3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(5):500-507. doi:10.7150/ijms.7849 This issue Cite
Research Paper
Differential Expression Profile of Long Non-coding RNAs during Differentiation of Cardiomyocytes
1. State key Laboratory of Reproductive Medicine, Department of Pediatrics, Nanjing Maternity and Child Health Care Hospital Affiliated to Nanjing Medical University, Nanjing 210029, China.
2. Department of Cardiology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu 210029, China
* The authors have contributed equally to this study and they should be regarded as joint first authors.
Received 2013-10-9; Accepted 2014-3-7; Published 2014-3-28
Abstract
Many long non-coding RNAs (lncRNAs) are species specific and seem to be less conserved than protein-coding genes. Some of them are involved in the development of the lateral mesoderm in the heart and in the differentiation of cardiomyocytes. The purpose of the study was to investigate the expression profiles of lncRNAs during the differentiation of P19 cells into cardiomyocytes, with a view to studying the biological function of lncRNAs and their involvement in the mechanism of heart development. First, we observed the morphology of P19 cells during differentiation using an inverted microscope. Then, cardiac troponin T (cTnT) expression was detected to validate that the cells had successfully differentiated into cardiac myocytes by real-time reverse transcriptase polymerase chain reaction (real-time RT-PCR) and western blotting. Lastly, the expression profile of lncRNA genes was obtained using an lncRNA microarray and real-time RT-PCR analyses. The microarray results showed that 40 lncRNAs were differentially expressed, of which 28 were upregulated and 12 were downregulated in differentiated cardiomyocytes. The differentially expressed lncRNAs were further validated. Our results illustrated a critical role of lncRNAs during the differentiation of P19 cells into cardiac myocytes, which will provide the foundation for further study of the biological functions of lncRNAs and the mechanism of heart development.
Keywords: lncRNAs, differentiation, caridiomyocytes, microarrays.
Introduction
The heart is the first functional organ that is developed in the process of embryonic development. It is very important for development that the heart is healthy. Numerous studies have revealed the accurate regulation of key molecular pathways during embryonic development, particularly in the cardiovascular system. Haploinsufficiency of essential genes often leads to cardiac malformations [1], which are the most common major congenital defects, with a prevalence of approximately eight in every 1,000 newborn infants [2].
The human transcriptome is composed of not only a large set of protein-coding messenger RNAs (mRNA), but also many non-protein coding transcripts that have structural, regulatory or unknown functions. Over the last decade, much attention focused on the microRNAs (miRNAs), a class of small non-coding RNAs that are involved in various biological and pathological processes [3, 4]. More recently, long non-coding RNAs (lncRNAs), generally defined as non-coding RNAs of more than 200nt in length without known protein-coding function [5], have risen to prominence, with central roles in a diverse range of functions in cell biology [6,7]. In contrast to miRNAs, lncRNAs have not been fully investigated. A handful studies have indicated that dysregulation of lncRNAs result in aberrant gene expression associated with cancers [8-10]. Although an increasing number of lncRNAs have been characterized, the role of lncRNAs in the differential of cardiomyocytes has not been investigated.
The P19 mouse embryonal carcinoma cell line is multipotent and can differentiate into cardiac myocytes with embryoid body formation in the presence of dimethylsulfoxide (DMSO) [11]. Thus, P19 cells have been used to study cardiac-specific transcription factors and upstream signaling pathways in cardiac differentiation [12-14]. Therefore, P19 cells are a suitable model for studying cardiac differentiation at the molecular and functional levels [15].
In this study, we initially identified differentially expressed lncRNAs during the differentiation of P19 cells using an lncRNA microarray. We subsequently validated the microarray results by real-time quantitative reverse transcription PCR (real-time qRT-PCR) for specific differentially expressed lncRNAs.
