohanarji S, Dharmalingam S, Kalusalingam A. Screening of Lignosus rhinocerus extracts as antimicrobial agents against selected human pathogens. J Pharm Biomed Sci. 2012;18:1-4
7. Phan CW, David P, Naidu M, Wong KH, Sabaratnam V. Neurite outgrowth stimulatory effects of culinary-medicinal mushrooms and their toxicity assessment using differentiating Neuro-2a and embryonic fibroblast BALB/3T3. BMC Complement Altern Med. 2013;13:261
8. Eik LF, Naidu M, David P, Wong KH, Tan YS, Sabaratnam V. Lignosus rhinocerus (Cooke) Ryvarden: A Medicinal Mushroom That Stimulates Neurite Outgrowth in PC-12 Cells. Evid Based Complement Alternat Med. 2012;2012:320308. doi:10.1155/2012/320308
9. Lai CKM, Wong KH, Cheung PCK. Antiproliferative Effects of Sclerotial Polysaccharides from Polyporus rhinocerus Cooke (Aphyllophoromycetideae) on Different Kinds of Leukemic Cells. Int J Med Mushrooms. 2008;10:255-64 doi:10.1615/IntJMedMushr.v10.i3.60
10. Lee ML, Tan NH, Fung SY, Tan CS, Ng ST. The Antiproliferative Activity of Sclerotia of Lignosus rhinocerus (Tiger Milk Mushroom). Evid Based Complement Alternat Med. 2012;2012:697603. doi:10.1155/2012/697603
11. Suziana Zaila CF, Farida Zuraina MY, Norfazlina MN, Lek Mun L. et al. Antiproliferative effect of Lignosus rhinocerotis, the Tiger Milk Mushroom on HCT 116 human colorectal cancer cells. TOPROCJ. 2013;4:65-70
12. Wong KH, Lai CKM, Cheung PCK. Immunomodulatory activities of mushroom sclerotial polysaccharides. Food Hydrocolloids. 2011;25:150-8 doi:10.1016/j.foodhyd.2010.04.008
13. Gao S, Lai CKM, Cheung PCK. Nondigestible Carbohydrates Isolated from Medicinal Mushroom Sclerotia as Novel Prebiotics. Int J Med Mushrooms. 2009;11:1-8 doi:10.1615/IntJMedMushr.v11.i1.10
14. Yap YH, Tan N, Fung S, Aziz AA, Tan C, Ng S. Nutrient composition, antioxidant properties, and anti-proliferative activity of Lignosus rhinocerus Cooke sclerotium. J Sci Food Agric. 2013;93:2945-52 doi:10.1002/jsfa.6121
15. Lee SS, Enchang FK, Tan NH, Fung SY, Pailoor J. Preclinical toxicological evaluations of the sclerotium of Lignosus rhinocerus (Cooke), the Tiger Milk mushroom. J Ethnopharmacol. 2013;147:157-63 doi:10.1016/j.jep.2013.02.027
16. Wong JH, Ng TB, Cheung RC, Ye XJ, Wang HX, Lam SK. et al. Proteins with antifungal properties and other medicinal applications from plants and mushrooms. Appl Microbiol Biotechnol. 2010;87:1221-35 doi:10.1007/s00253-010-2690-4
17. Xu X, Yan H, Chen J, Zhang X. Bioactive proteins from mushrooms. Biotechnol Adv. 2011;29:667-74 doi:10.1016/j.biotechadv.2011.05.003
18. Lau BF, Aminudin N, Abdullah N. Comparative SELDI-TOF-MS profiling of low-molecular-mass proteins from Lignosus rhinocerus (Cooke) Ryvarden grown under stirred and static conditions of liquid fermentation. J Microbiol Methods. 2011;87:56-63 doi:10.1016/j.mimet.2011.07.005
19. Yap HY, Chooi YH, Firdaus-Raih M, Fung SY, Ng ST, Tan CS. et al. The genome of the Tiger Milk mushroom, Lignosus rhinocerotis, provides insights into the genetic basis of its medicinal properties. BMC Genomics. 2014;15:635. doi:10.1186/1471-2164-15-635
20. Horie K, Rakwal R, Hirano M, Shibato J, Nam HW, Kim YS. et al. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum provides insight into their numerous functional protein components and diversity. J Proteome Res. 2008;7:1819-35 doi:10.1021/pr070369o
21. Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255-62 doi:10.1002/elps.1150090603
22. Paaventhan P, Joseph JS, Seow SV, Vaday S, Robinson H, Chua KY. et al. A 1.7A structure of Fve, a member of the new fungal immunomodulatory protein family. J Mol Biol. 2003;332:461-70
