Impact Factor
ISSN: 1449-1907
Int J Med Sci 2026; 23(1):313-324. doi:10.7150/ijms.124473 This issue Cite
Review
Role of Circular RNAs in Liver Diseases
Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, P.O. Box 2014, Jouf University, Aljouf Province, Saudi Arabia.
Received 2025-8-30; Accepted 2025-11-24; Published 2026-1-1
Abstract
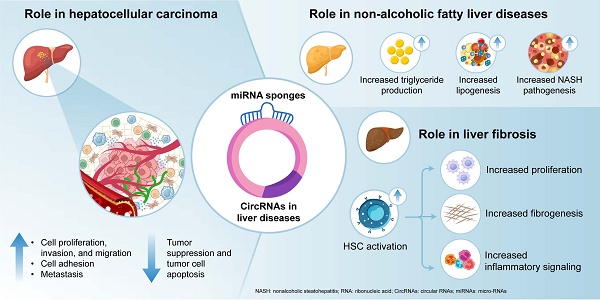
Circular RNAs (circRNAs) are a subclass of noncoding RNAs characterized by their closed-loop structure without terminal 3′ or 5′ ends. Studies have shown that circRNAs play pivotal roles in the regulation of various cellular processes. These molecules function as microRNA (miRNA) sponges, interact with RNA-binding proteins, and modulate gene transcription. CircRNAs are vital for regulating liver homeostasis, and dysregulation of their expression is correlated with liver diseases such as hepatic fibrosis, steatosis, inflammation, and liver cancer. Elucidating the functional significance of circRNAs in liver diseases is crucial, as this knowledge may facilitate the identification of novel diagnostic biomarkers and therapeutic targets for conditions that contribute significantly to global morbidity and mortality. This review aimed to highlight current research underscoring the functional roles of circRNAs in the molecular pathogenesis and progression of liver diseases, including hepatocellular carcinoma, nonalcoholic fatty liver disease, and liver fibrosis. To provide an updated and comprehensive overview, a literature search was conducted across major scientific databases. This review reveals that circRNAs perform multifaceted functions in liver homeostasis and disease by regulating gene expression through miRNA sponging, interacting with signaling pathways, and influencing cellular processes, including vascularization, metastasis, the cell cycle, apoptosis, cellular stress, metabolic activity, inflammatory responses, and cellular senescence. Despite their pivotal involvement in liver diseases, translating circRNA-based research into clinical practice remains challenging. In conclusion, circRNAs represent an emerging frontier in liver disease research, offering considerable promise for future clinical applications.
Keywords: circular RNA, liver, NAFLD, liver fibrosis, HCC
Introduction
Circular RNAs (circRNAs) represent a distinct subclass of noncoding RNAs that exist in a covalently closed-loop form. Their closed-loop structure, devoid of enzyme recognition elements, confers resistance to exonuclease degradation; thus, circRNAs exhibit greater stability and longer half-lives than linear RNAs [1]. Although predominantly located in the cytoplasm, circRNAs are also present in the nucleus; however, the mechanisms governing their export from the nucleus remain poorly understood. CircRNAs were first discovered in plant viroids in 1976 and later in the hepatitis delta virus. Two principal models have been proposed to explain circRNA biogenesis: exon skipping (lariat) and back splicing [2]. Based on their biogenesis, several types of circRNAs have been identified, including sense overlapping, antisense, intronic, exonic, and intergenic circRNAs [3].
CircRNAs affect the expression of many mammalian genes both transcriptionally and post-transcriptionally through transcriptional regulation, collaboration with microRNAs (miRNAs), and splicing interference. During transcription, nuclear circRNAs interact with RNA polymerase II and the U1 small nuclear ribonucleoprotein particle complex [4]. A subset of circRNAs is produced through the back-splicing of pre-mRNAs to generate circular transcripts. CircRNAs serve as binding sites for miRNAs and RNA-binding proteins, facilitating their role in post-transcriptional gene regulation. In addition, circRNAs can inhibit linear RNA splicing and regulate their expression. For example, Drosophila muscle-blind circRNAs drive their expression through the alternative splicing of precursor RNA [2].
Although circRNAs are considered non-coding RNAs, they can also encode proteins. For example, the groundbreaking research by van Heesch et al. on 80 human heart translatomes identified at least 40 circRNAs that encode proteins [5]. Another example is the hepatitis delta virus circRNA, which translates into a 122-amino acid protein within infected cells. In protein-coding circRNAs, cap-independent translation occurs via internal ribosome entry sites, N6-methyladenosine, or rolling circle amplification [6].
Under physiological conditions, circRNAs are involved in tissue and organ growth and development, and they regulate numerous cellular processes, including the cell cycle, cell stress, cellular senescence, metabolic activity, apoptosis, and inflammatory responses. Since circRNAs are resistant to standard RNA degradation pathways, cells eliminate them primarily via exocytosis. CircRNAs or their complexes are packaged into vesicles that are secreted into the extracellular space for removal from the cytoplasm [7]. Furthermore, the endoribonuclease RNase L degrades circRNAs [8].
CircRNAs accumulate primarily in slowly proliferating cells. Their expression varies across developmental stages and is altered in many diseases. Their low expression has hampered their characterization; however, because of their distinctive tissue-specific expression, they are useful as diagnostic biomarkers and novel therapeutic agents [3]. Next-generation sequencing and genome mapping tools have positioned circRNAs as a key focus of RNA research.
Understanding the functions of circRNAs in liver diseases is essential, as it may facilitate the discovery and validation of novel diagnostic biomarkers and therapeutic agents for disorders with substantial global health burdens. This review aimed to critically examine the evolving functions of circRNAs in the progression and pathogenesis of liver diseases.
CircRNA in the Liver
The liver is comprised of various cells that work together to regulate essential functions. Hepatic cells mediate lipid homeostasis, glucose metabolism, energy balance, immune responses, and detoxification. Globally, liver diseases are major causes of morbidity and impose a substantial economic burden on healthcare systems. They contribute to over two million deaths each year: one million due to complications from fibrosis and cirrhosis, and one million attributed to hepatocellular carcinoma (HCC) and viral hepatitis [9]. The mechanisms driving liver disease development are not fully understood, and treatment options for end-stage liver conditions remain limited, highlighting the need for early interventions and novel therapeutic strategies.
CircRNAs are important regulators of liver homeostasis and disease. One study demonstrated that 668 circRNAs are expressed exclusively in liver tissues [10]. In addition, the RAISE pipeline detected circRNAs in RNA-seq data from 61 human liver samples after rRNA depletion. In total, 59,128 circRNA candidates were observed in both adjacent non-tumor and HCC tissues [11].
CircRNAs in Liver Diseases
HCC
In 2020, HCC ranked sixth in global cancer incidence and third in cancer-related mortality. It accounts for 75‒85% of all primary liver cancer cases. Primary contributors to the risk of developing HCC include metabolic syndrome, viral infection, prolonged alcohol consumption, and conditions related to obesity and diabetes. Despite advances in treatments, including surgery and systemic drug therapies, the overall survival rate for patients with HCC remains poor, largely due to late diagnosis and the high degree of tumor heterogeneity in this cancer type [12].
