Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(7):969-975. doi:10.7150/ijms.84364 This issue Cite
Research Paper
WWOX Polymorphisms as Predictors of the Biochemical Recurrence of Localized Prostate Cancer after Radical Prostatectomy
1. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan.
2. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
3. School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan.
4. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
5. Department of Family Medicine, Taichung Veterans General Hospital, Taichung, Taiwan.
6. Department of Applied Chemistry, National Chi Nan University, Nantou, Taiwan.
7. Department of Medicine and Nursing, Hungkuang University, Taichung, Taiwan.
8. Department of Urology, Tung's Taichung MetroHarbor Hospital, Taichung, Taiwan.
9. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
Received 2023-3-16; Accepted 2023-5-11; Published 2023-5-21
Abstract
The downregulation of WW domain-containing oxidoreductase (WWOX), a tumor suppressor gene, is associated with the tumorigenesis and poor prognosis of various cancers. In this study, we investigated the associations between the polymorphisms of WWOX, clinicopathologic features of prostate cancer (PCa), and risk of postoperative biochemical recurrence (BCR). We evaluated the effects of five single-nucleotide polymorphisms (SNPs) of WWOX on the clinicopathologic features of 578 patients with PCa. The risk of postoperative BCR was 2.053-fold higher in patients carrying at least one “A” allele in WWOX rs12918952 than in those with homozygous G/G. Furthermore, patients with at least one polymorphic “T” allele in WWOX rs11545028 had an elevated (1.504-fold) risk of PCa with seminal vesicle invasion. In patients with postoperative BCR, the risks of an advanced Gleason grade and clinical metastasis were 3.317- and 5.259-fold higher in patients carrying at least one “G” allele in WWOX rs3764340 than in other patients. Our findings indicate the WWOX SNPs are significantly associated with highly aggressive pathologic features of PCa and an elevated risk of post-RP biochemical recurrence.
Keywords: WWOX, prostate cancer, polymorphism, biochemical recurrence
Introduction
Prostate cancer (PCa) is a common and fatal malignancy. In 2022, the estimated incidence and mortality rate of PCa were 268490 and 34500 in the United States, respectively [1]. Although radical prostatectomy (RP) is the treatment of choice for localized PCa, the rate of postoperative recurrence within 10 years is high at 20% to 40% [2]. Prostate-specific antigen (PSA) is used as a biomarker for not only the screening and detection of PCa but also the diagnosis of postoperative tumor recurrence. After RP, the serum level of PSA is expected to decrease to an undetectable level because of the complete removal of the prostatic tissue. The PSA threshold that is indicative of biochemical recurrence (BCR) has been a subject of debate [3, 4]. The American Urological Association and the European Association of Urology recommend a postoperative PSA level of ≥0.2 ng/mL in addition to a second confirmatory test for the diagnosis of BCR. However, the delineation of post-RP PSA values may not accurately reflect the nature of individual diseases. To overcome these limitations, some studies suggest for the inclusion of genetic adjustments of serum PSA level in risk assessments [5].
The WW-domain-containing oxidoreductase (WWOX) gene is identified on human chromosome 16q23.3-24.1, which contains the Common Chromosomal Fragile Site FRA16D. WWOX frequently undergoes chromosomal breaks and is frequently rearranged in various cancers [6-8]. The WWOX protein is involved in several cellular processes, such as metabolism, DNA damage response, and tumor suppression [9-12]. WWOX may interact with p53 and p73, accelerating apoptosis [13-15]. The abnormal transcription and low expression of WWOX have been reported in various cancers, including breast cancer, ovarian cancer, gastric cancer, oral cancer, and PCa [16, 17]. Several in vivo and in vitro studies have indicated that WWOX suppresses PCa tumorigenesis and progression by arresting the cell cycle in the G1 phase [17-20].
A single-nucleotide polymorphism (SNP) is a variation of at least 1% in a single nucleotide of the shared sequence of a gene within a population. Several genetic polymorphisms have been associated with PSA level, tumor susceptibility, tumor grade, and mortality in PCa [21-27]. Moreover, WWOX SNPs have been reported in numerous cancers [28-32]. However, the association between the polymorphism of WWOX and the risk of the postoperative BCR of PCa remains unclear. Therefore, in this study, we investigated the aforementioned association among patients with PCa in Taiwan.
Materials and Methods
Ethics and Patients
This study was conducted using data from a prospectively developed database of PCa. The study protocol was approved by the Institutional Review Broad of Taichung Veterans General Hospital, Taiwan (approval number: CE19062A-2). Written informed consent was obtained from all participants. We included 578 patients with prostate adenocarcinoma who underwent robotic RP with bilateral pelvic lymph node dissection at Taichung Veterans General Hospital between 2012 and 2017. The following medical data were collected: PSA level at diagnosis, Gleason score at the initial biopsy, Gleason grade group, TNM stage, other pathologic features [33], and D'Amico classification [34]. To investigate the genetic influence on the risk of the post-RP BCR of PCa, we divided the cohort according to PCa recurrence and analyzed the association between the polymorphisms of WWOX and the clinicopathologic features of PCa.
SNP Selection and Genomic DNA Extraction
In this study, we focused on the selection of five commonly observed polymorphisms (rs11545028, rs12918952, rs3764340, rs73569323, and rs383362) within the WWOX gene. These specific polymorphisms were chosen based on their well-established associations with cancer development, as demonstrated in previous studies [35, 36]. Among the selected polymorphisms, rs11545028, located in the 5' untranslated region (UTR) of the WWOX gene, was included in our analysis. Additionally, we examined rs12918952 and rs3764340, which are situated within the gene's exonic regions. These two SNPs are of particular interest as they have the potential to lead to amino acid changes, thereby potentially compromising the tumor suppression function of WWOX [35].
The genotypic frequencies of these WWOX SNPs were evaluated using StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and TaqMan. The results were analyzed using SDS software (version 3.0; Applied Biosystems). The details of methods, probes, and primer sequences used in the analysis of WWOX variants have been reported previously [30].
Blood samples were collected from the participants before surgery. The samples (whole blood) were collected in ethylenediaminetetraacetate-containing tubes and centrifuged immediately. Genomic DNA was extracted using QIAamp DNA Blood Mini Kits (Qiagen, Valencia, CA, USA) following the manufacturer's instructions.
Statistical Analysis
The chi-square test and Student's t tests were used to analyze between-group differences in demographic characteristics. Multivariate logistic regression models were used to estimate the odds ratios (ORs) and adjusted ORs (AORs) and with 95% confidence intervals (CIs) for the association between genotypic frequencies and clinicopathologic features. All analyses were performed using SAS (version 9.1, 2005, for Windows; SAS Institute, Cary, NC, USA). The statistical significance was set at p < 0.05.
Results
Patient Demographics
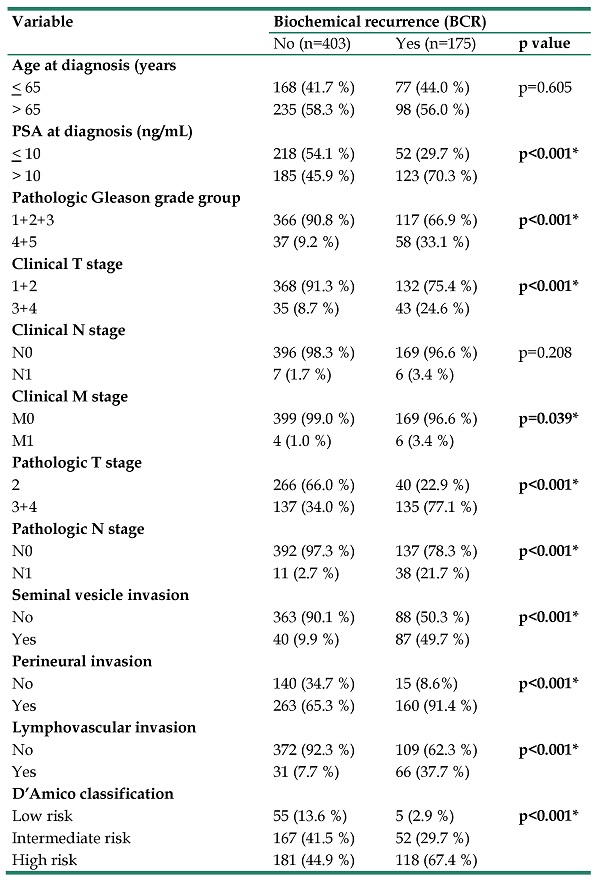
Table 1 presents the demographic characteristics of patients with postoperative BCR of PCa and those without PCa recurrence (175 and 405 patients, respectively). Patients with postoperative BCR exhibited the following clinicopathologic characteristics. They had significantly elevated PSA level at diagnosis (>10 ng/dL), a pathologic Gleason grade group (4+5); TNM stages T3+4, N1, and M1; and poor prognostic features, namely seminal vesicle, perineural, and lymphovascular invasion. The rates of postoperative BCR per the D'Amico classification were 67.4%, 29.7%, and 2.9% in high-, intermediate-, and low-risk patients, respectively.
Association of WWOX Polymorphism with Postoperative BCR
Table 2 presents the distribution frequencies of the 5 WWOX genotypes (rs11545028, rs12918952, rs3764340, rs73569323, and rs383362) in 578 patients. No patient carried the homologous A/A genotype in rs12918952. However, the proportion of patients carrying heterozygous G/A in WWOX rs12918952 was significantly higher among patients with postoperative BCR than among those without PCa recurrence (AOR: 2.053; 95% CI: 1.088 to 3.872; p= 0.025). No prominent trend was noted in the polymorphism frequency of rs11545028, rs3764340, rs73569323, or rs383362. AORs with 95% CIs were estimated using logistical regression models after adjusting for the effects of all possible confounders (Table 2).
The distributions of demographical characteristics in 578 patients with prostate cancer.
| Variable | Biochemical recurrence (BCR) | ||
|---|---|---|---|
| No (n=403) | Yes (n=175) | p value | |
| Age at diagnosis (years | |||
| ≤ 65 | 168 (41.7 %) | 77 (44.0 %) | p=0.605 |
| > 65 | 235 (58.3 %) | 98 (56.0 %) | |
| PSA at diagnosis (ng/mL) | |||
| ≤ 10 | 218 (54.1 %) | 52 (29.7 %) | p<0.001* |
| > 10 | 185 (45.9 %) | 123 (70.3 %) | |
| Pathologic Gleason grade group | |||
| 1+2+3 | 366 (90.8 %) | 117 (66.9 %) | p<0.001* |
| 4+5 | 37 (9.2 %) | 58 (33.1 %) | |
| Clinical T stage | |||
| 1+2 | 368 (91.3 %) | 132 (75.4 %) | p<0.001* |
| 3+4 | 35 (8.7 %) | 43 (24.6 %) | |
| Clinical N stage | |||
| N0 | 396 (98.3 %) | 169 (96.6 %) | p=0.208 |
| N1 | 7 (1.7 %) | 6 (3.4 %) | |
| Clinical M stage | |||
| M0 | 399 (99.0 %) | 169 (96.6 %) | p=0.039* |
| M1 | 4 (1.0 %) | 6 (3.4 %) | |
| Pathologic T stage | |||
| 2 | 266 (66.0 %) | 40 (22.9 %) | p<0.001* |
| 3+4 | 137 (34.0 %) | 135 (77.1 %) | |
| Pathologic N stage | |||
| N0 | 392 (97.3 %) | 137 (78.3 %) | p<0.001* |
| N1 | 11 (2.7 %) | 38 (21.7 %) | |
| Seminal vesicle invasion | |||
| No | 363 (90.1 %) | 88 (50.3 %) | p<0.001* |
| Yes | 40 (9.9 %) | 87 (49.7 %) | |
| Perineural invasion | |||
| No | 140 (34.7 %) | 15 (8.6%) | p<0.001* |
| Yes | 263 (65.3 %) | 160 (91.4 %) | |
| Lymphovascular invasion | |||
| No | 372 (92.3 %) | 109 (62.3 %) | p<0.001* |
| Yes | 31 (7.7 %) | 66 (37.7 %) | |
| D'Amico classification | |||
| Low risk | 55 (13.6 %) | 5 (2.9 %) | p<0.001* |
| Intermediate risk | 167 (41.5 %) | 52 (29.7 %) | |
| High risk | 181 (44.9 %) | 118 (67.4 %) | |
Association of WWOX Polymorphism and PCa Clinical Status with Postoperative BCR
We further investigated the association between WWOX polymorphism and PCa clinicopathologic features in our patients (Table 3), even in those with postoperative BCR (Table 4). Patients with at least one polymorphic “T” allele in rs11545028 had a significantly higher risk of PCa with seminal vesicle invasion than did the other patients (AOR: 1.504; 95% CI: 1.012 to 2.236; p = 0.043; Table 3). Among patients with postoperative BCR, those carrying at least one “G” allele in rs3764340 had increased risks of an advanced Gleason grade (AOR: 3.317; 95% CI: 1.479 to 7.438; p= 0.003) and clinical metastasis (AOR: 5.259, 95% CI: 1.008 to 27.450; p = 0.030) (Table 4). There were no other significant associations between the remaining SNPs and clinicopathologic features of PCa in our patient cohort (Supplementary Tables 1-4).
Discussion
Several studies have consistently revealed prominent associations between the polymorphisms of WWOX and the aggressive, poor prognosis of various malignancies, such as urothelial, esophageal squamous cell, hepatocellular, lung, pancreatic, and thyroid cancers [30, 31, 36-41]. To the best of our knowledge, the present study is the first to investigate the association between the polymorphisms of WWOX and the clinicopathologic features of resectable PCa in Taiwanese men. We found a significant association between the WWOX SNP rs12918952 and the risk of postoperative BCR. The WWOX SNP rs11545028 was associated with an elevated risk of seminal invasive disease. Furthermore, WWOX rs3764340 was associated with increased PCa aggressiveness (high Gleason score and clinical metastasis) in patients with postoperative BCR.
Chromosomal breaks and rearrangements frequently occur in the WWOX of patients with cancer [6-8]. Abnormal transcription and low expression of WWOX have been reported in several cancers with an aggressive, poor prognosis. As mentioned, WWOX is involved in PCa tumorigenesis and tumor [17-20]. The SNP rs12918952 is located in the exon 5 of WWOX. In hepatocellular carcinoma, the “G”-to-“A” substitution of rs12918952 may accelerate vascular invasion by altering catalytic activity and downregulating the expression of WWOX mRNA [30]. In our study, the risk of postoperative BCR was 2.053-fold higher in patients carrying at least one “A” allele in rs12918952 than in those with homozygous G/G. The SNP rs11545028, which is located in the exon 1 of WWOX, was found to be associated with a significant increase in the risk of seminal vesicle invasion. Furthermore, the SNP rs3764340, which is located in the exon 7 of WWOX, was found to be associated with a significant increase in the Gleason score. These SNPs in addition to preoperative PSA levels are key predictors of postoperative BCR, as indicated by several nomogram studies [42-44].
Distribution frequency of WWOX genotypes in 578 patients with prostate cancer.
| Variable | Biochemical recurrence (BCR) | |||
|---|---|---|---|---|
| No (n=403) | Yes (n=175) | OR (95% CI) | AOR (95% CI) | |
| rs11545028 | ||||
| CC | 239 (59.3%) | 102 (58.3%) | 1.00 | 1.00 |
| CT | 144 (35.7%) | 64 (36.6%) | 1.041 (0.716-1.515) | 0.916 (0.586-1.432) |
| TT | 20 (5.0%) | 9 (5.1%) | 1.054 (0.464-2.394) | 0.671 (0.246-1.826) |
| CT+TT | 164 (40.7%) | 73 (41.7%) | 1.043 (0.727-1.495) | 0.884 (0.574-1.360) |
| rs12918952 | ||||
| GG | 365 (90.6%) | 148 (84.6%) | 1.00 | 1.00 |
| GA | 38 (9.4%) | 27 (15.4%) | 1.752 (1.032-2.974) p=0.038* | 2.053 (1.088-3.872) p=0.026* |
| AA | 0 (0%) | (0%) | --- | --- |
| GA+AA | 38 (9.4%) | 27 (15.4%) | 1.752 (1.032-2.974) p=0.038* | 2.053 (1.088-3.872) p=0.026* |
| rs3764340 | ||||
| CC | 337 (83.6%) | 145 (82.9%) | 1.00 | 1.00 |
| CG | 61 (15.1%) | 27 (15.4%) | 1.029 (0.628-1.684) | 0.972 (0.537-1.759) |
| GG | 5 (1.3%) | 3 (1.7%) | 1.394 (0.329-5.912) | 3.402 (0.638-18.146) |
| CG+GG | 66 (16.4%) | 30 (17.1%) | 1.056 (0.658-1.696) | 1.077 (0.610-1.902) |
| rs73569323 | ||||
| CC | 388 (96.3%) | 167 (95.4%) | 1.00 | 1.00 |
| CT | 15 (3.7%) | 8 (4.6%) | 1.239 (0.515-2.979) | 1.784 (0.675-4.719) |
| TT | 0 (0%) | 0 (0%) | --- | --- |
| CT+TT | 15 (3.7%) | 8 (4.6%) | 1.239 (0.515-2.979) | 1.784 (0.675-4.719) |
| rs383362 | ||||
| GG | 310 (76.9%) | 128 (73.1%) | 1.00 | 1.00 |
| GT | 86 (21.3%) | 45 (25.7%) | 1.267 (0.837-1.920) | 1.012 (0.618-1.658) |
| TT | 7 (1.8%) | 2 (1.2%) | 0.692 (0.142-3.376) | 0.732 (0.099-5.405) |
| GT+TT | 93 (23.1%) | 47 (26.9%) | 1.224 (0.815-1.838) | 0.997 (0.614-1.619) |
The odds ratios (ORs) and with their 95% confidence intervals (CIs) were estimated by logistic regression models. The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) were estimated by multiple logistic regression models after controlling for Age at diagnosis, PSA, pathologic Gleason grade group, clinical T stage, clinical N stage, clinical M stage, pathologic T stage, seminal vesicle invasion, perineural invasion, lymphovascular invasion and D'Amico classification. * p value < 0.05 as statistically significant.
Odds ratio (OR) and 95% confidence interval (CI) of clinical status and WWOX rs11545028 genotypic frequencies in 578 patients with prostate cancer.
| Variable | Genotypic frequencies | |||
|---|---|---|---|---|
| rs11545028 | CC (N=341) | CT+TT (N=237) | OR (95% CI) | p value |
| PSA at diagnosis (ng/mL) | ||||
| ≤ 10 | 156 (45.7 %) | 114 (48.1 %) | 1.00 | p=0.577 |
| > 10 | 185 (54.3 %) | 123 (51.9 %) | 0.910 (0.653-1.268) | |
| Pathologic Gleason grade group | ||||
| 1+2+3 | 289 (84.8%) | 194 (81.9%) | 1.00 | p=0.356 |
| 4+5 | 52 (15.2%) | 43 (18.1%) | 1.232 (0.791-1.919) | |
| Clinical T stage | ||||
| 1+2 | 298 (87.4%) | 202 (85.2%) | 1.00 | p=0.455 |
| 3+4 | 43 (12.6%) | 35 (14.8%) | 1.201 (0.743-1.942) | |
| Clinical N stage | ||||
| N0 | 333 (97.7 %) | 232 (97.9 %) | 1.00 | p=0.851 |
| N1 | 8 (2.3 %) | 5 (2.1 %) | 0.897 (0.290-2.777) | |
| Clinical M stage | ||||
| M0 | 336 (98.5 %) | 232 (97.9 %) | 1.00 | p=0.560 |
| M1 | 5 (1.5 %) | 5 (2.1 %) | 1.448 (0.415-5.059) | |
| Pathologic T stage | ||||
| 2 | 186 (54.5%) | 120 (50.6%) | 1.00 | p=0.354 |
| 3+4 | 155 (45.5%) | 117 (49.4%) | 1.170 (0.839-1.631) | |
| Pathologic N stage | ||||
| N0 | 315 (92.4%) | 214 (90.3%) | 1.00 | p=0.377 |
| N1 | 26 (7.6%) | 23 (9.7%) | 1.302 (0.724-2.343) | |
| Seminal vesicle invasion | ||||
| No | 276 (80.9%) | 175 (73.8%) | 1.00 | p=0.043* |
| Yes | 65 (19.1%) | 62 (26.2%) | 1.504 (1.012-2.236) | |
| Perineural invasion | ||||
| No | 92 (27.0%) | 63 (26.6%) | 1.00 | p=0.916 |
| Yes | 249 (73.0%) | 174 (73.4%) | 1.020 (0.702-1.484) | |
| Lymphovascular invasion | ||||
| No | 284 (83.3%) | 197 (83.1%) | 1.00 | p=0.959 |
| Yes | 57 (16.7%) | 40 (16.9%) | 1.012 (0.649-1.576) | |
| D'Amico classification | ||||
| Low/Intermediate risk | 161 (47.2%) | 118 (49.8%) | 1.00 | p=0.542 |
| High risk | 180 (52.8%) | 119 (50.2%) | 0.902 (0.647-1.257) | |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models. * p value < 0.05 as statistically significant.
Odds ratio (OR) and 95% confidence interval (CI) of clinical status and WWOX rs3764340 genotypic frequencies in 175 prostate cancer patients with biochemical recurrence.
| Variable | Genotypic frequencies | |||
|---|---|---|---|---|
| rs3764340 | CC (N=145) | CG +GG (N=30) | OR (95% CI) | p value |
| PSA at diagnosis (ng/mL) | ||||
| ≤ 10 | 41 (28.3 %) | 11 (36.7 %) | 1.00 | p=0.360 |
| > 10 | 104 (71.7 %) | 19 (63.3 %) | 0.681 (0.298-1.555) | |
| Pathologic Gleason grade group | ||||
| 1+2+3 | 104 (71.7%) | 13 (43.3%) | 1.00 | p=0.003* |
| 4+5 | 41 (28.3%) | 17 (56.7%) | 3.317 (1.479-7.438) | |
| Clinical T stage | ||||
| 1+2 | 108 (74.5%) | 24 (80.0%) | 1.00 | p=0.523 |
| 3+4 | 37 (25.5%) | 6 (20.0%) | 0.730 (0.277-1.924) | |
| Clinical N stage | ||||
| N0 | 139 (95.9 %) | 30 (100 %) | 1.00 | p=0.257 |
| N1 | 6 (4.1 %) | 0 (0 %) | --- | |
| Clinical M stage | ||||
| M0 | 142 (97.9 %) | 27 (90.0 %) | 1.00 | p=0.030* |
| M1 | 3 (2.1 %) | 3 (10.0 %) | 5.259 (1.008-27.450) | |
| Pathologic T stage | ||||
| 2 | 33 (22.8%) | 7 (23.3%) | 1.00 | p=0.946 |
| 3+4 | 112 (77.2%) | 23 (76.7%) | 0.968 (0.382-2.456) | |
| Pathologic N stage | ||||
| N0 | 115 (79.3%) | 22 (73.3%) | 1.00 | p=0.470 |
| N1 | 30 (20.7%) | 8 (26.7%) | 1.394 (0.565-3.440) | |
| Seminal vesicle invasion | ||||
| No | 75 (51.7%) | 13 (43.3%) | 1.00 | p=0.403 |
| Yes | 70 (48.3%) | 17 (56.7%) | 1.401 (0.634-3.094) | |
| Perineural invasion | ||||
| No | 11 (7.6%) | 4 (13.3%) | 1.00 | p=0.306 |
| Yes | 134 (92.4%) | 26 (86.7%) | 0.534 (0.158-1.806) | |
| Lymphovascular invasion | ||||
| No | 89 (61.4%) | 20 (66.7%) | 1.00 | p=0.587 |
| Yes | 56 (38.6%) | 10 (33.3%) | 0.795 (0.347-1.821) | |
| D'Amico classification | ||||
| Low/Intermediate risk | 49 (33.8%) | 8 (26.7%) | 1.00 | p=0.448 |
| High risk | 96 (66.2%) | 22 (73.3%) | 1.404 (0.583-3.382) | |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models. * p value < 0.05 as statistically significant.
The primary objective in the treatment of surgically amenable PCa is to ensure long-term disease-free survival. Since the introduction of the trifecta concept (BCR-free survival and the recovery of continence and potency) in 2003, it has become the most popular tool in the evaluation of RP outcomes [45]. However, the risk of postoperative BCR within 10 years is approximately 20% to 40% and may be up to >60% in high-risk patients with PCa [46]. The rates of postoperative BCR in our cohort (67.4%, 29.7%, 2.9% in high-, intermediate-, low-risk patients, respectively) are consistent with those reported by other studies. Adjuvant and deferred radiotherapies are the treatments of choice (salvage therapy) for preventing tumor progression in patients with postoperative BCR. Many studies have indicated an association between postoperative radiotherapy and poor treatment outcomes (reduced sexual, urinary, and bowel functions) [47-49]. These adverse effects of salvage therapy considerably reduce the quality of life of patients with postoperative BCR. Therefore, high accuracy must be maintained in the prediction of postoperative BCR risk and the selection of surgical intervention for these patients. Genetic adjustment has emerged as a key tool to avoid postoperative complications and maintain the normal quality of life; this tool improves the predication accuracy of postoperative BCR risk [42, 50-52].
In 2005, Andrew et al. developed and validated a robust predictive model to predict BCR after operation based on clinical and pathological features [44]. However, the model did not include genetic factors. Given the highly variable behavior and clinical course of prostate cancer (PCa), it is important to adopt a personalized approach to oncologic risk stratification. Novel genetic approaches offer additional information to improve clinical decision making [53]. Several studies have identified different gene signatures associated with postoperative BCR. For example, a case-control study found that a 10-gene molecular signature (HDDA10) demonstrated superior performance in predicting BCR in PCa patients after RP (AUC = 0.65) [54]. Another study developed an original gene signature model that predicted 3-year BCR-free survival in PCa patients after RP (AUC = 0.836) [55]. CDO1 promoter methylation has been proposed as a feasible predictive biomarker for BCR-free survival in PCa patients following RP [56]. Additionally, a genetic risk score has been developed that can predict BCR by time-dependent receiver operating characteristic (t-ROC) curves in the training set (3-year AUC = 0.820, 5-year AUC = 0.809) [57]. All these studies approved the significance of genetic factors in the risk of postoperative BCR. The WWOX genetic polymorphism should also be considered in genetic signature analysis, as it has been shown to be associated with prostate cancer recurrence.
Our study has some limitations. First, we only included patients with surgically amenable PCa; therefore, the findings may not be generalizable to patients with advanced PCa. Second, we lacked tumor specimens and information on the expression levels of WWOX in patients with PCa. Therefore, further translational studies on WWOX expression and its association with prostate cancer behavior are needed to validate the effect of genetic signature on the prognosis of tumor recurrence. Furthermore, studies on the mechanisms of WWOX variation and their involvement in tumor progression and recurrence are warranted.
In conclusion, our findings indicate the polymorphisms of WWOX are associated with highly aggressive clinicopathologic features of PCa and an elevated risk of post-RP BCR. This study may serve as a reference for future studies aimed at preventing the postoperative BCR of PCa.
Supplementary Material
Supplementary tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022;72:7-33
2. Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. The Journal of urology. 2003;169:517-23
3. Cookson MS, Aus G, Burnett AL, Canby-Hagino ED, D'Amico AV, Dmochowski RR. et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. The Journal of urology. 2007;177:540-5
4. Cornford P, Bellmunt J, Bolla M, Briers E, De Santis M, Gross T. et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part II: Treatment of Relapsing, Metastatic, and Castration-Resistant Prostate Cancer. European urology. 2017;71:630-42
5. Gudmundsson J, Besenbacher S, Sulem P, Gudbjartsson DF, Olafsson I, Arinbjarnarson S. et al. Genetic correction of PSA values using sequence variants associated with PSA levels. Sci Transl Med. 2010;2:62ra92
6. Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer research. 2000;60:2140-5
7. Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK. et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651-63
8. Lee CS, Choo A, Dayan S, Richards RI, O'Keefe LV. Molecular Biology of the WWOX Gene That Spans Chromosomal Fragile Site FRA16D. Cells. 2021 10
9. Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem. 2009;108:737-45
10. Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. Journal of cellular physiology. 2007;212:307-10
11. Abu-Remaileh M, Joy-Dodson E, Schueler-Furman O, Aqeilan RI. Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells. The Journal of biological chemistry. 2015;290:30728-35
12. Tanna M, Aqeilan RI. Modeling WWOX Loss of Function in vivo: What Have We Learned? Front Oncol. 2018;8:420
13. Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T. et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4401-6
14. Zawacka-Pankau J, Kostecka A, Sznarkowska A, Hedström E, Kawiak A. p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell cycle (Georgetown, Tex). 2010;9:720-8
15. Chang NS, Doherty J, Ensign A, Schultz L, Hsu LJ, Hong Q. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. The Journal of biological chemistry. 2005;280:43100-8
16. Nunez MI, Ludes-Meyers J, Abba MC, Kil H, Abbey NW, Page RE. et al. Frequent loss of WWOX expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Res Treat. 2005;89:99-105
17. Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S. et al. A role for the WWOX gene in prostate cancer. Cancer research. 2006;66:6477-81
18. Hong Q, Sze CI, Lin SR, Lee MH, He RY, Schultz L. et al. Complement C1q activates tumor suppressor WWOX to induce apoptosis in prostate cancer cells. PloS one. 2009;4:e5755
19. Choi HJ, Park JH, Park JH, Lee KB, Oh SM. Pc2-mediated SUMOylation of WWOX is essential for its suppression of DU145 prostate tumorigenesis. FEBS Lett. 2015;589:3977-88
20. Lin JT, Li HY, Chang NS, Lin CH, Chen YC, Lu PJ. WWOX suppresses prostate cancer cell progression through cyclin D1-mediated cell cycle arrest in the G1 phase. Cell cycle (Georgetown, Tex). 2015;14:408-16
21. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC. et al. Genetic polymorphism and carbonic anhydrase 9 expression can predict nodal metastatic prostate cancer risk in patients with prostate-specific antigen levels </=10 ng/ml at initial biopsy. Urologic oncology. 2019
22. Chou YE, Hsieh MJ, Wang SS, Lin CY, Chen YY, Ho YC. et al. The impact of receptor of advanced glycation end-products polymorphisms on prostate cancer progression and clinicopathological characteristics. