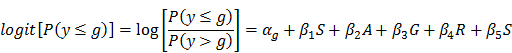3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(7):748-753. doi:10.7150/ijms.8770 This issue Cite
Research Paper
A Novel Catechol-O-Methyltransferase Variant Associated with Human Disc Degeneration
1. Department of Orthopaedic Surgery, Carolinas Medical Center, Charlotte, N.C.;
2. Department of Bioinformatics and Genomics, University of North Carolina at Charlotte, Charlotte, N.C.;
3. Bioinformatics Services Division, North Carolina Research Campus, Kannapolis, N.C.;
4. Molecular Core and Microarray Labs, Cannon Research Center, Carolinas Medical Center, Charlotte, N.C.
Received 2014-2-6; Accepted 2014-5-2; Published 2014-5-15
Abstract
Background: Disc degeneration and its associated low back pain are a major health care concern causing disability with a prominent role in this country's medical, social and economic structure. Low back pain is devastating and influences the quality of life for millions. Low back pain lifetime prevalence approximates 80% with an estimated direct cost burden of $86 billion per year. Back pain patients incur higher costs, greater health care utilization, and greater work loss than patients without back pain.
Methods: Research was performed following approval of our Institutional Review Board. DNA was isolated, processed and amplified using routine techniques. Amplified DNA was hybridized to Affymetrix Genome-Wide Human SNP Arrays. Quality control and genotyping analysis were performed using Affymetrix Genotyping Console. The Birdseed v2 algorithm was used for genotyping analysis. 2589 SNPs were selected a priori to enter statistical analysis using lotistic regression in SAS.
Results: Our objective was to search for novel single nucleotide polymorphisms (SNPs) associated with disc degeneration. Four SNPs were found to have a significant relationship to disc degeneration; three are novel. Rs165656, a new SNP found to be associated with disc degeneration, was in catechol-O-methyltransferase (COMT), a gene with well-recognized pain involvement, especially in female subjects (p=0.01). Analysis confirmed the previously association between COMT SNP rs4633 and disc degeneration. We also report two novel disc degeneration-related SNPs (rs2095019 and rs470859) located in intergenic regions upstream to thrombospondin 2.
Conclusions: Findings contribute to the challenging field of disc degeneration and pain, and are important in light of the high clinical relevance of low back pain and the need for improved understanding of its fundamental basis.
Keywords: SNP, catechol-O-methyltransferase (COMT), SNP, low back pain.
Introduction
Single nucleotide polymorphisms (SNPs) are variations in the DNA sequence wherein a single nucleotide (A, C, G or T) in the sequence is altered. SNPs can occur in both coding (exon) and non-coding (intron or intergenic) regions, but even non-coding SNPs may have a functional effect on gene expression by influencing transcription factors, miRNA binding, or splicing sites [1;2]. Prediction of potentially functional SNPs in potential drug-response genes continues to be a pharmacologic research area of great interest [3;4].
There is strong interest in genetic polymorphisms which are associated with human intervertebral disc degeneration, and several recent reviews have provided updates and overviews [5-7]. There is also a related high interest in the role of genes and their variants which are associated with low back pain. Variants in genes related to inflammation and matrix components have been reported to show a relation with disc degeneration and also peripheral pain [8].
COMT (catechol-O-methyltransferase) is an enzyme which inactivates catechols, including the significant neurotransmitters dopamine, noradrenaline and adrenaline [9]. In clinical studies of chronic pain, the relationship between COMT variants and pain conditions, including low back pain, is an area of considerable research interest and importance [9-12].
Materials and methods
Clinical Study Population
Study of human disc specimens was approved prospectively by the Human Subjects Institutional Review Board at Carolinas Medical Center. The need for informed consent was waived by the ethical board since disc tissue was removed as part of routine surgical practice (and discarded). Scoring of disc degeneration utilized a modification of the Thompson scoring system [13] which incorporated author ENH's radiologic, MRI and surgical findings. The Thompson system scores disc degeneration over the spectrum from a healthy disc (Thompson grade I) to discs with advanced degeneration (grade V, the most advanced stage of degeneration) [13]. Patient specimens were derived from surgical disc procedures performed on individuals with degenerative disc disease. All surgical patients presented with back pain. Surgical specimens were transported to the laboratory in sterile tissue culture medium. Annulus cells were established in monolayer culture [14], and expanded for use in as previously described [15]. During primary culture and expansion, cells were maintained in a 37oC incubator with 5% CO2 and 95% relative humidity.