Results and Discussion
P19 cells differentiation
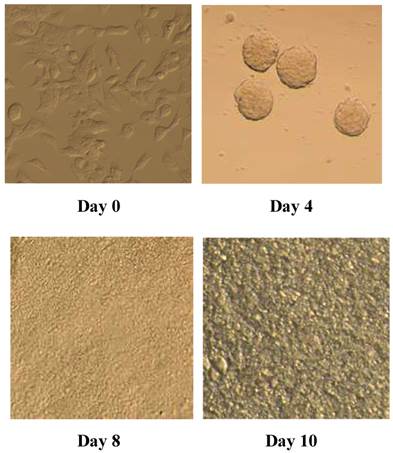
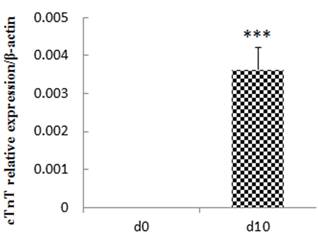
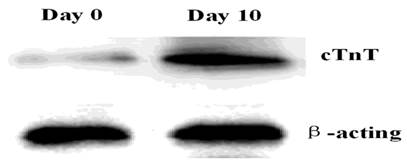
P19 cells differentiate into cardiac myocytes in the presence of DMSO. The efficiency of differentiation depends on the prior formation of non-adhering aggregates [17]. We observed and photographed the morphological changes in P19 cells using an inverted microscope to investigate the process of P19 cell differentiation. We discovered that P19 cells aggregated during the first 4 days and there were beating cell colonies on day 10 (Figure 1). The myocyte differentiation marker cTnT was detected on day 0 and day10, respectively, to validate that the cells had differentiated into cardiac myocytes. As shown in Figure 2 and Figure 3, mRNA expression and protein expression of cTnT were much higher on day 10 compared to day 0. Thus, the beating cell colonies generated on day 10 of P19 cell differentiation, and the high expression of cTnT, demonstrated that the cells had differentiated into cardiac myocytes.
lncRNA microarray
LncRNA microarrays are powerful tools for studying the biological function of lncRNAs. We conducted lncRNA microarray analysis on P19 cells at day 0 and day 10 of DMSO exposure. According to the microarray data, we selected lncRNAs that were upregulated by more than five-fold and downregulated by more than three-fold. In addition, poorly conserved lncRNAs were excluded. The conservation of lncRNAs was determined using the online Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Ultimately, 40 differentially expressed lncRNAs that were highly conserved in cardiac myocytes (day 10) compared to normal P19 cells (day 0) were identified (Table 1). Among them, 28 were upregulated and 12 were downregulated. We randomly chose five upregulated lncRNAs (ENSMUST00000159006, uc009byc.1, AK089560, ENSMUST00000101005, ENSMUST00000124503) and three downregulated lncRNAs (uc007keu.1, AK028257, BC030682) for qRT-PCR validation (Table 2).
Morphology of P19 cells during differentiation into cardiac myocytes (day 0, day 4, day 8, day 10). P19 cells were aggregated for 4 days and colonies of beating cells were observed on day 10 under an inverted microscope, as described in Materials and methods.
Relative expression of cTnT at day 10 compared with day 0. The experiment was repeated three times with consistent results. ***p<0.001
Expression of the cTnI protein in P19 cells. Total proteins were isolated from P19 cells and analyzed by western blotting. Lane 1, day 0; Lane 2, day 10. The experiment was repeated three times with consistent results.
40 differentially expressed lncRNAs.