Oncogenic circRNAs in HCC
circRNAs are crucial for regulating the initiation and progression of HCC [13]. For example, hsa_circ_0005075 interacts with miR-93-3p, miR-23b-5p, miR-23a-5p, and miR-581 to regulate cell adhesion during HCC development [14]. In hepatitis B-related HCC, upregulation of circRNA_100338, which targets miR-141-3p, is associated with poor survival and metastatic progression [15]. Additional circRNAs implicated in liver cancer include circFUT8, circZFR, and circIOP11. Specifically, circFUT8 competitively binds to miR-17-3p, miR-570-3p, and miR-552-3p; circZFR potentially interacts with miR-130b-5p, miR-511-5p, miR-642a-5p, miR-329-5p, and miR-532-3p; and circIPO11 targets miR-659-3p, miR-106a-3p, and miR-424-5p [16]. Circ-HOMER1, which is upregulated in HCC cells and tissues, stimulates HCC growth by targeting the miR-1322/CXCL6 axis [17]. RhoA and circ_000839 levels are elevated in HCC, and miR-200b expression is negatively correlated with circ_000839 levels [18]. Circβ-catenin is upregulated in liver cancer tissues, and its downregulation reduces β-catenin protein concentrations without affecting its mRNA concentration. Circβ-catenin also plays a role in stabilizing full-length β-catenin by counteracting GSK3β-mediated phosphorylation and subsequent β-catenin degradation, thereby contributing to Wnt pathway activation [19]. Elevated levels of circACVR2A have been detected in HCC cell lines. CircACVR2A interacts with miR-511-5p to regulate the signaling axis [20]. Both HCC tissues and cells exhibit high levels of circPIAS1, and suppression of circPIAS1 reduces cell proliferation and migration. Overexpression of circPIAS, which binds to miR-455-3p, inhibits ferroptosis and upregulates nuclear protein 1, which subsequently activates FTH1 transcription, promoting iron storage [21]. In HCC, circESYT2 is upregulated and promotes tumor growth and metastasis by interacting with the miR-665/enolase 2 axis [22]. Elevated levels of circ_0067934 in HCC tissues, which correlate with increased tumor metastasis and growth, are associated with the miR-1324 and Wnt/β-catenin pathway [23]. In HCC, SCD-circRNA2 expression, which is regulated by RNA-binding protein 3 (RBM3), is upregulated. Modulation of SCD-circRNA2 and RBM3 levels increases HCC cell proliferation. Furthermore, RBM3-SCD-circRNA2 regulates p-ERK activation [24]. Hsa_circ_0000092, which targets miRNA-338-3p, is upregulated in HCC, and its downregulation correlates with reduced cell invasion, proliferation, and angiogenesis by decreasing HN1 expression [25]. Circ-PRMT5 overexpression potentially plays a critical role in HCC cell glycolysis, migration, and proliferation by targeting miR-188-5p/HK2 [26]. Mechanistically, circMAT2B stimulates the expression of the glycolytic enzyme PKM2 by interacting with miR-338-3p [27]. Glycolysis plays a pivotal role in HCC owing to its role in metabolic reprogramming, which is a hallmark of cancer. In the presence of sufficient oxygen, HCC cells preferentially utilize aerobic glycolysis rather than mitochondrial oxidative phosphorylation to generate ATP and biosynthetic macromolecules essential for rapid proliferation. Accelerated HCC growth usually exceeds angiogenesis, resulting in hypoxic conditions that further reinforce glycolytic flux to sustain anabolic demands [26, 27]. CircASAP1, which regulates miR-532-5p- and miR-326-mediated signaling, increases in HCC tissues. CircASAP1 stimulates HCC cell invasion and proliferation, and regulates macrophage infiltration into tumor tissues [28]. CircUHRF1, secreted by HCC cells, suppresses the immune response by inhibiting natural killer cell function via upregulation of TIM-3 expression [29]. CircRHOT1, which modulates TIP60 recruitment to the NR2F6 promoter, contributes to the activation of NR2F6 transcription and is upregulated in HCC [30]. Circ_0000105 is overexpressed in liver cancer and enhances phosphoinositide-3-kinase regulatory subunit 1 expression by targeting miR-498; its overexpression correlates with increased HCC proliferation and reduced apoptosis [31]. Circ_0091579, which targets miRNA-490-3p, is upregulated in HCC. Its downregulation strongly inhibits cell proliferation and metastasis [32]. High levels of circPRKCI in HCC suppress apoptosis and promote invasion by interacting with miRNA-545 to regulate E2F7 and reduce AKT3 protein expression [33]. HCC migration, proliferation, and invasion increase via the upregulation of hsa_circRNA_100084, which may stimulate insulin-like growth factor 2 (IGF2) by sequestering miR-23a-5p [34]. CircSOD2 overexpression correlates with cell migration, cell growth, and the cell cycle. CircSOD2 inhibits miR-502-5p, thereby suppressing SOCS3 expression and activating the Janus kinase 2 (JAK2)/STAT3 pathway [35]. MUC1, which is elevated in tumor cells, is repressed by miRNA-485-5p. Downregulation of MUC1 in cells is correlated with decreased cell viability, invasion, and migration but increased apoptosis. Upregulation of circHECTD1, which interacts with miRNA-485-5p, increases MUC1 expression and promotes HCC progression [36]. In HCC, circ_0016788 is also elevated [37,38]. Loss of circ_0016788 inhibits tumor growth in vivo; in vitro, HCC cell proliferation, invasion, colony formation, cell vitality, and glycolysis are suppressed, whereas apoptosis is enhanced. Circ_0016788 mediates its effects through themiR-506-3p/ poly(ADP-ribose) polymerase family member 14 (PARP14) and miR-486/CDK4 pathways [37,38]. CircZNF566 is implicated in HCC metastasis and tumorigenesis; in vitro, it targets the tryptophan 2,3-dioxygenase axis via miR-4738-3p to promote cell invasion, proliferation, and migration [39]. CircTMEM45A, which acts via the miR-665/IGF2 axis, is upregulated in HCC and correlates with tumorigenesis and the progression of cell mobility [40]. Tumor stage, size, and vascular invasion are correlated with elevated circ-0046600 expression, and suppression of circ-0046600 inhibits cell migration. Most hsa-circ-0046600 is located in the cell cytoplasm, where it stimulates HIF-1α expression through binding miR-640 [41]. The knockdown of circMAN2B2, which is markedly elevated in HCC, suppresses cell proliferation by sponging the miR-217 and regulating the mitogen-activated protein kinase 1 pathway [42]. Increased circPTGR1 expression, which has three isoforms, in serum exosomes of patients with HCC is indicative of advanced tumor stage and worse prognosis. Loss of circPTGR1 is correlated with reduced migration and invasion of 97L and HepG2 cells. The circPTGR1 isoforms and MET compete to specifically bind miR449a [43]. Elevated circPVT1 expression in HCC is associated with reduced miR-377 levels. Knockdown of circPVT1 impairs HCC tumor growth, suppresses glycolysis and proliferation, and enhances apoptosis. CircPVT1 in HCC is regulated by binding to miR-377 [44]. Circ_0008450 overexpression has been detected in HCC [45,46]. Downregulation of hsa_circ_0008450 results in reduced migration, proliferation, and invasion, while enhancing apoptosis. Hsa_circ_0008450 upregulates zeste homolog 2 by sponging miR-214-3p and miR-548p [45,46]. CircRNA-104718, which is overexpressed in HCC, binds to miR-218-5p, thereby increasing the expression of thioredoxin domain-containing protein 5. Thus, higher circRNA-104718 levels accelerate invasion, proliferation, and migration, while downregulating apoptosis in HCC cells. In a mouse model, upregulation of circRNA-104718 increased tumor size and promoted HCC metastasis [47]. Circ-ZNF652 is elevated in both the serum and tumor cells of patients with HCC. Circ-ZNF652 influences cell glycolysis, invasion, proliferation, and migration by interacting with miR-29a-3p, thereby modulating guanylyl cyclase domain-containing 1 [48]. Increased circ_0000267 expression correlates with poor prognosis and increased severity of HCC, and its upregulation stimulates cell growth by binding to miR-646 [49]. Circ-FOXP1 is significantly upregulated in the serum and tissues of patients with HCC. Circ-FOXP1 overexpression upregulates the oncogenic transcription factor sex-determining region Y-box 9 through miR-875-3p and miR-421. Its overexpression leads to accelerated tumor growth and reduced apoptosis in HCC cells [50]. Upregulation of hsa_circ_101280 in HCC, which is associated with enhanced tumor cell proliferation and decreased apoptosis, is mediated by miR-375 sponging and JAK2 activation [51]. In HCC cells, knockdown of circFBLIM1 levels inhibits invasion and proliferation while enhancing apoptosis. CircFBLIM1 sponges miR-346 to regulate FBLIM1 expression [52]. In HCC cells, knocking down high circABCC2 levels reduces invasion and proliferation while enhancing apoptosis. CircABCC2 upregulates ABCC2 expression by interacting with miR-665 [53].