DNA isolation and preparation
DNA was isolated according to the manufacturer's protocol for the Wizard®Genomic DNA Purification Kit (Promega, Madison, WI). For SNP chip analysis, the DNA was further cleaned following the manufacturer's protocol for the kit Genomic DNA Clean & Concentrator (Zymo Research, Irvine, CA).
SNP microarray processing
The Affymetrix Genome-Wide Human SNP Nsp/Sty Assay Kit 6.0 (Affymetrix, Santa Clara, CA, USA) was used to fragment and label the DNA sample. Briefly, 250 ng of total genomic DNA was digested with Nsp I and Sty I restriction enzymes and ligated to adaptors that recognize the cohesive 4 bp overhangs. All fragments resulting from restriction enzyme digestion served as substrates for adaptor ligation regardless of size. Adaptor-ligated DNA fragments were amplified using a generic primer that recognizes the adaptor sequence. Fragments in the 200 to 1,100 bp size range were preferentially PCR amplified. PCR amplification products for each restriction enzyme digest were combined and purified using the isopropanol precipitation method. The amplified DNA was then hybridized to Genome-Wide Human SNP Arrays 6.0 (Affymetrix) containing 1.8 million genetic markers, including more than 906,600 single nucleotide polymorphisms (SNPs).
SNP Data analysis methods
Quality control and genotyping analysis were performed using Affymetrix Genotyping Console 4.1 (Affymetrix, Inc., Santa Clara, CA). Affymetrix recommended thresholds of contrast QC>=0.4 and SNP call rate > 90% were used to select samples and SNPs that qualified for downstream analysis. The Birdseed v2 algorithm was used for genotyping analysis. Due to the relatively small sample size, only 2589 SNPs on the SNP array were selected to enter statistical analysis. SNPs were selected a priori based on a literature review (Linguamatics Ltd, Cambridge, UK) and pathway analysis (Ingenuity Systems, Inc., Redwood City, CA), and were selected based on QC criteria. Selected SNPs included: 1) SNPs reported to have association with disc degeneration disease, 2) SNPs that were not reported to have association with the disease, but were on the genes, or in the intergenic regions next to the genes, that were reported to have association, 3) SNPs that were on the pathways with reported association with the disease.
Statistical analyses
Statistical analyses were performed in SAS (SAS Institute Inc., Cary, NC). Logistic regression was used to examine the association between each SNP and disc degeneration grade, where disc degeneration grade was the response variable, and SNP was the predictor variable. Age, gender, ethnicity, and spine site were considered as potential confounders and adjusted in the model. Genotypes were coded 11, 12 or 22, where allele 1 and 2 indicate the major and minor allele, respectively: “11” represents the genotype that has two major alleles; “12” represents the genotype that has one major allele and one minor allele; and “22” represents the genotype that has two minor alleles. The homozygous genotype (11 or 22) that had smaller sample size was grouped together with the heterozygous genotype (12) to increase sample size. Logistic regression was performed separately for each SNP to address the following question: “Does having one or two minor alleles increase (or decrease) the likelihood of having a greater disc degeneration grade?” The following logistic regression equation was employed:
where αg is the intercept for each level of g; g is the disc degeneration grade (1-5); β1 to β5 are coefficients of predictor variables: , S = SNP (two categories after grouping the heterozygous genotype with one of the homozygous genotypes), A = age, G = gender, R = race, and S = spinal site.
The simple M method [16] was used to calculate the effective number of independent tests, followed by applying the Bonferroni correction for multiple testing corrections.
The SNPs that were found to be significantly associated with disc degeneration grade by the analysis described above were further analyzed by a gender-stratified logistic regression to determine if the SNP effect was different between the two genders. Stratified logistic regression for age, race and site was not performed due to small sample size (i.e., n < 10 in at least one stratum).
We recognized that the allele-dose effect could be missed due to the combination of the heterozygous group with one of the homozygous groups in the analysis described above. Therefore, for the SNPs that were found to be significantly associated with disc degeneration grade, the average disc degeneration grade was calculated for each of the three genotypes (11, 12 or 22) to determine if an allele-dose effect exists in these SNPs.
Results
The clinical study population was composed of 40 subjects for whom the Affymetrix recommended threshold (contrast QC>=0.4) was met. The study population consisted of one Grade I patient (17 yr old male), 6 grade II patients (mean age 38.6 yr ± 5.9 (mean ± s.e.m.; 1 male, 5 females), 12 Grade III patients (mean age 39.9 ± 2.8; 5 males, 7 females); 17 Grade IV patients (mean age 51.1 ± 3.2; 6 males, 11 females), and 4 Grade V patients (mean age 60.0 ± 9.1; 2 males, 2 females). All surgical patients presented with back pain. Additional demographic features shown by disc grade are presented in Table 1. As shown in this table, some sample heterogeneity in age, gender, ethnicity and spinal site was observed; therefore these factors were adjusted in the logistic regression model to remove potential confounding effects. Because the sample size in this study is small, three steps in data analysis were taken to increase statistical power. These are discussed in detail in the Discussion section below.
Based on the literature review, pathway analysis, and quality control (SNP call rate >90%), 2589 SNPs were selected a priori. These SNPs were then analyzed by logistic regression. Four SNPs (SNPs rs2095019 and rs4708592, located upstream of the THBS2 gene, and SNPs rs165656 and rs4633, located within the COMT gene) were found to be significantly associated with disc degeneration disease after adjusting for confounders, and correction for multiple testing. Note that the Bonferroni correction for multiple testing used here was based on 38 (not 2589) tests, because the Simple M method [16] revealed that there were only 38 independent tests among the 2589 tests. The Bonferroni corrected P values for association with disc degeneration grade are: 0.032, 0.049, 0.043, and 0.043, respectively. SNPs rs2095019, rs4708592 and rs165656 are novel SNPs reported here for the first time in patients with degenerative disc disease. Detailed results are shown in Table 2.
The novel pain-related SNP reported here for the first time in association with disc degeneration and pain, rs165656, lies within the COMT gene region. SNPs rs2095019 and rs4708592, novel findings in the present study, both lie in intergenic regions located upstream to THBS2 (thrombospondin 2), a matricellular protein which our laboratory has previously shown to be present in the human disc [17]. Rs2095019 is positioned 51,000 base pairs from the starting position of THBS2, and rs4708592 lays 106,329 base pairs from the starting position.
For SNPs that were found to be significantly associated with disc degeneration, the average disc degeneration grade of each genotype was calculated. For rs2095019, allele A was found to be significantly associated with lower disc degeneration grade, and an allele-dose effect was observed. The average disc degeneration grade was 2.4 for AA (n=5), 3.14 for AC (n=14), and 3.86 for CC (n=21). No clear allele-dose effect was observed for the other three SNPs. For example, for the two COMT SNPs, the highest disc degeneration grade was observed in the heterozygote group, but not in one of the homozygous groups (result not shown).
Stratified logistic regression analyses were also performed. For both the novel COMT SNP rs165656 reported here, and the previously recognized COMT SNP rs4633, a significant effect was present for female gender (p = 0.0104) (but not for male gender (p = 0.3783)).
Demographic Features of Study Population by Disc Grade (% Incidence).
| Grade I (n = 1) | Grade II (n = 6) | Grade III (N = 12) | Grade IV (n = 17) | Grade V (n = 4) | ||
|---|---|---|---|---|---|---|
| Age (years) | ≤35 | 100% | 33.33% | 25% | 17.65% | 0 |
| 36-59 | 0 | 66.67% | 75% | 58.82% | 50% | |
| ≥60 | 0 | 0 | 0 | 23.52% | 50% | |
| Gender | Male | 100% | 33.33% | 41.67% | 35.29% | 50% |
| Female | 0 | 66.67% | 58.33% | 64.71% | 50% | |
| Race | Black | 0 | 33.33% | 33.33% | 23.53% | 25% |
| White (not of Hispanic origin) | 0 | 66.67% | 66.67% | 76.47% | 75% | |
| Unknown | 100% | 0 | 0 | 0 | 0 | |
| Spinal Site* | L2-3 | 0 | 0 | 0 | 5.88% | 0 |
| L3-4 | 0 | 16.67% | 0 | 0 | 25% | |
| L4-5 | 0 | 33.33% | 41.67% | 35.29% | 50% | |
| L5-S1 | 100% | 50% | 58.33% | 58.82% | 25% | |
*L, lumbar; S, sacral
SNPs Significantly Associated with Disc Degeneration.
| Gene | COMT | COMT | Intergenic | Intergenic | ||||
|---|---|---|---|---|---|---|---|---|
| SNP | Rs165656 | Rs4633 | Rs2095019 | Rs4708592 | ||||
| Major allele (allele 1) | G | C | C | T | ||||
| Minor allele (allele 2) | C | T | A | C | ||||
| Association with disc degeneration previously reported in the literature | No | Yes [10;24] | No | No | ||||
| Raw P-value | 0.00113 | 0.001133 | 0.000833 | 0.001281 | ||||
| P-value (after multiple testing correction) | 0.043054 | 0.043054 | 0.031653 | 0.048662 | ||||
| Genotype Incidence (# (%)) | GG or GC | CC | CC or CT | TT | CC | CA or AA | TT | TC or CC |
| Grade I * | 1 (100%) | 0 (0%) | 1 (100%) | 0 (0%) | 0 (0%) | 1 (100%) | 0 (0%) | 1 (100%) |
| Grade II | 2 (33%) | 4 (67%) | 2 (33%) | 4 (67%) | 1 (17%) | 5 (83%) | 2 (33%) | 4 (67%) |
| Grade III | 9 (75%) | 3 (25%) | 9 (75%) | 3 (25%) | 4 (33%) | 8 (67%) | 7 (58%) | 5 (42%) |
| Grade IV | 13 (76%) | 4 (24%) | 13 (76%) | 4 (24%) | 13 (76%) | 4 (24%) | 16 (94%) | 1 (6%) |
| Grade V | 4 (100%) | 0 (0%) | 4 (100%) | 0 (0%) | 3 (75%) | 1 (25%) | 4 (100%) | 0 (0%) |
* Note the small sample size in the Grade I analysis (n=1) which was present here.
Discussion
The lifetime prevalence of disc degeneration and its associated low back pain approximates 80% with estimated health care costs exceeding $100-200 billion/year. Chronic low back pain takes a high socio-economic toll in terms of lost wages, reduced productivity; chronic, long term low back pain also takes a high personal toll, also, and can lead to disability and job loss [18]. Our identification here of a new SNP associated with a major pain gene is especially interesting in light of the high clinical relevance of low back pain and the need for an improved understanding of the fundamental basis of low back pain.
This study investigated the association between SNPs and disc degeneration. Due to the small sample size (N=40), several steps were taken to increase the power of this study. First, instead of analyzing 906,600 SNPs genotyped by the array, we analyzed 2589 SNPs that were known or have the potential to be associated with disc degeneration based on literature review and pathway analysis. Detailed SNP selection criteria are described in the Methods section. Secondly, for each SNP, we combined the heterozygote group with one of the homozygote group to increase sample size in each subgroup. Thirdly, we used the simple M method [16](which was well cited by genetic association studies) to find the actual number of independent tests, which was found to be 38. Therefore, the Bonferroni correction was based on 38 (not 2589) independent tests. Bonferroni correction assumes that the test for each SNP is independent. This assumption is severely violated in SNP studies, because many SNPs are highly correlated through linkage disequilibrium. Therefore the search for the actual number of independent tests was necessary, and it greatly increased statistical power. With the three steps discussed above, we were able to detect significant associations with a relative small sample size. Additional details on these three steps are described in the Methods section.
In a recent genome-wide analysis of pain- and neurotrophin-related gene expression in the degenerating human disc (which did not include SNPs), our laboratory identified significant differences in expression of COMT in more degenerated (grades IV and V) discs vs. less degenerated discs (grades I-III) (p = 0.002), and also in surgical compared to control human discs (p = 0.035) [19]. Other pain-related genes identified in our analyses were bradykinin receptor B1 and calcitonin gene-related peptide (CGRP).
Kim and Schwartz have presented a review of pain and spinal disease [20]. Chronic low back pain is a complicated event which is related to a number of factors, including structural, psychosocial, genetic and occupational factors [7]. As noted by Tegeder and Lotsch [7], genetic profiles of individual patients are of interest since this may open an opportunity for identifying patients at high risk for chronic pain, and in the future potentially opening the opportunity for individualized pain management.
Catechol-O-methyltransferase (COMT) codes for a protein which is important in catabolic pathways of a number of pain-relevant neurotransmitters, including noradrenalin, adrenaline and dopamine [21], and is a protein which has been attributed to related issues including pain perception, mood, cognition, and responses to physical/emotional stressors [22]. Patients with a specific COMT polymorphism identified by Zubieta et al. showed higher sensory and effective pain ratings [23], and Dai et al. have identified an association between specific COMT SNPs associated with pain sensitivity and outcomes following surgery for low back pain [24], thus pointing to the potential importance of further exploration of COMT SNPs in patients with disc degeneration.
Identification of the COMT SNP rs165656 in association with disc degeneration was a novel finding in this report. This SNP has previously been reported in association with mental retardation in a Chinese population in the Qinba region [25]. Although not always included in reviews of low back pain and SNPs, (such as reviews of Eskola et al. [6] and of Mayer et al. [5]), Tegeder and Lotsch have provided a recent 2009 review of COMT genetic variants related to back pain [7]. Connections between the COMT SNP rs4633, identified in our present study, and pain have been previously reported and provide evidence for modulation of low back pain by COMT variants [7], and the relationship to disc degeneration and pain [10;24]. The most common and well-studied COMT polymorphism associated with pain appears to be V158M (val158met; rs4680). This SNP was not identified in the present study population.
In humans, associations have been reported between COMT functional haplotypes and thermal pain perception and capsaicin-induced pain [22]. COMT variants have been seen to influence pain phenotypes, especially in women with chronic pain reminding us that in the future COMT genetic differences may be of clinical relevance in tailored pain management [26]. COMT variants have also been noted to influence the efficacy of morphine therapy in cancer patients with pain [27].
Our finding of a significant effect related to female gender status in both the novel COMT SNP reported here and the COMT SNP rs4633 is similar to the significant relationship seen with female gender in the work of Belfer et al. in an average pain sensitivity COMT haplotype [22]. These authors commented that such findings reflect a fundamental contribution of COMT to the complex area of pain processes. Belfer and Segall also remind us that the pain experience by humans has contributions from both genetic and environmental factors [26].
For the two COMT SNPs, it is interesting that the highest disc degeneration grade was observed in the group of patients with one minor allele (CG for rs165656 and CT for rs4633), but not in the group with two minor alleles (CC for rs165656 and TT for rs4633). The lack of allele-dose effect could be due to small sample size in this study, but it is also important to note that previous studies have also observed a lack of allele-dose effect in COMT SNPs [10;28;29].
Note should also be made concerning the two novel disc degeneration-associated SNPs reported here (rs2095019 and rs4708592) which lie in the intergenic region near the THBS2 gene. Genes in noncoding regions, and even silent SNPs, can have effects on cellular processes, including DNA transcription, RNA splicing, mRNA stability or mRNA transport/translation [9]. THBS2 is an important matricellular protein which is present within the disc [17], and which is known to modulate collagen fibrillogenesis [30] and matrix metalloproteinase 2 interactions [31;32], biologic functions critically important to the disc. It is also interesting to note that our previous work with THBS2 null mice showed disruption of the lamellar morphology in the disc [33]. Related important observations include the previous work by Hirose et al. which identified a THBS2 SNP associated with disc degeneration [34], and the work of Valdes et al which showed an association of a thrombospondin SNP with the radiographic progression of disc degeneration [35]. These observations increase our interest in the relationship of disc degeneration to the two intergenic SNPs near the THBS2 gene.