23. Barros L, Venturini BA, Baptista P, Estevinho LM, Ferreira IC. Chemical composition and biological properties of portuguese wild mushrooms: a comprehensive study. J Agric Food Chem. 2008;56:3856-62 doi: 10.1021/jf8003114
24. Peumans WJ, Van Damme EJ. Lectins as plant defense proteins. Plant Physiol. 1995;109:347-52
25. Pusztai A, Grant G. Assessment of lectin inactivation by heat and digestion. Methods Mol Med. 1998;9:505-14 doi:10.1385/0-89603-396-1:505
26. Wang H, Ng TB, Ooi VEC. Lectins from mushrooms. Mycol Res. 1998;102:897-906 doi:10.1017/S0953756298006200
27. Kellens JTC, Peumans WJ. Developmental accumulation of lectin in Rhizoctonia solani: a potential role as a storage protein. J Gen Microbiol. 1990;136:2489-95 doi:10.1099/00221287-136-12-2489
28. Kabir S. Jacalin: a jackfruit (Artocarpus heterophyllus) seed-derived lectin of versatile applications in immunobiological research. J Immunol Methods. 1998;212:193-211
29. Sankaranarayanan R, Sekar K, Banerjee R, Sharma V, Surolia A, Vijayan M. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold. Nat Struct Biol. 1996;3:596-603
30. Müntz K, Belozersky MA, Dunaevsky YE, Schlereth A, Tiedemann J. Stored proteinases and the initiation of storage protein mobilization in seeds during germination and seedling growth. JExB. 2001;52:1741-52 doi:10.1093/jexbot/52.362.1741
31. Praxedes-Garcia P, Cruz-Silva I, Gozzo AJ, Abreu Nunes V, Torquato RJ, Tanaka AS. et al. Biochemical aspects of a serine protease from Caesalpinia echinata Lam. (Brazilwood) seeds: a potential tool to access the mobilization of seed storage proteins. ScientificWorldJournal. 2012;2012:562715. doi:10.1100/2012/562715
32. Fungal serine protease and use thereof. Juntunen K, Valtakari L, Makinen S, Kallio J, Vehmaanpera J, Ojapalo P, et al. http://www.google.com/patents/US20110028375
33. Song KM, Okuyama M, Kobayashi K, Mori H, Kimura A. Characterization of a glycoside hydrolase family 31 alpha-glucosidase involved in starch utilization in Podospora anserina. Biosci Biotechnol Biochem. 2013;77:2117-24
34. Bannister JV, Bannister WH, Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit Rev Biochem. 1987;22:111-80
35. del Rio LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ. Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic Biol Med. 1992;13:557-80
36. Leaver MJ, George SG. A piscine glutathione S-transferase which efficiently conjugates the end-products of lipid peroxidation. Mar Environ Res. 1998;46:71-4 doi:10.1016/S0141-1136(97)00071-8
37. Douglas KT. Mechanism of action of glutathione-dependent enzymes. Adv Enzymol Relat Areas Mol Biol. 1987;59:103-67
38. Cho HY, Lee HJ, Kong KH. A phi class glutathione S-transferase from Oryza sativa (OsGSTF5): molecular cloning, expression and biochemical characteristics. J Biochem Mol Biol. 2007;40:511-6
39. van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936-46 doi:10.1002/bies.1134
40. Fothergill-Gilmore LA, Watson HC. The phosphoglycerate mutases. Adv Enzymol Relat Areas Mol Biol. 1989;62:227-313
41. Pazzagli L, Pantera B, Carresi L, Zoppi C, Pertinhez TA, Spisni A. et al. Cerato-platanin, the first member of a new fungal protein family: cloning, expression, and characterization. Cell Biochem Biophys. 2006;44:512-21 doi:10.1385/cbb:44:3:512
42. Berne S, Lah L, Sepcic K. Aegerolysins: structure, function, and putative biological role. Protein Sci. 2009;18:694-706 doi:10.1002/pro.85
Author contact
![]() Corresponding author: yean_ny_niecom.
Corresponding author: yean_ny_niecom.

 Global reach, higher impact
Global reach, higher impact