3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(3):230-234. doi:10.7150/ijms.5278 This issue Cite
Research Paper
Detection of Hypermethylated Spastic Paraplegia-20 in Stool Samples of Patients with Colorectal Cancer
Department of Surgical Oncology the First Affiliated Hospital, Xi'an Jiaotong University College of Medicine, Xi'an, Shaanxi 710061, China.
Received 2012-9-22; Accepted 2013-1-7; Published 2013-1-13
Abstract
Background. Analysis of aberrant hypermethylation in stool DNA might provide a novel strategy for noninvasive detection of colorectal cancer.
Aims. To explore the feasibility of detecting hypermethylation in Spastic paraplegia-20 promoter as a stool-based DNA marker for detection of colorectal cancer.
Methods. We collected 96 tissue and stool samples from patients with colorectal cancer and 30 stool samples healthy individuals.
Results. Hypermethylated Spastic paraplegia-20 occurs in 85.4% (82/96) of patients with colorectal cancer in the tissue samples. In the stool samples, the results indicate 80.2% (77/96) sensitivity and 100% (30/30) specificity of the test for detecting colorectal cancer by using the stool samples as a noninvasive method.
Conclusions. The study reveals that hypermethylation in Spastic paraplegia-20 promoter is a highly specific and sensitive biomarker for screening colorectal cancer in stool samples as a noninvasive method.
Keywords: Colorectal cancer, Screening, SPG20, Stool DNA, Hypermethylation, Biomarker.
Introduction
Colorectal cancer (CRC) is estimated the fourth common invasive cancer newly diagnosed in the United States of America in 2011[1]. Fortunately, the CRC mortality rate can be decreased by the early detection of cancer[2]. Till now, there is variety kinds of methods to screen the early CRC, such as guaiac-based fecal occult blood test, fecal immunochemical test, colonoscopy, CT and stool DNA test[3, 4]. Stool DNA test is supposed to be a possible superior sensitivity and specificity compared to the guaiac-based fecal occult blood test and fecal immunochemical test[5, 6], and it can be a cost-effective alternative for CRC screening if the cost of the test substantially decreased or if its availability would entice a large fraction of otherwise unscreened persons to receive screening[7].
Till now, a number of genes are found with different sensitivities and specificities for diagnose in CRC[8-10]. Spastic paraplegia-20 (SPG20) encodes Spartin, a multifunctional protein, which has previously been found to be involved in intracellular epidermal growth factor recept or trafficking[11]. Prior studies have demonstrated that the promoter of the SPG20 gene was hypermethylated in colon cancer cell lines[12]. However, to date, SPG20 hypermethylation has not been assessed thoroughly in stool samples of CRC patients. In the present study, we studied the feasibility of detecting hypermethylation in SPG20 promoter as a stool-based DNA marker for detection of CRC using methylation-specific polymerase chain reaction (MSP).
Methods
Patients and collection of tissue and stool samples
In order to avoid bias, we performed this research as a blinded assay. Samples were collected and stored by a unique person who did not perform the downstream experiments. All the excretive stool samples were collected using the 15ml collection tubes before colonoscopy from subjects referred for colonoscopy at the First Affiliated Hospital of Xi'an Jiaotong University. All colonoscopies were performed by experienced gastroenterologists using the same preps. Stool samples from 96 CRC patients through endoscopy and biopsy was collected. Paired colorectal tissue samples were routinely collected from 96 patients with sporadic CRC undergoing surgery in our hospital. Additionally, normal mucosa samples taken in distance from the carcinomas were analyzed. Meanwhile, another 30 stool samples from the endoscopically healthy patients were analyzed. Samples were sent to the laboratory within 30 minutes after defecation, aliquoted (200-220 mg each) and stored at liquid nitrogen until analysis. The investigation was approved by the ethical committee of the First Affiliated Hospital of Xi'an Jiaotong University.