In summary, the findings reported here contribute to the challenging field of disc pain by identifying a new COMT SNP associated with disc degeneration. COMT variants continue to be of high interest, especially in women with chronic pain. The present work also identified two novel disc degeneration-related SNPs located upstream of the THBS2 gene. Findings are important in light of the high clinical relevance of low back pain and the need for improved understanding of the fundamental basis of low back pain.
Acknowledgements
We thank Synthia Bethea for expert technical assistance in DNA isolation. HEG and ENH wish to thank the Brooks Back Pain Research Endowment for general laboratory support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform. 2005;6:44-56
2. Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276-287
3. Pang GSY, Wang J, Wang Z, Lee CGL. Predicting potentially functional SNPs in drug-response genes. Pharmacogenomics. 2009;10:639-653
4. Shastry BS. SNPs in disease gene mapping, medicinal drug development and evolution. J Human Genetics. 2007;52:871-880
5. Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, Hecht AC. Genetic polymorphisms associated with intervertebral disc degeneration. The Spine Journal. 2013;13:299-317
6. Eskola PJ, Lemmelä S, Kjaer P, Solovieva S, Männikkö M, Tommerup N. et al. Genetic association studies in lumbar disc degeneration: A systematic review. PLoS ONE. 2012;7:e49995
7. Tegeder I, Lötsch J. Current evidence for a modulation of low back pain by human genetic variants. J Cell Mol Med. 2009;13:1605-1619
8. Omair A, Holder M, Lie BA, Reikeras O, Brox JI. Treatment outcome of chronic low back pain and radiographic lumbar disc degeneration are associated with inflammatory and matrix degrading gene variants: a prospective genetic association study. BMC Musculoskeletal Dis. 2013;14:105
9. Andersen S, Skorpen F. Variation in the COMT gene: implications for pain perception and pain treatment. Pharmacogenomics. 2009;10:669-684
10. Omair A, Lie BA, Reikeras O, Holden M, Brox JI. Genetic contribution of catechol-o-methyltransferase variants in treatment outcome of low back pain: a prospective genetic association study. BMC Musculoskeletal Dis. 2012;13:76
11. Jacobsen LM, Schistad EI, Storesund A, Pedersen LM, Rygh LJ, Roe C. et al. The COMT rs4680 Met allele contributes to long-lasting low back pain, sciatica and disability after lumbar disc herniation. Eur J Pain. 2012;16:1064-1069
12. Kambur O, Männistö PT. Catechol-O-methyltransferase and pain. Internatl Rev Neurobiol. 2010;95:227-279
13. Thompson JP, Pearce RH, Schechter MT, Adams ME, Tsang IKY, Bishop PB. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine. 1990;15:411-415
14. Gruber HE, Fisher EC Jr, Desai B, Stasky AA, Hoelscher G, Hanley EN. Human intervertebral disc cells from the annulus: Three- dimensional culture in agarose or alginate and responsiveness to TGF-β1. Exp Cell Res. 1997;235:13-21
15. Gruber HE, Hoelscher GL, Leslie K, Ingram JA, Hanley ENJr. Three-dimensional culture of human disc cells within agarose or a collagen sponge: assessment of proteoglycan production. Biomaterials. 2006;27:371-376
16. Gao X, Starmer J, Martin ER. A genetic testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361-369
17. Gruber HE, Ingram JA, Hanley EN Jr. Immunolocalization of thrombospondin in the human and sand rat intervertebral disc. Spine. 2006;31:2556-2561
18. Katz JN. Lumbar disc disorders and low-back pain: Socioeconomic factors and consequences. J Bone Joint Surg [Am ]. 2006 88A, Suppl 2:21-24
19. Gruber HE, Hoelscher GL, Ingram JA, Hanley EN. Genome-wide analysis of pain-, nerve- and neurotrophin-related gene expression in the degenerating human annulus. Molecular Pain. 2012;8:63
20. Kim DH, Schwartz CE. The genetics of pain: implications for evaluation and treatment of spinal disease. The Spine Journal. 2010;10:827-840
21. Anderson S, Skorpen F. Variation in the COMT gene: implications for pain perception and pain treatment. Pharmacoeconomics. 2009;10:669-684
22. Belfer I, Segall SK, Laviviere WR, Smith SB, Dai F, Slade GD. et al. Pain modality- and sex-specific effects of COMT genetic functional variants. Pain. 2013;154:1368-1376
23. Zubieta J-K, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y. et al. COMT val158 met genotype affects υ-opioid neurotransmitter responses to a pain stressor. Science. 2003;299:1240-1243
24. Dai F, Belfer I, Schwartz CE, Banco R, Martha JF, Tighioughart H. et al. Association of catechol-O-methyltransferase genetic variants with outcome in patients undergoing surgical treatment for lumbar degenerative disc disease. The Spine Journal. 2010;10:949-957
25. Zhang K, Gao J, An C, Gao X, Zheng Z, Li R. et al. An association study between cathecol-O-methyltransferase gene and mental retardation in the Chinese Han population. Neurosci Lett. 2007;419:83-87
26. Belfer I, Segall S. COMT genetic variants and pain. Drugs of Today. 2011;47:457-467
27. Rakvåg TT, Ross JR, Sato H, Skorpen F, Kaasa S, Klepstad P. Genetic variation in the catechol-O-methyltransferase (COMT) gene and morphine requirements in cancer patients with pain. Molecular Pain. 2008;4:64
28. Gürsoy S, Erdal E, Herken H, Madenci E, Alsehirli B, Erdal N. Significance of catechol-O-methyltransferase gene polymorphism in fibromyalgia syndrome. Rheumatol Int. 2002;23:104-107
29. Kim H, Mittal DP, Iadarola MJ, Dionne RA. Genetic predictors for acute experimental cold and heat pain sensitivity in humans. J Med Genet. 2006;43:e40
30. Bornstein P, Kyriakides TR, Yang Z, Armstrong LC, Birk DE. Thrombospondin 2 modulates collagen fibrillogenesis and angiogenesis. J Invest Dermatol. 2000;5:61-66
31. Yang Z, Kyriakides TR, Bornstein P. Matricellular proteins as modulators of cell-matrix interactions: Adhesive defect in thrombospondin 2-null fibroblasts is a consequence of increased levels of matrix metalloproteinase-2. Mol Biol Cell. 2000;11:3353-3364
32. Bachmeier BE, Nerlich A, Mittermaier N, Weiler C, Lumenta C, Wuertz K. et al. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur Spine J. 2009;18:1573-1586
33. Gruber HE, Bornstein P, Sage HE, Ingram JA, Norton HJ, Hanley ENJr. Disruption of the thrombospondin-2 gene alters lamellar morphology but does not effect vascularization of the adult mouse lumbar disc. Biomed Central Arthritis Research & Therapy. 2008;10:R96
34. Hirose Y, Chiba K, Karasugi T, Nakajima M, Kawaguchi Y, Mikami Y. et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. Amer J Human Gen. 2008;82:1122-1129
35. Valdes AM, Hassett G, Hart DJ, Spector TD. Radiographic progression of lumbar spine disc degeneration is influenced by variation at inflammatory genes. A candidate SNP association study in the Chingford cohort. Spine. 2005;30:2445-2451
Author contact
![]() Corresponding author: Helen E. Gruber, Ph.D., Orthopaedic Research Biology, Cannon Research., Room 304, Carolinas Medical Center, P.O. Box 32861, Charlotte, N.C. 28232. 704/355-5665; fax: 704/355-5620; e-mail: helen.gruberorg.
Corresponding author: Helen E. Gruber, Ph.D., Orthopaedic Research Biology, Cannon Research., Room 304, Carolinas Medical Center, P.O. Box 32861, Charlotte, N.C. 28232. 704/355-5665; fax: 704/355-5620; e-mail: helen.gruberorg.

 Global reach, higher impact
Global reach, higher impact