Tumor-suppressor circRNAs in HCC
Lower hsa_circ_0001649 expression in HCC is associated with the presence of a tumor embolus and larger tumor size [54,55]. Both in vitro and in vivo studies have demonstrated that the upregulation of circ-0001649 reduces HCC migration and proliferation. Hsa_circ_0001649 mediates its effects through activation of SNF2 histone linker PHD RING helicase by sponging miR-4688, miR-127-5p, and miR-612 [54]. Circ-ITCH expression is markedly reduced in cancerous tissues compared to controls [56]. Downregulation of hsa_circ_0005986 accelerates HCC cell proliferation, and higher levels of hsa_circ_0005986 correlate with better survival outcomes in patients with cancer. Circ_0005986 downregulation results in increased miR-129-5p expression, which reduces Notch1 mRNA levels [57]. Hsa_circ_0004018, which is suppressed in HCC relative to adjacent non-tumorous tissues, is involved in HCC metastasis and carcinogenesis via the miR-626/miR-30e-5p-MYC pathway [58]. Patients with HCC and reduced circMTO1 expression (hsa_circRNA_104135/ hsa_circRNA_0007874) have shorter survival times. CircMTO1 is associated with miR-9, and its silencing in HCC decreases p21 levels, which promotes tumor growth and invasion [59]. Decreased hsa-circ-0000221 expression correlates with reduced PTPN11 mRNA expression in the serum of patients with HCC. PTPN11 inhibits proliferation in various human cancers, while these patients exhibit higher miR-661 expression. Overexpression of hsa-circ-0000221 reduces cell viability and increases apoptosis, correlating with the accumulation of G1 and upregulation of CCDN1 in HCC. Hsa-circ-0000221 regulates PTPN11 expression by inhibiting miR-661 [60]. The zinc-finger family gene ZKSCAN1 and its circRNA, circZKSCAN, are downregulated in HCC. Inhibition of circZKSCAN1 induces cell proliferation, invasion, and migration. CircZKSCAN1 modulates cancer-associated pathways, including integrin β4, transforming growth factor-β1 (TGF-β1), and chemokine receptor 4 [61]. Hsa_circ_0067531, which is critical for HCC development via phosphoinositide 3-kinase, is reduced in human HCC tissues in vitro [62]. In HCC, circTRIM33-12 expression is reduced and correlates with increased invasion, migration, proliferation, and immune evasion. CircTRIM33-12 promotes Ten-eleven translocation methylcytosine dioxygenase 1 expression and reduces 5-hydroxymethylcytosine levels through miR-191 [63]. CircHIAT1 suppresses tumor growth by acting as a sponge for miR-3171, thereby upregulating PTEN, and is downregulated in patients with HCC [64]. CircLARP4 is also downregulated in HCC. CircLARP, which mediates cell cycle arrest in vitro, reduces HCC proliferation and promotes senescence. HCC progression is inhibited by circLARP through miR-761, thereby activating the RUNX3 and p53/p21 pathway [65]. The altered expression of circRNAs and their implicated roles in HCC are summarized in Table 1.
Expression and roles of dysregulated circular RNAs in hepatocellular carcinoma
| Reference | Biological Function | Sponge Target | Expression Pattern | Circular RNA |
|---|---|---|---|---|
| [14] | Hsa_circ_0005075 regulates cell adhesion. | miR-93-3p miR-23b-5p miR-23a-5p miR-581 | Increased | Hsa_circ_0005075 |
| [15] | Elevated circRNA_100338 levels are linked to decreased survival and metastatic advancement. | miR-141-3p | Increased | CircRNA_100338 |
| [16] | Not identified | miR-570-3p miR-17-3p miR-552-3p | Increased | CircFUT8 Hsa_circ_0003028 Hsa_circ_101368 |
| [16] | Not identified. | miR-511-5p miR-532-3p miR-130b-5p miR-329-5p miR-642a-5p | Increased | CircZFR Hsa_circ_10072088 Hsa_circRNA_103809 |
| [16] | Not identified. | miR-659-3p miR-106a-3p miR-424-5p | Increased | CircIOP11 Hsa_circ_0007915 Hsa_circ_103847 |
| [17] | Circ-HOMER1 stimulates migration, proliferation, and invasion, while reducing apoptosis. HOMER1 targets CXCL6. | miR-1322 | Increased | Circ-HOMER1 |
| [18] | Not identified. | Not identified | Increased | Circ_000839 |
| [19] | Circβ-catenin activates Wnt/β-catenin. | Not identified | Increased | Circβ-catenin |
| [20] | CircACVR2A regulates invasion, proliferation, and migration. CircACVR2A regulates PI3K-Akt. | miR-511-5p | Increased | CircACVR2A |
| [21] | CircPIAS1 knockdown reduces migration and proliferation. Its overexpression suppresses ferroptosis and upregulates NUPR1. | miR-455-3p | Increased | CircPIAS1 |
| [22] | CircESYT2 mediates metastasis and growth through its interaction with ENO2. | miR-665 | Increased | CircESYT2 |
| [23] | Circ_0067934 overexpression promotes growth and metastasis. It regulates FZD5/Wnt/β-catenin. | miR-1324 | Increased | Circ_0067934 |
| [24] | SCD-circRNA2 upregulation is mediated by RBM3. RBM3-SCD-circRNA2 promotes proliferation and p-ERK activation. | Not identified | Increased | SCD-circRNA2 |
| [25] | Hsa_circ_0000092 increases progression by upregulating HN1 expression. | miR-338-3p | Increased | Hsa_circ_0000092 |
| [26] | Circ-PRMT5 mediates migration, proliferation, and glycolysis through the HK2 axis. | miR-188-5p | Increased | Circ-PRMT5 |
| [27] | CircMAT2B stimulates PKM2 gene expression, which encodes a glycolysis enzyme. | miR-338-3p | Increased | CircMAT2B |
| [28] | CircASAP1 stimulates invasion and proliferation and regulates the infiltration of macrophages. | miR-326 miR-532-5p | Increased | CircASAP1 |
| [29] | CircUHRF1 regulates immunosuppression by inhibiting natural killer cell functions through upregulation of TIM-3 expression. | Not identified | Increased | CircUHRF1 |
| [30] | CircRHOT1 promotes HCC proliferation and metastasis. It recruits TIP60 to the promoter of NR2F6 to initiate the transcription of NR2F6. | Not identified | Increased | CircRHOT1 |
| [31] | Circ_0000105 promotes proliferation and reduces apoptosis in HCC. Circ_0000105 promotes PIK3R1 expression. | miR-498 | Increased | Circ_0000105 |
| [32] | Circ_0091579 promotes HCC proliferation and metastasis. | miRNA-490-3p | Increased | Circ_0091579 |
| [33] | circ-PRKCI significantly inhibits HCC cell apoptosis and promotes cell invasion. | miRNA-545 | Increased | Circ-PRKCI |
| [34] | Hsa_circRNA_100084 stimulates IGF2. | miR-23a-5p | Increased | Hsa_circRNA_100084 |
| [35] | CircSOD2 promotes growth, cycle, and migration. Increased DNMT3a downregulates SOCS3 and enhances JAK2/STAT3. | miR-502-5p | Increased | CircSOD2 |
| [36] | CircHECTD1 facilitates MUC1 expression and promotes progression. | miRNA-485-5p | Increased | CircHECTD1 |
| [37,38] | Hsa_circ_0016788 mediates CDK4 and PARP14. | miR-506-3p miR-486 | Increased | Hsa_circ_0016788 |
| [39] | CircZNF566 upregulation increases tumorigenesis, metastasis, invasion, migration, and proliferation. CircZNF566 regulates the TDO2 axis. | miR-4738-3p | Increased | CircZNF566 |
| [40] | CircTMEM45A regulates cell mobility and tumorigenesis. Its action is mediated by the IGF2 axis. | miR-665 | Increased | CircTMEM45A |
| [41] | Hsa-circ-0046600 mediates migration. | miR-640 | Increased | Hsa-circ-0046600 |
| [42] | CircMAN2B2 regulates proliferation via MAPK1. | miR-217 | Increased | CircMAN2B2 |
| [43] | CircPTGR1 isoforms stimulate migration by targeting MET. | miR449a | Increased | CircPTGR1 |
| [44] | CircPVT1 regulates proliferation, glycolysis, and apoptosis by targeting TRIM23. | miR-377 | Increased | CircPVT1 |
| [45,46] | Hsa_circ_0008450 regulates proliferation, invasion, and apoptosis by promoting EZH2. | miR-548p miR-214-3p | Increased | Hsa_circ_0008450 |
| [47] | CircRNA-104718 overexpression increases tumor size, promotes metastasis, migration, invasion, and proliferation, and inhibits apoptosis. | miR-218-5p | Increased | CircRNA-104718 |
| [48] | Circ-ZNF652 regulates glycolysis, proliferation, invasion, and migration through GUCD1. | miR-29a-3p | Increased | Circ-ZNF652 |
| [49] | Circ_0000267 upregulation promotes invasion, migration, and proliferation, and ameliorates apoptosis. | miR-646 | Increased | Circ_0000267 |
| [50] | Overexpression of circ-FOXP1 increases oncogenic transcription factor SOX9 expression, cell growth, and invasion, and suppresses apoptosis. | miR-421 miR-875-3p | Increased | Circ-FOXP1 |
| [51] | Hsa_circ_101280 mediates tumorigenesis and proliferation, while reducing apoptosis by JAK2. | miR-375 | Increased | Hsa_circ_101280 |
| [52] | CircFBLIM1 knockdown increases apoptosis and decreases cell proliferation and invasion. | miR-346 | Increased | CircFBLIM1 |
| [53] | CircABCC2 inhibition reduces proliferation and invasion and increases apoptosis by regulating ABCC2. | miR-665 | Increased | CircABCC2 |
| [54,55] | Circ-0001649 upregulation reduces proliferation and migration. It mediates SHPRH activation. | miR-127-5p miR-4688 miR-612 | Decreased | Hsa_circ_0001649 |
| [56] | Polymorphisms (rs4911154 and rs10485505) in circ-ITCH increase the risk of HCC. | Not identified | Decreased | Circ-ITCH |
| [57] | Downregulation of hsa_circ_0005986 stimulates proliferation and reduces Notch1 expression. | miR-129-5p | Decreased | Hsa_circ_0005986 |
| [58] | Hsa_circ_0004018 regulates metastasis and carcinogenesis by targeting MYC. | miR-626 miR-30e-5p | Decreased | Hsa_circ_0004018 |
| [59] | CircMTO1 silencing reduces p21 expression as well as proliferation and invasion. | miR-9 | Decreased | CircMTO1 Hsa_circRNA_104135 Hsa_circRNA_0007874 |
| [60] | Low hsa-circ-0000221 expression reduces PTPN11 expression. Its overexpression lowers cell viability, increases apoptosis, and upregulates G1 protein and CCDN1. | miR-661 | Decreased | Hsa-circ-0000221 |
| [61] | Silencing circZKSCAN1 in HCC induces proliferation, invasion, and migration. CircZKSCAN1 influences TGF-β1, ITGB4, and CXCR4 expression. | Not identified | Decreased | CircZKSCAN1 |
| [62] | Hsa_circ_0067531 regulates the PI3K pathway. | Not identified | Decreased | Hsa_circ_0067531 |
| [63] | CircTRIM33-12 reduction increases migration, invasion, proliferation, and immune evasion by promoting TET1 expression and reducing 5hmC expression. | miR-191 | Decreased | CircTRIM33-12 |
| [64] | CircHIAT1 inhibits growth and upregulates PTEN expression. | miR-3171 | Decreased | CircHIAT1 |
| [65] | CircLARP4 reduces proliferation, mediates cell cycle arrest, and promotes senescence by increasing RUNX3 expression and activating p53/p21. | miR-761 | Decreased | CircLARP4 |
Abbreviations: HCC: hepatocellular carcinoma; circRNA: circular RNA; miR / miRNA: microRNA; MAPK1: mitogen-activated protein kinase 1; JAK2: Janus kinase 2; EZH2: enhancer of zeste homolog 2; TXNDC5: thioredoxin domain-containing protein 5; MET: mesenchymal-epithelial transition factor; PTPN11: protein tyrosine phosphatase non-receptor type 11; CCDN1: cyclin D1; SOX9: SRY-box 9; FZD5: frizzled class receptor 5; PKM2: pyruvate kinase M2; HKL2: hexokinase-like 2; DNMT3a: DNA methyltransferase 3 alpha; SOCS3: suppressor of cytokine signaling 3; STAT3: signal transducer and activator of transcription 3; IGF2: insulin-like growth factor 2; CDK4: cyclin-dependent kinase 4; PARP14: poly (ADP-ribose) polymerase family member 14; TDO2: tryptophan 2,3-dioxygenase; GUCD1: guanylyl cyclase domain containing 1; SHPRH: SNF2 histone linker PHD RING helicase; ITGB4: integrin beta 4; TGF-β1: transforming growth factor beta 1; CXCR4: chemokine receptor 4; PI3K: phosphoinositide 3-kinase; TET1: ten-eleven translocation methylcytosine dioxygenase 1; 5hmC: 5-hydroxymethylcytosine; PTEN: phosphatase and tensin homolog; RUNX3: runt-related transcription factor 3; p-ERK: phosphorylated extracellular signal-regulated kinase; TRIM23: tripartite motif containing 23; FBLIM1: filamin-binding LIM protein 1.
Nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH)
NAFLD, the primary cause of chronic liver disease worldwide, is characterized by abnormal fat accumulation in hepatocytes. Triglyceride accumulation, lipid peroxidation, and mitochondrial dysfunction are the hallmarks of liver steatosis [66]. With the global prevalence of NAFLD at approximately 30%, it may increase over the next two decades. Therefore, raising awareness and promoting the understanding of NAFLD remain crucial. The inadequate response of healthcare organizations places a substantial strain on healthcare systems and economies [66].
NAFLD is a multisystem condition associated with metabolic disorders. NASH, a more advanced form of NAFLD, can progress to hepatic fibrosis, cirrhosis, and eventually liver cancer. The hallmark features of NASH include hepatocyte ballooning and hepatic inflammation [66]. NAFLD affects both children and adults, with its prevalence increasing with age in males between 45 and 65 years [67]. Genetic factors, including polymorphisms in PNPLA3 and TM6SF2 genes, elevate the risk of NAFLD [68].
CircRNAs are increasingly recognized for their distinctive expression patterns in NAFLD. Significant differences in circRNA expression have been observed in NASH and NAFLD animal models [69]. In one NAFLD animal model, 231 circRNAs were upregulated and 165 were downregulated, as quantified using a circRNA microarray [70]. Another study revealed that the expression of 57 circRNAs was upregulated and that of 36 circRNAs was downregulated in a high-fat diet-fed mouse model. CircRNAs also regulate the expression of DDAH1 and VAV3 genes in NAFLD [71].
Circ_0057558 levels were elevated in NAFLD models. This circRNA acts as a sponge for miR-206 and enhances triglyceride production and lipogenesis by relieving the repression of AMP-activated protein kinase (AMPK) and Rho-kinase 1 signaling pathways [72]. CircRNA_002581 is also highly expressed in NASH. Knockdown of circRNA_002581 reduces hepatic inflammation, oxidative stress, and lipid accumulation, whereas its overexpression alleviates the suppression of cytoplasmic polyadenylation element-binding protein by miR-122. CircRNA_002581 contributes to NASH pathogenesis by suppressing autophagy through the PTEN-AMPK-mTOR axis [73]. CircRNA-homeodomain-interacting protein kinase 3 is induced in patients with NASH compared to controls, suggesting that its interaction with miRNA-29a regulates NASH pathogenesis. miRNA-29a may contribute to NASH pathogenesis by decreasing the activity of the Wnt-β-catenin pathway [74].
CircRNA_0046367 expression is diminished in hepatocellular steatosis models; however, its restoration interferes with miR-34a suppression of peroxisome proliferator-activated receptor α (PPARα) [75]. Restoration of PPARα activity improves hepatocellular steatosis by modulating genes involved in fatty acid oxidation, transport, and lipid metabolism [75]. CircRNA_0046366 plays a critical role in lipid metabolism, as its levels decrease in free fatty acid (FFA)-induced hepatocellular steatosis. CircRNA_0046366 counteracts the inhibitory impacts of miR-34a on PPARα. Enhanced gene expression of triglyceride-specific lipolysis, such as solute carrier family 27A, correlates with restored PPARα expression, thereby decreasing triglyceride content and alleviating hepatocellular steatosis [76]. CircScd1 expression is reduced in NAFLD tissues and regulates the degree of lipid accumulation. CircScd1 also reduces steatosis by activating the JAK2/STAT5 pathway [77]. In addition, hsa_circ_0048179 levels are suppressed in NAFLD models induced by oleate and palmitate, along with decreased glutathione peroxidase 4 (GPX4) expression, an antioxidant enzyme that protects cells by preventing peroxidation of membrane lipids. Overexpression of Hsa_circ_0048179 stimulates GPX4 expression via miR-188-3p, attenuating oleate/palmitate-induced reactive oxygen species (ROS), lipid accumulation, steatosis, and mitochondrial dysfunction in HepG2 cells [78]. In patients with NASH, the mitochondrial steatohepatitis-associated circRNA ATP5B Regulator (SCAR) limits fibroblast activation and mitochondrial ROS production. A reduction in its expression correlates with the transition from steatosis to NASH and insulin resistance. In vivo, modulation of circRNA SCAR attenuates insulin resistance and cirrhosis associated with a high-fat diet [79]. In rodents with NAFLD, a high-fat, high-cholesterol diet disrupts the hepatic circRNA profile, notably reducing 28 circRNAs. LNCPINT-derived circRNAs are critical regulators of circRNA, miRNA, and mRNA interactions. Deficiency of these circRNAs relieves the inhibition of miR-669c-3p and miR-466i-3p, resulting in AMPK deactivation. Downregulation of the AMPK pathway promotes lipogenic gene expression and drives hepatic steatosis [80]. CircRNA_0001805 expression is decreased in high-fat diet-fed mice, FFA-treated hepatocytes, and patients with NAFLD. Enhanced expression of circRNA_0001805 correlates with the attenuation of lipid metabolism abnormalities and inflammatory activity. CircRNA_0001805 targets miR-106a-5p and miR-320a, both of which act as upstream suppressors of ABCA1/CPT1 [81]. In hepatic steatosis, circRNA_021412 modulates the miR-1972/LPIN1 pathway, which regulates the expression of steatosis-related genes through PPARα activation [82]. Table 2 presents the correlation between circRNAs and NAFLD.
Hepatic Fibrosis
Liver fibrosis results from excessive buildup of extracellular matrix components triggered by ongoing liver damage, which promotes wound healing. Fibrosis-related liver injuries result from various factors, including chronic hepatitis B or C infections, drug use, genetic conditions, excessive alcohol intake, metabolic disorders, autoimmune diseases, and cholestasis. The rising incidence of type 2 diabetes has led to an increase in liver fibrosis due to NASH. Liver fibrosis can be reversed at early stages if the underlying causes are addressed. However, progression of fibrosis can cause cirrhosis, liver failure, portal hypertension, and HCC [83]. Hepatic fibrosis involves various cell types and mediators. Among these, hepatic stellate cells (HSCs), which are considered the main producers of fibrous matrices, are crucial for the initiation and progression of liver fibrosis and serve as primary effector cells during fibrosis development. Following liver injury, quiescent HSCs are activated by cytokines—such as interleukin 6 (IL-6), IL-17, and IL-22—which are secreted by adjacent cells. Once activated, HSCs transdifferentiate into myofibroblasts, which are involved in fibrosis development [84]. HSC activation is also triggered by lipopolysaccharide (LPS) and TGF-β, both of which increase the production of fibrotic markers [84]. Excessive production of the extracellular matrix disrupts the liver's structure, leading to the replacement of hepatocytes with scar tissue. Examples of extracellular matrix include fibronectin, laminin, collagen I, and collagen III. Since many circRNAs are involved in advancing or suppressing hepatic fibrosis, they represent promising biological markers for tracking the progression of liver fibrosis. In addition, these circRNAs provide critical insights into the mechanisms underlying hepatic fibrosis and highlight promising targets for diagnostic and therapeutic applications [84].
In irradiated HSCs, 179 circRNAs were highly expressed, whereas 630 circRNAs showed reduced expression relative to normal conditions. Among these altered circRNAs, hsa_circ_0072765, hsa_circ_0054345, and hsa_circ_0071410 were significantly upregulated, whereas hsa_circ_0070963, hsa_circ_0013255, and hsa_circ_0061893 were significantly downregulated. Silencing hsa_circ_0071410 elevated miR-9-5p expression, which attenuated HSC activation. Downregulation of miR-9-5p mitigates the effect of hsa_circ_0071410 inhibition, thereby reducing HSC activation [85]. CircUbe2k is upregulated in mice treated with carbon tetrachloride (CCl4) to induce liver fibrosis, as well as in LX-2 cells stimulated with TGF-β1. CircUbe2k increases TGF-β2 activity by targeting miR-149-5p, and suppression of circUbe2k inhibits the expression of fibrotic markers such as alpha smooth muscle actin (α-SMA) and collagen, type I, alpha 1 (Col1α1) [86]. Circ-PWWP2A expression promotes HSC activation and proliferation after LPS and TGF-β treatment, with MiR-223 and miR-203 identified as its downstream targets [87]. CircRNA-0067835 is markedly upregulated in LX-2 cells lacking thymosin β4, and silencing of this circRNA suppresses cell proliferation and enhances apoptosis. Bioinformatic analyses predict that circRNA-0067835 interacts with miR-155, thereby modulating forkhead box O3 expression through its sponging activity [88]. In LX2 cells exposed to radiation, CircRSF1 expression is increased and predicted to bind to miR-146a-5p. The circRSF1-miR-146a-5p complex increases cell viability, fibrosis, and inflammation via RAC1 activation [89]. CircTUBD1 promotes liver fibrosis by modulating miRNA-203a-3p and Smad signaling. In a radiation-induced liver fibrosis model, reducing circTUBD1 activity suppresses hepatic fibrosis biomarkers [90]. Hsa_circ_0072765 is upregulated in HSCs exposed to TGF-β1, and its knockdown ameliorates HSC activation and migration through the transient receptor potential vanillin 3 pathway [91]. In CCl4-treated mice and RAW264.7 cells stimulated with IFN-γ and LPS, circMcph1 expression is elevated. CircMcph1 regulates IL-1 receptor-associated kinase 2 (Irak2) activity during liver fibrosis by sponging miR-370-3p [92]. CircRNA-007371 exhibits angiogenic effects in a mouse fibrosis model induced by thioacetamide. Overexpression of circRNA-007371 promotes cell proliferation. Acting as a miRNA sponge, circRNA-007371 enhances angiogenesis [93]. CircRNA cVIM facilitates the upregulation of TGF-β receptor subtypes through miR-122-5p and miR-9-5p, resulting in the activation of the TGF-β/Smad signaling axis [94]. Circ_0008494 functions as a sponge for miR-185-3p in human fibrotic tissues, and knockdown of this interaction reduces HSC proliferation, activation, and migration while promoting apoptosis [95].
Expression and roles of dysregulated circular RNAs in nonalcoholic fatty liver disease
| Reference | Biological Function | Sponge Target | Expression Pattern | Circular RNA |
|---|---|---|---|---|
| [72] | Circ_0057558 promotes lipogenesis and triglyceride secretion by relieving the repression of ROCK1 and AMPK. | miR-206 | Increased | Circ_0057558 |
| [73] | CircRNA_002581 regulates CPEB1. CircRNA_002581 knockdown reduces oxidative stress, lipid accumulation, and inflammation. | miR-122 | Increased | CircRNA_002581 |
| [74] | CircRNA-HIPK3 overexpression downregulates Wnt/β-catenin, contributing to NASH pathogenesis. | miRNA-29a | Increased | CircRNA-HIPK3 |
| [75] | Normal circRNA_0046367 concentrations abolish the inhibitory effects of PPARα. | miR-34a | Decreased | CircRNA_0046367 |
| [76] | CircRNA_0046366 reduces triglycerides and ameliorates hepatocellular steatosis. Normal circRNA_0046366 concentrations abolish the inhibition of PPARα. | miR-34a | Decreased | CircRNA_0046366 |
| [77] | CircScd1 reduces the severity of lipid accumulation by JAK2/STAT5 activation. | Not identified | Decreased | CircScd1 |
| [78] | Hsa_circ_0048179 accelerates GPX4 and reduces reactive oxygen species and lipid accumulation. | miR-188-3p | Decreased | Hsa_circ_0048179 |
| [79] | CircRNA SCAR suppresses reactive oxygen species and fibroblast activation. It binds to ATP5B and suppresses mPTP activity by preventing CypD and mPTP linkage. | ATP5B | Decreased | CircRNA SCAR |
| [80] | Circ_0001452 suppression inhibits AMPK and promotes lipogenic gene expression. | miR-669c-3p miR-466i-3p | Decreased | Circ_0001452 Circ_0001453 Circ_0001454 |
| [81] | CircRNA_0001805 overexpression decreases lipid accumulation and mitigates lipid metabolism disorders and inflammation. It acts as ABCA1/CPT1 suppressor | miR-106a-5p miR-320a | Decreased | CircRNA_0001805 |
| [82] | CircRNA_021412 regulates LPIN1, inducing steatosis-related genes via PPARα activation. | miR-1972 | Decreased | CircRNA_021412 |
Abbreviations: AMPK: AMP-activated protein kinase; ROCK1: Rho-kinase 1; CPEB1: cytoplasmic polyadenylation element-binding protein 1; NASH: nonalcoholic steatohepatitis; PPARα: peroxisome proliferator-activated receptor alpha; JAK2: Janus kinase 2; STAT5: signal transducer and activator of transcription 5; GPX4: glutathione peroxidase 4; ATP5B: ATP synthase subunit beta; mPTP: mitochondrial permeability transition pore; CypD: cyclophilin D; ABCA1: ATP-binding cassette transporter A1; CPT1: carnitine palmitoyltransferase 1; miR / miRNA: microRNA.
CircPSD3, derived from the pleckstrin and Sec7 domain-containing 3 (PSD3) gene, is reduced in CCl4-treated mouse livers and primary HSCs. Furthermore, overexpression of circPSD3 is associated with decreased collagen deposition, reduced liver enzyme and hydroxyproline levels, and lower expression of profibrogenic and proinflammatory cytokines. CircPSD3 acts as a miR-92b-3p sponge, enhancing Smad7 expression [96]. CircFBXW4 expression is reduced during liver fibrosis. Increased circFBXW4 suppresses HSC proliferation and activation, promotes apoptosis, mitigates hepatic fibrotic damage in mice, and exhibits anti-inflammatory properties. Mechanistically, circFBXW4 interacts with miR-18b-3p during hepatic fibrosis [97]. Hsa_circ_0004018 expression decreases during liver fibrogenesis, and its overexpression suppresses fibrosis progression. By sponging hsa-miR-660-3p, hsa_circ_0004018 may indirectly reduce TEP1 expression [98]. CircCREBBP expression is also reduced during liver fibrosis. Overexpression of circCREBBP prevents liver fibrosis progression by inhibiting HSC proliferation and activation. Mechanistically, circCREBBP enhances LEFTY2 expression by interacting with hsa-miR-1291 [99]. Hsa_circ_0070963 reduces fibrotic damage by modulating miR-223-3p, which interacts with LEMD3. This circRNA is downregulated during fibrosis; however, restoring its normal expression suppresses HSC activation and attenuates fibrotic biomarkers [100]. In silico analysis indicated that the regulatory role of mmu_circ_34116 in HSC activation was mediated through the miR-22-3P/BMP7 signaling pathway. Suppression of mmu_circ_34116 stimulates α-SMA expression [101]. CircDIDO1 inhibits fibrosis by acting as an miR-141-3p sponge. CircDIDO1 overexpression downregulates profibrotic markers, suppresses proliferation, and enhances apoptosis and cell cycle arrest in HSCs by suppressing the PTEN/AKT pathway [102]. Hsa_circ_0007874 (circMTO1), derived from the MTO1 gene, is downregulated in fibrotic mouse livers and activated HSCs. Enhancing circMTO1 expression inhibits the activation of HSCs triggered by TGF-β1. CircMTO1 targets PTEN and Smad7 via miR-17-5p and miR-181b-5p [103,104]. Table 3 summarizes the circRNAs associated with the regulation of liver fibrosis.
Expression and roles of dysregulated circular RNAs in liver fibrosis
| Reference | Biological Function | Sponge Target | Expression Pattern | Circular RNA |
|---|---|---|---|---|
| [85] | Hsa_circ_0071410 downregulation reduces HSCs activation. | miR-9-5p | Increased | Hsa_circ_0071410 |
| [86] | CircUbe2k increases TGF-β2 activity, and its suppression reduces α-SMA and Col1α1. | miR-149-5p | Increased | CircUbe2k |
| [87] | Circ-PWWP2A regulates HSC activation and proliferation. | miR-223 miR-203 | Increased | Circ-PWWP2A |
| [88] | CircRNA-0067835 regulates fibrosis progression by modulating FOXO3a expression. Its silencing suppresses HSC proliferation and enhances apoptosis. | miR-155 | Increased | CircRNA-0067835 |
| [89] | CircRSF1 increases cell viability, cell fibrosis, and inflammation by activating RAC1. | miR-146a-5p | Increased | CircRSF1 |
| [90] | CircTUBD1 induces liver fibrosis by regulating Smad3. A reduction in circTUBD1 suppresses the expression of COL1A1, COL3A1, and CTGF. | miR-203a-3p | Increased | CircTUBD1 |
| [91] | Hsa_circ_0072765 suppression inhibits activation, migration, and proliferation by targeting TRPV3. | miR-197-3p | Increased | Hsa_circ_0072765 |
| [92] | A reduction in circMcph1 decreases fibrosis and inflammation. It modulates Irak2 expression. | miR-370-3p | Increased | CircMcph1 |
| [93] | CircRNA-007371 overexpression increases angiogenesis, proliferation, and migration. | Not identified | Increased | CircRNA-007371 |
| [94] | CircRNA cVIM activates HSCs and regulates TGFBR, resulting in TGF-β/Smad activation. | miR-9-5p miR-122-5p | Increased | CircRNA cVIM |
| [95] | Circ_0008494 downregulation decreases HSC proliferation, activation, and migration and stimulates apoptosis. Col1a1 is its target. | miR-185-3p | Increased | Circ_0008494 |
| [96] | CircPSD3 overexpression decreases collagen, profibrogenic and proinflammatory markers, liver enzymes, and liver hydroxyproline by enhancing Smad7. | miR-92b-3p | Decreased | CircPSD3 |
| [97] | CircFBXW4 suppresses hepatic inflammation, HSC proliferation, and activation, and promotes apoptosis by controlling FBXW7. | miR-18b-3p | Decreased | CircFBXW4 |
| [98] | Hsa_circ_0004018 suppresses fibrosis by inhibiting TEP1. | miR-660-3p | Decreased | Hsa_circ_0004018 |
| [99] | CircCREBBP reduces the activation and proliferation of HSCs. It is involved in LEFTY2 expression. | miR-1291 | Decreased | CircCREBBP |
| [100] | Hsa_circ_0070963 reduces HSC activation and α-SMA and type I collagen expression. Its antifibrotic effect is mediated by LEMD3 regulation. | miR-223-3p | Decreased | Hsa_circ_0070963 |
| [101] | Mmu_circ_34116 regulates HSC activation. Its inhibition is associated with increased α-SMA expression. | miR-22-3P | Decreased | Mmu_circ_34116 |
| [102] | CircDIDO1 decreases profibrotic markers and HSC proliferation and induces apoptosis by inhibiting the PTEN/AKT pathway. | miR-141-3p | Decreased | CircDIDO1 |
| [103,104] | CircMTO1 decreases HSC proliferation as well as the expression of α-SMA and type I collagen by PTEN and Smad7 regulation. | miR-17-5p miR-181b-5p | Decreased | CircMTO1 |
Abbreviations: HSCs: hepatic stellate cells; TGF-β1: transforming growth factor beta 1; α-SMA: alpha-smooth muscle actin; Col1α1 / COL1A1: collagen type I alpha 1 chain; COL3A1: collagen type III alpha 1 chain; CTGF: connective tissue growth factor; TRPV3: transient receptor potential vanilloid 3; Irak2: interleukin-1 receptor-associated kinase 2; TGFBR: transforming growth factor beta receptor; Smad3 / Smad7: mothers against decapentaplegic homolog 3 / 7; FBXW7: F-box and WD repeat domain containing 7; TEP1: telomerase-associated protein 1; LEFTY2: left-right determination factor 2; LEMD3: LEM domain containing 3; PTEN: phosphatase and tensin homolog; AKT: protein kinase B; miR / miRNA: microRNA
Conclusions
This review highlights the biogenesis, function, and importance of circRNAs, emphasizing their essential roles as hallmarks of HCC, NAFLD, NASH, and liver fibrosis. However, further research is needed to elucidate the mechanisms underlying their synthesis, biological functions, and clearance. The effects of circRNAs on liver disease through their interactions with miRNAs indicate their potential as biomarkers for prognosis, diagnosis, and targeted therapy. Despite this promise, translating the mechanisms identified by basic research into clinical applications for early detection and prognosis of liver disease remains a considerable challenge. Understanding the dysregulation of circRNA expression in liver diseases may clarify their roles in regulating various molecules and signaling pathways. Nonetheless, their contributions to liver homeostasis, disease development, and the molecular mechanisms regulating circRNAs remain poorly understood. The distinct functions of various liver cell types in liver pathogenesis complicate efforts to determine the precise roles of circRNAs in liver disease. In addition, studies linking circRNAs with liver diseases have focused primarily on HCC, highlighting the need to expand investigations to other liver diseases. Finally, the regulatory effects of circRNA polymorphisms on circRNA expression and function remain to be elucidated.
Abbreviations
miRNA: microRNA; circRNAs: Circular RNAs; HCC: hepatocellular carcinoma; NAFLD: nonalcoholic fatty liver diseases; Mbl: muscle-blind; NUPR1: nuclear protein 1; ENO2: enolase 2; MAPK1: mitogen-activated protein kinase 1; EZH2: zeste homolog 2; TXNDC5: thioredoxin domain-containing protein 5; GUCD1: guanylyl cyclase domain containing 1; SOX9: SRY (sex determining region Y)-box 9; JAK2: Janus kinase 2; ITGB4: integrin β4; TGF-β1: transforming growth factor-β1; CXCR4: chemokine receptor 4; SHPRH: SNF2 histone linker PHD RING helicase; PI3K: phosphoinositide 3-kinase; TET1: ten-eleven translocation methylcytosine dioxygenase 1; 5hmC: 5-hydroxymethylcytosine; NASH: nonalcoholic steatohepatitis; ROCK1: Rho-kinase 1; AMPK: AMP-activated protein kinase; CPEB: cytoplasmic polyadenylation element-binding protein; HIPK3: homeodomain interacting protein kinase 3; PPARα: peroxisome proliferator-activated receptor α; FFA: free fatty acid; SLC27A: solute carrier family 27A; GPX4: glutathione peroxidase 4; ROS: reactive oxygen species; SCAR: circRNA ATP5B regulator; mROS: mitochondrial ROS; HSCs: hepatic stellate cells; IL: interleukin; LPS: lipopolysaccharide; α-SMA: alpha smooth muscle actin; Col1α1: collagen, type I, alpha 1; Tβ4: thymosin β4; FOXO3a: forkhead box O3; TRPV3: transient receptor potential vanillin 3; IRAK2: IL-1 receptor-associated kinase 2; TAA: thioacetamide; PSD3: pleckstrin and Sec7 domain-containing 3.
Acknowledgements
The author thanks the Dean of Graduate Studies and Scientific Research, Jouf University, Al Jouf Province, Saudi Arabia, for funding this project (no. DGSSR-2025-01-01630).
Funding
Jouf University Research Grant No. DGSSR-2025-01-01630 funded this work.
Author contributions
The author participated in the conception, literature review, data analysis, and writing of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cao X, Cai Z, Zhang J, Zhao F. Engineering circular RNA medicines. Nat Rev Bioeng. 2025;3(4):270-287
2. Greene J, Baird AM, Brady L. et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38
3. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428-442
4. Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185(12):2016-2034
5. van Heesch S, Witte F, Schneider-Lunitz V. et al. The Translational Landscape of the Human Heart. Cell. 2019;178(1):242-260.e29
6. Nokkeaw A, Thamjamrassri P, Tangkijvanich P, Ariyachet C. Regulatory Functions and Mechanisms of Circular RNAs in Hepatic Stellate Cell Activation and Liver Fibrosis. Cells. 2023;12(3):378
7. Zeng X, Yuan X, Cai Q, Tang C, Gao J. Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases. Cells. 2021;10(8):1945
8. Liu CX, Li X, Nan F. et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177(4):865-880.e21
9. Wang P, Zhang Y, Deng L. et al. The function and regulation network mechanism of circRNA in liver diseases. Cancer Cell Int. 2022;22(1):141
10. Song M, Xia L, Sun M, Yang C, Wang F. Circular RNA in Liver: Health and Diseases. Adv Exp Med Biol. 2018;1087:245-257
11. Li L, Zheng YC, Kayani MUR. et al. Comprehensive analysis of circRNA expression profiles in humans by RAISE. Int J Oncol. 2017;51(6):1625-1638
12. Shen H, Liu B, Xu J. et al. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14(1):134
13. Louis C, Leclerc D, Coulouarn C. Emerging roles of circular RNAs in liver cancer. JHEP Rep. 2022;4(2):100413
14. Shang X, Li G, Liu H. et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine (Baltimore). 2016;95(22):e3811
15. Huang XY, Huang ZL, Xu YH. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7(1):5428
16. Ren S, Xin Z, Xu Y, Xu J, Wang G. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle. 2017;16(22):2204-2211
17. Zhao M, Dong G, Meng Q, Lin S, Li X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J Cell Biochem. 2020;121(11):4440-4449
18. Wang BG, Li JS, Liu YF, Xu Q. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol. 2017;39(7):1010428317719577
19. Liang WC, Wong CW, Liang PP. et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84
20. Fei D, Wang F, Wang Y. et al. Circular RNA ACVR2A promotes the progression of hepatocellular carcinoma through mir-511-5p targeting PI3K-Akt signaling pathway. Mol Cancer. 2024;23(1):159
21. Zhang XY, Li SS, Gu YR. et al. CircPIAS1 promotes hepatocellular carcinoma progression by inhibiting ferroptosis via the miR-455-3p/NUPR1/FTH1 axis. Mol Cancer. 2024;23(1):113
22. Du W, Li Y, Wang X. et al. Circular RNA circESYT2 serves as a microRNA-665 sponge to promote the progression of hepatocellular carcinoma through ENO2. Cancer Sci. 2024;115(8):2659-2672
23. Zhu Q, Lu G, Luo Z. et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626-632
24. Dong W, Dai ZH, Liu FC. et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155-167
25. Pu J, Wang J, Li W. et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. 2024;28(6):e15010
26. Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19(3):269-279
27. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology. 2019;70(4):1298-1316
28. Hu ZQ, Zhou SL, Li J. et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2020;72(3):906-922
29. Zhang PF, Gao C, Huang XY. et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110
30. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1):119
31. Sun Y, Sun X, Huang Q. Circ_0000105 promotes liver cancer by regulating miR-498/PIK3R1. J Gene Med. 2020;22(11):e3256
32. Niu WY, Chen L, Zhang P, Zang H, Zhu B, Shao WB. Circ_0091579 promotes proliferative ability and metastasis of liver cancer cells by regulating microRNA-490-3p. Eur Rev Med Pharmacol Sci. 2019;23(23):10264-10273
33. Qi SX, Sun H, Liu H, Yu J, Jiang ZY, Yan P. Role and mechanism of circ-PRKCI in hepatocellular carcinoma. World J Gastroenterol. 2019;25(16):1964-1974
34. Yang J, Li Y, Yu Z. et al. Circular RNA Circ100084 functions as sponge of miR-23a-5p to regulate IGF2 expression in hepatocellular carcinoma. Mol Med Rep. 2020;21(6):2395-2404
35. Zhao Z, Song J, Tang B. et al. CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J Exp Clin Cancer Res. 2020;39(1):259
36. Jiang QL, Feng SJ, Yang ZY, Xu Q, Wang SZ. CircHECTD1 up-regulates mucin 1 expression to accelerate hepatocellular carcinoma development by targeting microRNA-485-5p via a competing endogenous RNA mechanism. Chin Med J (Engl). 2020;133(15):1774-1785
37. Chen M, Hu G, Zhou X, Peng Z, Wen W. Hsa_circ_0016788 regulates hepatocellular carcinoma progression via miR-506-3p/poly-adenosine diphosphate-ribose polymerase. J Gastroenterol Hepatol. 2021;36(12):3457-3468
38. Guan Z, Tan J, Gao W. et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234(1):500-508
39. Li S, Weng J, Song F. et al. Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death Dis. 2020;11(6):452
40. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A Acts as the Sponge of MicroRNA-665 to Promote Hepatocellular Carcinoma Progression. Mol Ther Nucleic Acids. 2020;22:285-297
41. Zhai Z, Fu Q, Liu C. et al. Emerging Roles Of hsa-circ-0046600 Targeting The miR-640/HIF-1α Signalling Pathway In The Progression Of HCC. Onco Targets Ther. 2019;12:9291-9302
42. Fu X, Zhang J, He X. et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer. 2020;11(11):3318-3326
43. Wang G, Liu W, Zou Y. et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432-445
44. Bu N, Dong Z, Zhang L, Zhu W, Wei F, Zheng S. CircPVT1 Regulates Cell Proliferation, Apoptosis and Glycolysis in Hepatocellular Carcinoma via miR-377/TRIM23 Axis. Cancer Manag Res. 2020;12:12945-12956
45. Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Cell Biochem. 2019;120(6):9487-9494
46. Lin T, Dai Y, Guo X. et al. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manag Res. 2019;11:9133-9143
47. Yu J, Yang M, Zhou B. et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;133(13):1487-1503
48. Li Y, Zang H, Zhang X, Huang G. Exosomal Circ-ZNF652 Promotes Cell Proliferation, Migration, Invasion and Glycolysis in Hepatocellular Carcinoma via miR-29a-3p/GUCD1 Axis. Cancer Manag Res. 2020;12:7739-7751
49. Pan H, Tang L, Jiang H. et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem. 2019;120(7):11350-11357
50. Wang W, Li Y, Li X. et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother. 2020;121:109517
51. Cao S, Wang G, Wang J, Li C, Zhang L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol Cell Biol. 2019;97(2):218-228
52. Bai N, Peng E, Qiu X. et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37(1):172
53. Bai N, Peng E, Xia F, Wang D, Li X, Li X. CircABCC2 Regulates Hepatocellular Cancer Progression by Decoying MiR-665. J Cancer. 2019;10(17):3893-3898
54. Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (Albany NY). 2019;11(10):3362-3375
55. Qin M, Liu G, Huo X. et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161-169
56. Guo W, Zhang J, Zhang D. et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8(29):48169-48177
57. Fu L, Chen Q, Yao T. et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8(27):43878-43888
58. Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8(35):58405-58416
59. Han D, Li J, Wang H. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151-1164
60. Matboli M, Hassan MK, Ali MA. et al. Impact of circ-0000221 in the Pathogenesis of Hepatocellular via Modulation of miR-661-PTPN11 mRNA Axis. Pharmaceutics. 2022;14(1):138
61. Yao Z, Luo J, Hu K. et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422-437
62. Zhang K, Che S, Su Z. et al. CD90 promotes cell migration, viability and sphere-forming ability of hepatocellular carcinoma cells. Int J Mol Med. 2018;41(2):946-954
63. Zhang PF, Wei CY, Huang XY. et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18(1):105
64. Wang Z, Zhao Y, Wang Y, Jin C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed Pharmacother. 2019;116:108932
65. Chen Z, Zuo X, Pu L. et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110(2):568-581
66. Zeng Q, Liu CH, Wu D, Jiang W, Zhang N, Tang H. LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Biomolecules. 2023;13(3):560
67. Khalifa O, Errafii K, Al-Akl NS, Arredouani A. Noncoding RNAs in Nonalcoholic Fatty Liver Disease: Potential Diagnosis and Prognosis Biomarkers. Dis Markers. 2020;2020:8822859
68. Yepmo M, Potier JB, Pinget M, Grabarz A, Bouzakri K, Dumond Bourie A. Discussing the role of circular RNA in the pathogenesis of non-alcoholic fatty liver disease and its complications. Front Endocrinol (Lausanne). 2022;13:1035159
69. Zeng Q, Liu CH, Ampuero J. et al. Circular RNAs in non-alcoholic fatty liver disease: Functions and clinical significance. RNA Biol. 2024;21(1):1-15
70. Guo J, Zhou Y, Cheng Y. et al. Metformin-Induced Changes of the Coding Transcriptome and Non-Coding RNAs in the Livers of Non-Alcoholic Fatty Liver Disease Mice. Cell Physiol Biochem. 2018;45(4):1487-1505
71. Yuan X, Diao J, Du A, Wen S, Zhou L, Pan Y. Circular RNA expression profiles and features in NAFLD mice: a study using RNA-seq data. J Transl Med. 2020;18(1):476
72. Chen X, Tan QQ, Tan XR, Li SJ, Zhang XX. Circ_0057558 promotes nonalcoholic fatty liver disease by regulating ROCK1/AMPK signaling through targeting miR-206. Cell Death Dis. 2021;12(9):809
73. Jin X, Gao J, Zheng R. et al. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. 2020;11(2):123
74. Abdelgwad M, Zakaria R, Marzouk S. et al. The Emerging Role of Circular RNA Homeodomain Interacting Protein Kinase 3 and Circular RNA 0046367 through Wnt/Beta-Catenin Pathway on the Pathogenesis of Nonalcoholic Steatohepatitis in Egyptian Patients. Rep Biochem Mol Biol. 2023;11(4):614-625
75. Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxid Med Cell Longev. 2017;2017:3960197
76. Guo XY, Sun F, Chen JN, Wang YQ, Pan Q, Fan JG. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol. 2018;24(3):323-337
77. Li P, Shan K, Liu Y, Zhang Y, Xu L, Xu L. CircScd1 Promotes Fatty Liver Disease via the Janus Kinase 2/Signal Transducer and Activator of Transcription 5 Pathway. Dig Dis Sci. 2019;64(1):113-122
78. Yang W, Zhao J, Zhao Y. et al. Hsa_circ_0048179 attenuates free fatty acid-induced steatosis via hsa_circ_0048179/miR-188-3p/GPX4 signaling. Aging (Albany NY). 2020;12(23):23996-24008
79. Zhao Q, Liu J, Deng H. et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell. 2020;183(1):76-93.e22
80. Xie Y, Cao Y, Guo CJ. et al. Profile analysis and functional modeling identify circular RNAs in nonalcoholic fatty liver disease as regulators of hepatic lipid metabolism. Front Genet. 2022;13:884037
81. Li J, Qi J, Tang Y. et al. A nanodrug system overexpressed circRNA_0001805 alleviates nonalcoholic fatty liver disease via miR-106a-5p/miR-320a and ABCA1/CPT1 axis. J Nanobiotechnology. 2021;19(1):363
82. Guo XY, He CX, Wang YQ. et al. Circular RNA Profiling and Bioinformatic Modeling Identify Its Regulatory Role in Hepatic Steatosis. Biomed Res Int. 2017;2017:5936171
83. Li QY, Gong T, Huang YK. et al. Role of noncoding RNAs in liver fibrosis. World J Gastroenterol. 2023;29(9):1446-1459
84. Wang G, Tong J, Li Y. et al. Overview of CircRNAs Roles and Mechanisms in Liver Fibrosis. Biomolecules. 2023;13(6):940
85. Chen Y, Yuan B, Wu Z, Dong Y, Zhang L, Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. 2017;629:35-42
86. Zhu S, Chen X, Wang JN. et al. Circular RNA circUbe2k promotes hepatic fibrosis via sponging miR-149-5p/TGF-β2 axis. FASEB J. 2021;35(6):e21622
87. Liu W, Feng R, Li X, Li D, Zhai W. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging (Albany NY). 2019;11(21):9569-9580
88. Zhu L, Ren T, Zhu Z. et al. Thymosin-β4 Mediates Hepatic Stellate Cell Activation by Interfering with CircRNA-0067835/miR-155/FoxO3 Signaling Pathway. Cell Physiol Biochem. 2018;51(3):1389-1398
89. Chen Y, Yuan B, Chen G. et al. Circular RNA RSF1 promotes inflammatory and fibrotic phenotypes of irradiated hepatic stellate cell by modulating miR-146a-5p. J Cell Physiol. 2020;235(11):8270-8282
90. Niu H, Zhang L, Wang B. et al. CircTUBD1 Regulates Radiation-induced Liver Fibrosis Response via a circTUBD1/micro-203a-3p/Smad3 Positive Feedback Loop. J Clin Transl Hepatol. 2022;10(4):680-691
91. Jin Y, Guo X, Zhang R, Yan C. Hsa_circ_0072765 knockdown inhibits proliferation, activation and migration in transforming growth factor-beta (TGF-β)-induced hepatic stellate cells (HSCs) by the miR-197-3p/TRPV3 axis. Histol Histopathol. 2023;38(11):1295-1306
92. Xu JJ, Chen X, Zhu S. et al. Myc-mediated circular RNA circMcph1/miR-370-3p/Irak2 axis is a progressive regulator in hepatic fibrosis. Life Sci. 2023;312:121182
93. Zhao C, Qian S, Tai Y. et al. Proangiogenic role of circRNA-007371 in liver fibrosis. Cell Prolif. 2023;56(6):e13432
94. Zhou Z, Zhang R, Li X. et al. Circular RNA cVIM promotes hepatic stellate cell activation in liver fibrosis via miR-122-5p/miR-9-5p-mediated TGF-β signaling cascade. Commun Biol. 2024;7(1):113
95. Li B, Zhou J, Luo Y. et al. Suppressing circ_0008494 inhibits HSCs activation by regulating the miR-185-3p/Col1a1 axis. Front Pharmacol. 2022;13:1050093
96. Bu FT, Zhu Y, Chen X. et al. Circular RNA circPSD3 alleviates hepatic fibrogenesis by regulating the miR-92b-3p/Smad7 axis. Mol Ther Nucleic Acids. 2021;23:847-862
97. Chen X, Li HD, Bu FT. et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics. 2020;10(11):4851-4870
98. Li S, Song F, Lei X, Li J, Li F, Tan H. hsa_circ_0004018 suppresses the progression of liver fibrosis through regulating the hsa-miR-660-3p/TEP1 axis. Aging (Albany NY). 2020;12(12):11517-11529
99. Yang YR, Hu S, Bu FT. et al. Circular RNA CREBBP Suppresses Hepatic Fibrosis Via Targeting the hsa-miR-1291/LEFTY2 Axis. Front Pharmacol. 2021;12:741151
100. Ji D, Chen GF, Wang JC. et al. Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging (Albany NY). 2020;12(2):1643-1655
101. Zhou Y, Lv X, Qu H. et al. Preliminary screening and functional analysis of circular RNAs associated with hepatic stellate cell activation. Gene. 2018;677:317-323
102. Ma L, Wei J, Zeng Y. et al. Mesenchymal stem cell-originated exosomal circDIDO1 suppresses hepatic stellate cell activation by miR-141-3p/PTEN/AKT pathway in human liver fibrosis. Drug Deliv. 2022;29(1):440-453
103. Wang W, Dong R, Guo Y. et al. CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J Cell Mol Med. 2019;23(8):5486-5496
104. Jin H, Li C, Dong P, Huang J, Yu J, Zheng J. Circular RNA cMTO1 Promotes PTEN Expression Through Sponging miR-181b-5p in Liver Fibrosis. Front Cell Dev Biol. 2020;8:714
Author contact
![]() Corresponding author: Badr Alzahrani, Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, P.O. Box 2014, Jouf University, Aljouf Province, Saudi Arabia. ORCID: https://orcid.org/0000-0003-4158-640X. Tel: +966553139444, E-mail: baalzahraniedu.sa.
Corresponding author: Badr Alzahrani, Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, P.O. Box 2014, Jouf University, Aljouf Province, Saudi Arabia. ORCID: https://orcid.org/0000-0003-4158-640X. Tel: +966553139444, E-mail: baalzahraniedu.sa.

 Global reach, higher impact
Global reach, higher impact