| Regulation | lncRNA | chromosomal localization | RNA length | Start locus | Stop locus |
|---|---|---|---|---|---|
| 28 up-regulated lncRNAs | AK158639 | chr2 | 417 | 169458512 | 169458928 |
| uc007vie.1 | chr15 | 4168 | 12959401 | 12963569 | |
| ENSMUST00000159006 | chr6 | 253 | 52108522 | 52112019 | |
| AK166199 | chr13 | 1351 | 16122089 | 16123440 | |
| AK052877 | chr19 | 1301 | 30638978 | 30640279 | |
| uc009biz.1 | chr6 | 606 | 36664928 | 36665534 | |
| uc009pal.1 | chr9 | 3270 | 41388823 | 41400910 | |
| uc009eqi.1 | chr6 | 2534 | 143777066 | 143779600 | |
| ENSMUST00000101005 | chr6 | 1302 | 119912384 | 119913686 | |
| ENSMUST00000124503 | chr11 | 454 | 35163570 | 35164287 | |
| AK020106 | chr11 | 802 | 69615637 | 69616437 | |
| AK142834 | chrX | 2388 | 118021841 | 118024227 | |
| uc007cpz.1 | chr1 | 2470 | 135197537 | 135200007 | |
| uc007prv.1 | chr13 | 2198 | 21925626 | 21929399 | |
| uc009byc.1 | chr6 | 545 | 52122879 | 52124051 | |
| AK078053 | chr18 | 1378 | 36459754 | 36461132 | |
| NR_024257 | chr2 | 4066 | 9802872 | 9808394 | |
| AK089560 | chr5 | 2683 | 13525726 | 13528408 | |
| AK142308 | chr18 | 1262 | 37965377 | 37966636 | |
| AK046177 | chr13 | 606 | 117639650 | 117640255 | |
| AK135062 | chr7 | 2464 | 104057136 | 104059598 | |
| AK138321 | chr11 | 2303 | 47744849 | 47747149 | |
| uc008sdp.1 | chr4 | 3039 | 22409926 | 22412965 | |
| uc008fug.1 | chr18 | 948 | 83172461 | 83173409 | |
| AK028129 | chr3 | 2428 | 96043315 | 96045742 | |
| uc008xbx.1 | chr5 | 802 | 34516538 | 34517340 | |
| uc008euf.1 | chr18 | 1629 | 43480650 | 43482279 | |
| ENSMUST00000127359 | chr14 | 344 | 47007193 | 47008957 | |
| 12 down-regulated lncRNAs | uc007keu.1 | chr11 | 1635 | 75565382 | 75579340 |
| AK033485 | chr1 | 2241 | 54532201 | 54534442 | |
| uc007pyj.1 | chr13 | 1179 | 28700386 | 28977221 | |
| uc008sac.1 | chr4 | 1060 | 11893711 | 11921427 | |
| AK137254 | chr7 | 5124 | 127773275 | 127778400 | |
| AK028257 | chr14 | 272 | 55735163 | 55735434 | |
| BC030048 | chr17 | 1092 | 35087185 | 35088238 | |
| BC030682 | chr7 | 1343 | 71031236 | 71032537 | |
| ENSMUST00000117553 | chr2 | 1125 | 111840336 | 111841461 | |
| ENSMUST00000172121 | chr6 | 291 | 64941211 | 64941502 | |
| AK010244 | chr2 | 1771 | 125082798 | 125084785 | |
| uc008mcn.1 | chr2 | 1771 | 125082798 | 125084785 |
lncRNAs differentially expressed between cardiomyocytes that differentiated from P19 cells (day 10) compared with normal cells (day 0).
| up-regulated lncRNA | fold change | GeneSymbol | down-regulated lncRNA | fold change | GeneSymbol |
|---|---|---|---|---|---|
| ENSMUST00000159006 | 46.21 | Gm15051 | uc007keu.1 | 8.07 | Ywhae |
| uc009byc.1 | 21.50 | AK142386 | AK028257 | 4.71 | |
| AK089560 | 15.47 | BC030682 | 3.4 | ||
| ENSMUST00000101005 | 6.29 | Wnk1 | |||
| ENSMUST00000124503 | 5.11 | Gm12122 |
Validation of differentially expressed lncRNAs
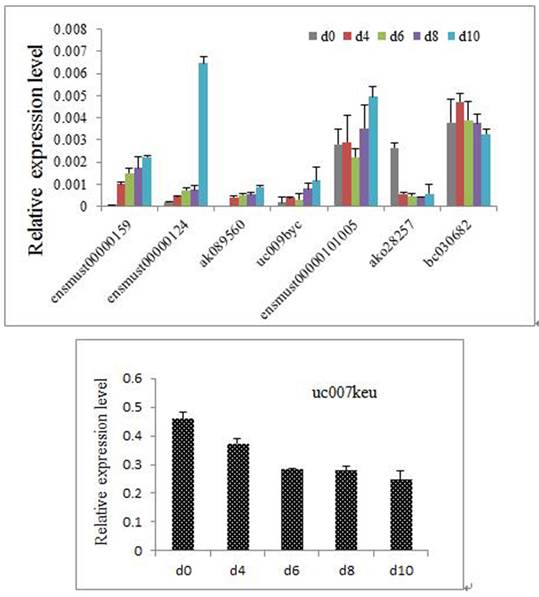
We performed real-time qRT-PCR expression analysis on P19 cells at day 0 and day 10 to confirm the microarray results. Using GAPDH as a normalization control, the statistics demonstrated that four out of the five upregulated lncRNAs (P=0.038, 0.000016, 0,022 and 0.017 for ENSMUST00000159006, uc009byc.1, AK089560, ENSMUST00000124503, respectively) and two of the three downregulated lncRNAs (P=0.00012 and 0.001 for AK028257 and uc007keu.1, respectively) showed significantly different expressions (Figure 4). In addition, we validated the eight differentiated expressed lncRNAs at different time points during the differentiation. D0, d4, d6, d8, d10 were chosen as the time points. As shown in the figure 5, the expressed trends during the differentiation are consistent with the comparison between d0 and d10. The major difference might be the beginning time of the change was different.
Discussion
Congenital heart defects (CHD) are the most common major congenital malformation, accounting for approximately 40% of perinatal deaths and more than one fifth of deaths in the first month of life [18]. Although many studies have focused on heart development in recent decades, details of the mechanism remain unclear [19, 20]. P19 cells are isolated from an experimental embryo-derived mouse teratocarcinoma and can differentiate into cardiac myocytes with embryoid body formation in the presence of DMSO [21]. Thus, we simulated heart development in vitro by differentiating P19 cells into cardiomyocytes.
This study focused on determining the lncRNAs expression profile during cardiomyocyte differentiation and explaining the differences between cardiomyocytes and undifferentiated P19 cells. We identified 40 differentially expressed lncRNAs (28 upregulated and 12 downregulated). Real-time qRT-PCR validated four of five upregulated and two of three downregulated lncRNAs.
Some researchers have demonstrated that the expression of many lncRNAs is different during development and that their functions range from the control of pluripotency to lineage specification [22, 23]. In theory, lncRNAs have intrinsic cis-regulatory capacity, which has been confirmed and whose mechanism has been described. Increasing numbers of reports show that lncRNAs can play a role in both cis and trans [24, 25], and more direct experimental studies are required to determine the precise proportion of cis regulators. Regulating the expression of some lncRNAs may influence the expression of their neighboring protein-coding genes, including several master regulators of cellular differentiation [26-28]. It is in the early stage that the role of lncRNAs in heart development has attracted much attention from researchers. Indeed, two lncRNAs, Fendrr and Braveheart (Bvht), were recently uncovered to be involved in the development of the lateral mesoderm in the heart and the differentiation of cardiac myocytes, respectively [29, 30]. Deficiency of Fendrr, particularly in the nascent lateral plate mesoderm, can result in a thin ventricular wall of the heart. Fendrr regulates the expression of certain core transcription factors in heart development by modulating the epigenetic profile of cells to generate cardiac hypoplasia [29]. In a similar way to Fendrr, Bvht interacts with SUZ12, a component of PRC2, to alter cardiomyocytes differentiation and retain the cardiac phenotype in neonatal cardiomyocytes [30].
Most of the four up-regulated and two down-regulated lncRNAs have no official Human Genome Nomenclature Committee symbol and their function is still unclear. However, some studies have been shown that Ywhae play a critical role in many diseases, such as HIV neurocognitive impairment [31], neuronal migrational defects [32], bipolar disorder [33] and endometrial stromal sarcoma [34].
Although we have identified some differentially expressed lncRNAs during the cardiac differentiation, it is too early for us to confirm their relationship with cardiac malformation. Therefore, subgroup analysis of lncRNAs should be performed to explore this relationship in the future. In addition, most lncRNAs have a distinct spatial and temporal specificity in the process of organismal differentiation and development. It has been shown that lncRNAs have different expression patterns in different parts of the brain [35]. We should sample more cells from different times in the process of differentiation, such as d4, d6 and d8, to examine alterations in lncRNA expression in the early stage of differentiation.
Validation of lncRNA microarray data using real-time RT-PCR. The real-time RT-PCR reactions were repeated three times for every lncRNA. * p < 0.05, **p<0.01, ***p<0.001.
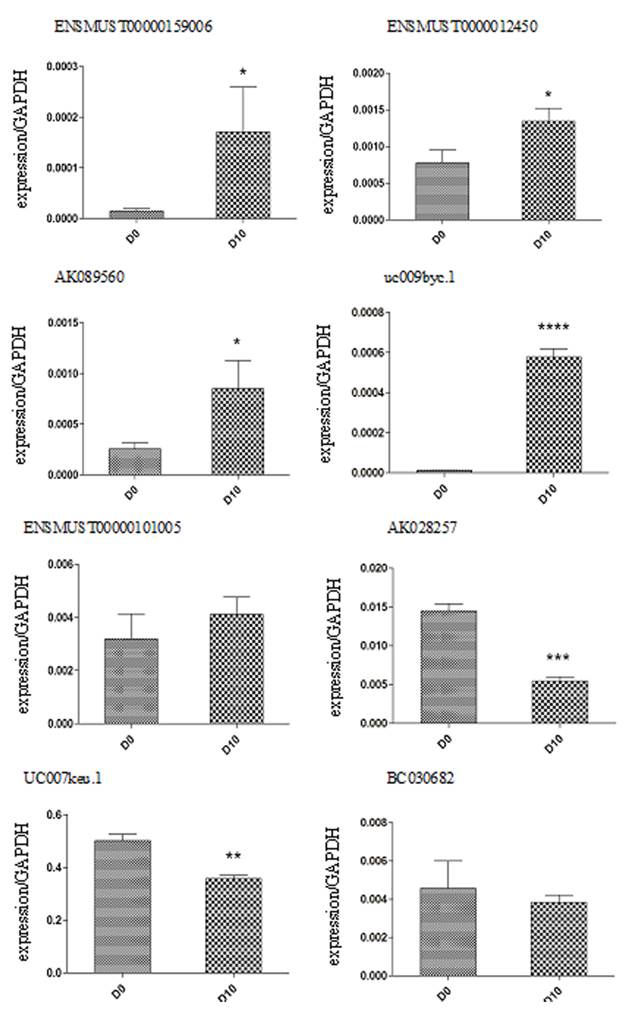
The 8 differentiated expressed lncRNAs at different time points of the differentiation. (Because the relative expression of uc007keu was much higher than the other lncRNAs, we performed two histograms for clarity and aesthetic feeling.)
Materials and methods
P19 cell culture and induction of differentiation
P19 cells were bought from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in complete medium (α-MEM + 10% fetal bovine serum, FBS + 100 U/ml penicillin + 100ug/ml streptomycin, pH 7.2-7.4) (α-MEM, FBS, penicillin and streptomycin, Gibco-BRL, Grand Island, NY, USA) in a 5% CO2 atmosphere at 37°C. During differentiation, P19 cells were maintained in suspension as aggregates for 4 days in complete medium containing 1% dimethylsulfoxide (DMSO, Sigma, St. Louis, MO, USA) in bacteriological dishes. On day 4, the cell aggregates were transferred to cell culture flasks and then adherently cultivated from the 5th to the 10th day without DMSO. The culture medium was replaced every 2 days. We harvested cells on differentiation day 0 and day 10. The morphological changes in P19 cells were observed under an inverted microscope (Nikon Eclipse TE300, Tokyo, Japan) equipped with phase-contrast objectives and a digital camera (Nikon E4500).
Quantitative real time-PCR (qPCR) and western blotting
Total RNA was extracted from the harvested cells using a mirVana extraction kit (Ambion, Austin, TX, USA), following the manufacturer's protocol. RNA was measured using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE, USA) to assess its quantity and quality, and stored at -80°C. In general, we simultaneously performed RNA extraction and cDNA transcription for all subjects. The total RNA was reverse-transcribed to cDNA with a High-Capacity cDNA reverse transcription kit (Applied Biosystems, Austin, USA.). According to the manufacturer's protocol, we used 1µg of mRNA to activate 20µl of the reverse transcription reaction. The reaction comprised 25°C for 10min, 37°C for 120min, 85°C for 5 min and a hold at 4°C. Subsequently, real-time PCR was performed in triplicate for each sample and included no-template negative controls. For the final volume of 20 µl reaction, 1 µl of synthesized cDNA was mixed with 8 µl of diethylpyrocarbonate (DEPC)-treated water, 10 µl of TaqMan Gene Expression Master Mix and 1 µl of cardiac troponin T (cTnT) / β-actin TaqMan Gene Expression Assay (Applied Biosystems, cTnT ID: Mm01290256_m, β-actin ID: Mm00607939_s1). The reaction conditions comprised 50°C for 2min, 95°C for 10min; followed by 40 cycles of 95°C for 15s and 60°C for 1min on the ABI 7500 Real-Time PCR system (Applied Biosystems). β-actin was used as a reference to obtain the relative expression of cTnT, which was determined with the comparative cycle threshold (CT) (2-△CT) method, in which △CT = CT cTnT -CT β-actin.
A monoclonal rabbit anti-cTnT antibody and a monoclonal rabbit anti-β-actin antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cells were broken using the lysis buffer provided in the total protein extraction kits (KeyGen, Inc., China). The lysate supernatant was obtained after centrifugation at 14000×g for 30min at 4°C. We then measured the protein concentration with a BCA protein detection kit (KeyGen, Inc., China). Western blotting was conducted as previously described [16].
Construction of the lncRNA microarray
We pooled three replicate samples of cells on day 0 and day 10 of DMSO exposure, respectively, to perform lncRNA microarray analysis. Total RNA was isolated from the two samples as above and was quantified using a NanoDrop spectrophotometer (NanoDrop). RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Each sample was then amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3' bias, using a random priming method. The labeled cRNAs were hybridized onto the Mouse LncRNA Array v2.0 (8 x 60K, Arraystar). The hybridized arrays were washed, fixed and scanned with using the Agilent DNA Microarray Scanner (part number G2505C). Data were extracted using Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). Differentially expressed LncRNAs between the two samples were identified by Fold Change filtering. The threshold set for upregulated lncRNAs was more than five-fold and for downregulated lncRNAs it was more than three-fold. The lncRNAs discussed in this article were carefully collected from the most authoritative databases, such as RefSeq, UCSC Knowngenes, Ensembl and many related literature.
Validation of differentially expressed lncRNAs
Total RNA extraction and cDNA transcription were conducted as above. For real-time PCR, we added 1µl of cDNA to 12.5µl of SYBR-Green Gene Expression Master Mix (Applied Biosystems, Inc), 10.5µl f DEPC-treated water and 0.5µl of reverse and forward primers. cDNA was amplified for 50 cycles on the ABI 7500 Real-Time PCR system (Applied Biosystems). The primers sequences used are listed in Table 3. GAPDH was used as a reference to obtain the relative expression of target lncRNAs which was determined with the comparative cycle threshold (CT) (2-△CT) method, in which △CT = CT target lncRNA - CT GAPDH.
Primers for real-time RT-PCR.
| Gene name | Primers | Tm (℃) |
|---|---|---|
| ENSMUST00000159006 | P5:GGAGCTGACTTGGAGCACTG | 60 |
| P3:AACAGACCTCTTGCCAGTTCA | ||
| uc009byc.1 | P5:AACTTGCGTCTGGAGTTGGG | 60 |
| P3:CCCAGAATAGCAGCACCTCA | ||
| AK089560 | P5:ATGCTTTCCCAGGGTGTGTT | 60 |
| P3:GGCTAGGATTTCCCGACGAG | ||
| ENSMUST00000101005 | P5:TGTTGATACAGCCTCAGTCCAT | 60 |
| P3:GTTGGAAGTGGCGAGTTTGG | ||
| ENSMUST00000124503 | P5:GACACGAAGAAGAACCACATCA | 60 |
| P3:GCCTGCGAGGATTCTATTTATT | ||
| uc007keu.1 | P5:AAAATGTGATTGGAGCCAGAAG | 60 |
| P3:GTCCTCTCCTCCCTTGTTTTCT | ||
| AK028257 | P5:CTCTCCTCTCCGCTTCTCTCT | 60 |
| P3:CATCCCAGCACAAATCAATGT | ||
| BC030682 | P5:GACCTGGCTCTTCCTCAT | 60 |
| P3:TTCCATCTGTCCGTTCTG | ||
| GAPDH | P5:ATTCAACGGCACAGTCAA | 60 |
| P3:CTCGCTCCTGGAAGATGG |
Statistical analysis
All statistical analyses were performed using the Student's t-test with SPSS software version 13.0 (SPSS, Inc, Chicago, IL, USA). P-values less than 0.05 were considered statistically significant, and all the statistical tests were two-sided.
Conclusion
In conclusion, we identified a set of lncRNAs that were aberrantly expressed in cardiomyocytes compared to undifferentiated P19 cells, which will provide the foundation for the further study of the biological function of lncRNAs and the mechanism of heart development.
Abbreviations
lncRNAs: long non-coding RNAs; cTnT: cardiac troponin T; RT-PCR: reverse transcriptase polymerase chain reaction; mRNA: messenger RNAs; miRNA: microRNA; DMSO: dimethylsulfoxide; CHD: congenital heart defects; DEPC: diethylpyrocarbonate.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81070500), the Key Medical Personnel Foundation of Jiangsu Province (Grant No. RC2011021), the Nanjing Medical Science and Technique Development Foundation (QRX11107), and the Science and Technology Development Foundation of Nanjing Medical University (Grant No. 2012NJMU195).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking mirna-1-2. Cell. 2007;129:303-317
2. Meberg A, Lindberg H, Thaulow E. Congenital heart defects: the patients who die. Acta Paediatr. 2005;94:1060-1065
3. Bartel DP. Micrornas: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
4. Ambros V. The functions of animal micrornas. Nature. 2004;431:350-355
5. Saxena A, Carninci P. Long non-coding rna modifies chromatin: epigenetic silencing by long non-coding rnas. Bioessays. 2011;33:830-839
6. Kaikkonen MU, Lam MT, Glass CK. Non-coding rnas as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430-440
7. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding rna as modular scaffold of histone modification complexes. Science. 2010;329:689-693
8. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. Malat-1, a novel noncoding rna, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-8041
9. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of hulc, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding rna. Gastroenterology. 2007;132:330-342
10. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding rna hotair reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076
11. Skerjanc IS. Cardiac and skeletal muscle development in p19 embryonal carcinoma cells. Trends Cardiovasc Med. 1999;9:139-143
12. van der Heyden MA, Defize LH. Twenty one years of p19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res. 2003;58:292-302
13. Hu DL, Chen FK, Liu YQ, Sheng YH, Yang R, Kong XQ, Cao KJ, Gu HT, Qian LM. Gata-4 promotes the differentiation of p19 cells into cardiac myocytes. Int J Mol Med. 2010;26:365-372
14. Han SP, Pan Y, Peng YZ, Gu XQ, Chen RH, Guo XR. Folbp1 promotes embryonic myocardial cell proliferation and apoptosis through the wnt signal transduction pathway. Int J Mol Med. 2009;23:321-330
15. van der Heyden MA, van Kempen MJ, Tsuji Y, Rook MB, Jongsma HJ, Opthof T. P19 embryonal carcinoma cells: a suitable model system for cardiac electrophysiological differentiation at the molecular and functional level. Cardiovasc Res. 2003;58:410-422
16. Kaczocha M, Glaser ST, Deutsch DG. Identification of intracellular carriers for the endocannabinoid anandamide. Proc Natl Acad Sci U S A. 2009;106:6375-6380
17. van der Heyden MA, Defize LH. Twenty one years of p19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res. 2003;58:292-302
18. Trojnarska O, Grajek S, Katarzynski S, Kramer L. Predictors of mortality in adult patients with congenital heart disease. Cardiol J. 2009;16:341-347
19. Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922-1927
20. Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943-948
21. Skerjanc IS. Cardiac and skeletal muscle development in p19 embryonal carcinoma cells. Trends Cardiovasc Med. 1999;9:139-143
22. Kung JT, Colognori D, Lee JT. Long noncoding rnas: past, present, and future. Genetics. 2013;193:651-669
23. Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91-98
24. Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist rna and the mechanism of x chromosome inactivation. Annu Rev Genet. 2002;36:233-278
25. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding rnas with enhancer-like function in human cells. Cell. 2010;143:46-58
26. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding rna induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-419
27. Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106-1113
28. Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding rna pairs in the developing brain. PLoS Genet. 2009;5:e1000617
29. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncrna fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206-214
30. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding rna required for cardiovascular lineage commitment. Cell. 2013;152:570-583
31. Morales D, Hechavarria R, Wojna V, Acevedo SF. Ywhae/14-3-3epsilon: a potential novel genetic risk factor and csf biomarker for hiv neurocognitive impairment. J Neurovirol. 2013
32. Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A. 14-3-3epsilon is important for neuronal migration by binding to nudel: a molecular explanation for miller-dieker syndrome. Nat Genet. 2003;34:274-285
33. Liu J, Li ZQ, Li JY, Li T, Wang T, Li Y, Xu YF, Feng GY, Shi YY, He L. Polymorphisms and haplotypes in the ywhae gene increase susceptibility to bipolar disorder in chinese han population. J Clin Psychiatry. 2012;73:e1276-e1282
34. Croce S, Hostein I, Ribeiro A, Garbay D, Velasco V, Stoeckle E, Guyon F, Floquet A, Neuville A, Coindre JM, Macgrogan G, Chibon F. Ywhae rearrangement identified by fish and rt-pcr in endometrial stromal sarcomas: genetic and pathological correlations. Mod Pathol. 2013
35. Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding rnas in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716-721
Author contact
![]() Corresponding author: SP Han, ZB Yu, Department of Pediatrics, Nanjing Maternity and Child Health Care Hospital of Nanjing Medical University, No. 123 Tian Fei Xiang, Mo Chou Road, Nanjing 21004, China. E-mail: shupinghanedu.cn (SP Han); zhangbinyuedu.cn (ZB Yu). Tel: +86-025-52226561, Fax: +86-025-52226561.
Corresponding author: SP Han, ZB Yu, Department of Pediatrics, Nanjing Maternity and Child Health Care Hospital of Nanjing Medical University, No. 123 Tian Fei Xiang, Mo Chou Road, Nanjing 21004, China. E-mail: shupinghanedu.cn (SP Han); zhangbinyuedu.cn (ZB Yu). Tel: +86-025-52226561, Fax: +86-025-52226561.

 Global reach, higher impact
Global reach, higher impact