DNA isolation from tissue and stool samples
Genomic DNA was isolated from colorectal tissues (10±1 mg) by using TissueGen DNA Kit (CWBIO) and from stool samples (200-220 mg) by using QIAamp DNA Stool Mini Kit (Qiagen) according to the manufacturer's protocol. The total yield of DNA was determined by ultraviolet absorbance at 260 nm, and the quality of DNA was verified by agarose gel electrophoresis and PCR amplification of the human β-actin.
Bisulfite modification
For all tissue and stool samples, 2 μg genomic DNA was chemically modified by sodium bisulphate to convert all unmethylated cytosines to uracils while leaving methylcytosines unaltered by using the EpiTect Bisulfite Kit (Qiagen), and eluted in 20 μL of elution buffer.
Methylation-specific PCR
The bisulfite-modified DNA was used as the template for PCR. SPG20 gene was examined, and template-free distilled water was included as a negative control for amplification. The primer sequences[12] for the methylated templates were 5´-TGGAACGTTTTGGTTGTTAC-3´(forward) and 5´-TACCTCGAAAACTCCCTACG-3´(reverse). And the primer sequences for the unmethylated templates were 5´-GTGGAATGTTTTGGTTGTTAT-3´(forward) and 5´-TTACCTCAAAAACTCCCTACA-3´(reverse). Each PCR reaction mix consisted of a total volume of 25 µl containing 1 × PCR buffer (Takara), 200 µM dNTPs, 0.4µM concentration of each primer (BGI), 1U of HotStar Taq enzyme (Takara) and 3 µl bisulfite-modified DNA. The thermocycler conditions were as follows: 95°C for 30 seconds, followed by 40 cycles at 94°C for 30 seconds, annealing temperature for both methylated primer pairs and unmethylated primer pairs was 50°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. PCR products were analyzed by 2% agarose gel electrophoresis and Gold View staining following by visualization with ultraviolet illumination using a gel imaging analyzing system.
Statistical analysis
To compare characteristics of the different groups of patients and samples, Chi-square test and Fisher exact test were used as appropriate. Statistical analyses were performed using SPSS 13.0. All statistical tests were conducted 2-sided, and P values < 0.05 were considered to be statistically significant.
Results
Stool samples were obtained from 30 endoscopically diagnosed healthy individuals and 96 CRC patients histologically diagnosed. There was no significant difference in age and gender between the subgroups (Table 1). MSP was performed on all 318 samples which included 96 stool samples from CRC patients with matched CRC tissue samples and 30 stool samples from healthy individuals.
The characteristics of patients and normal people.
| Cancer | Normal | P value | |
|---|---|---|---|
| Total number | 96 | 30 | |
| Age (year) | 62.2±9.5 | 62.9±8.9 | 0.716 |
| Male:Female | 3:1 | 7:3 | 0.587 |
82 of 96 (85.4%) CRC tissue samples contained hypermethylated SPG20 while only 3 of 96 (3.1%) normal tissue samples from the distant normal colonic epithelium contained hypermethylated SPG20 (Figure 1) indicated that the rate of hyper- methylation of SPG20 promoter DNA was significantly higher in the cancer mucosa than in the normal mucosa (P ˂ 0.001). So, based on these data, we did some further analysis of SPG20. We chose the hypermethylation of SPG20 promoter DNA in stool samples as a noninvasive biomarker for CRC detection, especially in patients with early tumors.
Detection of unmethylated (U) and hypermethylated (M) SPG20 in tissue samples of patients with colorectal cancer (C1-C4) and tissue samples of from the distant normal colonic epithelium (N1-N4).
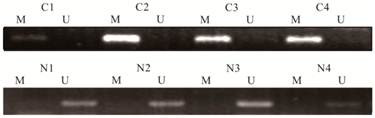
Following the performance of the MSP on the DNA extracted from the tissue samples, we further performed MSP on all 126 stool samples. We found that 91 of 96 (94.7%) CRC patients had concordant hypermethylated status of SPG20 in their tissue samples and stool samples. The five mismatching samples were all in hypermethylated SPG20 group that we can only found hypermethylated SPG20 in their tissue samples. This meant that 77 of 96 (80.2%) stool samples from CRC patients were SPG20 hypermethylation positive. While no methylated SPG20 promoter DNA was detected in the stool samples from the endoscopically healthy individuals (Figure 2). These results indicated a 80.2% sensitivity and 100% specificity of the test for detecting CRC by using the stool samples as a noninvasive method (Table 2). The rate of hypermethylated SPG20 between the two subgroups was significantly different (P ˂ 0.001). In our study, there were 21 stage I patients. In tissue samples, the methylated SPG20 promoter DNA was detected in 16 of the 21 stage I patients. The positive rate between stage I and the rest patients did not show a significant difference (P = 0.175). Meanwhile, in the stool sample analysis, the hypermethylated SPG20 were found in 14 stage I patients (71.4%). Compared with the rest patients, the positive rate did not show a significant difference (P = 0.307). As a screening method, the positive rate of hypermethylated SPG20 was significantly higher in the stage I patients than in the healthy individuals. The correlation between SPG20 methylation status and the clinicopathologic parameters of CRC patients was also examined. It did not show significant differences in age, gender and tumor location either (Table 3).
Detection of unmethylated (U) and hypermethylated (M) SPG20 in stool samples of patients with colorectal cancer (C1-C4) and stool samples of healthy individuals (N1-N4).

The positive rate of SPG20 hypermethylation in tissue and stool samples.
| Hypermethylation Status | |||
|---|---|---|---|
| Positive | Negative | P value | |
| Tissue samples | |||
| CRC | 82 | 14 | |
| Normal | 3 | 93 | <0.001 |
| Stool samples | |||
| CRC | 77 | 19 | |
| Normal | 0 | 30 | <0.001 |
Correlation between SPG20 methylation status in stool DNA of CRC patients and clinicopathological parameters.
| Parameter | Cases (N) | P value |
|---|---|---|
| Age (year) | ||
| <50 | 10 | |
| 50-60 | 19 | |
| 60-70 | 28 | |
| ≥70 | 20 | 0.946 |
| Gender | ||
| Male | 58 | |
| Female | 19 | 0.882 |
| Tumor location | ||
| Rectum | 32 | |
| Left hemicolon | 27 | |
| Right hemicolon | 18 | 0.947 |
| TNM stage | ||
| I | 14 | |
| II | 25 | |
| III | 35 | |
| IV | 3 | 0.307 |
Discussion
Stool DNA tests required the collection of only a single stool sample per round and no diet or medication restrictions. In some respects, it also manifested a satisfactory sensibility and specificity. But the challenge of detecting the stool DNA is to find the trace amounts of methylated or mutated human DNA located in a large amount of wild DNA(about 99.99% of stool DNA content is bacterial or dietary; human DNA is merely 0.01% of the total stool DNA with average concentrations of only 100 ng/g).[13] The multimarker panel of the PreGen-Plus, the first generation of stool DNA test, was found to be present in 25% (3/12) of CRC patients with a 25% sensitivity and 96% (2246/2340) specificity [14]. Within the past a few years, several groups and research centers had reported a large amount of clinical results about the sensitivity and specificity of stool DNA test by using different markers in colorectal cancer patient.[8, 10, 15-19] If such targets additionally showed high sensitivity and specificity for CRC, they would be suitable for early detection of CRC.
In our study, we have explored the feasibility of detecting hypermethylated SPG20 in stool DNA as a screening method for CRC. What we found have shown that the promoter of SPG20 gene is hypermethylated frequently in stool of patients with CRC with an especially high sensitivity and specificity by using the MSP technique.
The hypermethylation located in DNA promoter which is a defined region of the gene and this phenomenon can be easily analyzed in a single cost-effective reaction. SPG20 encodes the protein Spartin which has a role in cytokinesis that can subsequently cause aneuploidy has been shown to be associated with carcinogenesis.[20] Studies have shown that hypermethylation of SPG20 promoter is associated with the loss of gene expression and subsequent lack of Spartin protein [12].
At present, there is no institution which had studied the hypermethylated SPG20 in stool samples as a molecule marker for CRC screening. In this study, we simultaneously detected hypermethylated SPG20 in tissue samples and stool samples taken from the patients with CRC. We also take the stool samples from the healthy individuals. In the tissue samples, it indicated an 85.4% sensitivity and 96.9% specificity. This was similar with Lind et al's research, they found an 88% sensitivity and 100% specificity[12]. Further, we found that the hypermethylated SPG20 in stool samples had a high sensitivity and specificity (80.2% and 100%) for detecting CRC. Although there were only 21 stage I patients among the 96 individuals, the sensitivity and specificity between the stage I group and the rest patients did not show a significant difference. Therefore, using SPG20 hypermethylation as a marker in the stool samples appears to suggest a powerful screening method for detecting CRC including early stage cancer. Till now, fecal occult blood testing is the only available noninvasive screening method that reduces the risk of death for CRC, but the sensitivity is low and this method can be affect by the food patients eat. Just as the first generation multimarker panel of the PreGen-Plus, it is also important to choose a combination of hypermethylated markers which will display a high sensitivity and specificity in detecting CRC. Maybe secreted frizzled-related protein gene 2[21] and tissue factor pathway inhibitor 2[15] are the best choice. At present, some new techniques have taken off, such as such as the Quantitative allele-specific real-time target, signal amplification technology, Tem-PCR. These methods may be compatible for detecting multiple DNA methylated biomarkers in stool samples[22-24]. But still, it may be hard to reach 100 % sensitivity with methylated biomarkers to screening CRC only because a portion of CRC, the so-called CpG Island Methylator Phenotype (CIMP)-negative tumors has no or low frequencies of methylated genes[25, 26]. Base on this phenomenon, some combined test may be needed, such as fecal occult blood test. Another question is that incomplete bisulfite modification might result in the false positive. Also, the cost for detecting CRC by using the MSP method need to be solved.
In conclusion, detection of SPG20 promoter hypermethylation is a highly specific and sensitive biomarker for colorectal cancers. In stool samples, SPG20 also shows an equal sensitivity and specificity that it is very promising to choose SPG20 as a biomarker in the noninvasive screening for CRC.
Abbreviations
CRC: colorectal cancer; SPG20: Spastic paraplegia-20; MSP: methylation-specific polymerase chain reaction.
Acknowledgements
This work was supported in part by a grant from the Scientific and Technologic Bureau of Xi'an.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011. CA: A Cancer Journal for Clinicians. 2011
2. Hardcastle JD, Chamberlain JO, Robinson MHE, Moss SM, Amar SS, Balfour TW. et al. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. The Lancet. 1996;348:1472-7
3. de Wijkerslooth T, Bossuyt P, Dekker E. strategies in screening for colon carcinoma. Neth J Med. 2011;69:112-9
4. Burt RW, Barthel JS, Dunn KB, David DS, Drelichman E, Ford JM. et al. Colorectal cancer screening. Journal of the National Comprehensive Cancer Network. 2010;8:8-61
5. Kisiel JB, Ahlquist DA. Stool DNA screening for colorectal cancer: opportunities to improve value with next generation tests. Journal of clinical gastroenterology. 2011;45:301
6. van Dam L, Kuipers EJ, van Leerdam ME. Performance improvements of stool-based screening tests. Best Practice & Research Clinical Gastroenterology. 2010;24:479-92
7. Lansdorp-Vogelaar I, Knudsen AB, Brenner H. Cost-effectiveness of colorectal cancer screening. Epidemiologic reviews. 2011;33:88-100
8. Hellebrekers DMEI, Lentjes MHFM, Van Den Bosch SM, Melotte V, Wouters KAD, Daenen KLJ. et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clinical Cancer Research. 2009;15:3990-7
9. Mayor R, Casadomé L, Azuara D, Moreno V, Clark S, Capella G. et al. Long-range epigenetic silencing at 2q14. 2 affects most human colorectal cancers and may have application as a non-invasive biomarker of disease. British journal of cancer. 2009;100:1534-9
10. Melotte V, Lentjes MHFM, van den Bosch SM, Hellebrekers DMEI, de Hoon JPJ, Wouters KAD. et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. Journal of the National Cancer Institute. 2009;101:916-27
11. Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Molecular biology of the cell. 2007;18:1683-92
12. Lind G, Raiborg C, Danielsen S, Rognum T, Thiis-Evensen E, Hoff G. et al. SPG20, a novel biomarker for early detection of colorectal cancer, encodes a regulator of cytokinesis. Oncogene. 2011;30:3967-78
13. Klaassen CHW, Jeunink MAF, Prinsen CFM, Ruers TJM, Tan AC, Strobbe LJA. et al. Quantification of human DNA in feces as a diagnostic test for the presence of colorectal cancer. Clinical chemistry. 2003;49:1185-7
14. Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127-39
15. Glöckner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J. et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer research. 2009;69:4691
16. Kim MS, Louwagie J, Carvalho B, sive Droste JST, Park HL, Chae YK. et al. Promoter DNA methylation of oncostatin m receptor-β as a novel diagnostic and therapeutic marker in colon cancer. PLoS One. 2009;4:e6555
17. Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ. et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Annals of internal medicine. 2008;149:441-50
18. Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH, Park DI. Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Diseases of the Colon & Rectum. 2009;52:1452
19. Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L. et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. The American journal of gastroenterology. 2008;103:2862-70
20. Sagona AP, Stenmark H. Cytokinesis and cancer. FEBS letters. 2010;584:2652-61
21. Chang E, Park DI, Kim YJ, Kim BK, Park JH, Kim HJ. et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepato-gastroenterology. 2010;57:720-7
22. Berger BM, Ahlquist DA. Stool DNA screening for colorectal neoplasia: biological and technical basis for high detection rates. Pathology-Journal of the RCPA. 2012;44:80
23. Zou H, Allawi H, Cao X, Domanico M, Harrington J, Taylor WR. et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clinical chemistry. 2012;58:375-83
24. Deng L, Qi Z, Zou B, Wu H, Huang H, Kajiyama T. et al. Digital Detection of Multiple Minority Mutants in Stool DNA for Noninvasive Colorectal Cancer Diagnosis. Analytical chemistry. 2012;84:5645-52
25. Ostwald C, Linnebacher M, Weirich V, Prall F. Chromosomally and microsatellite stable colorectal carcinomas without the CpG island methylator phenotype in a molecular classification. International journal of oncology. 2009;35:321
26. Bosch LJW, Mongera S, sive Droste JST, Oort FA, van Turenhout ST, Penning MT. et al. Analytical sensitivity and stability of DNA methylation testing in stool samples for colorectal cancer detection. Cellular Oncology. 2012:1-7
Author contact
![]() Corresponding author: Cheng-Xue Dang PhD, The Department of surgical oncology the First Affiliated Hospital, Xi'an Jiaotong University College of Medicine, 277 W. Yanta Road, Xi'an, Shaanxi 710061, China, dangchengxuecom.cn.
Corresponding author: Cheng-Xue Dang PhD, The Department of surgical oncology the First Affiliated Hospital, Xi'an Jiaotong University College of Medicine, 277 W. Yanta Road, Xi'an, Shaanxi 710061, China, dangchengxuecom.cn.

 Global reach, higher impact
Global reach, higher impact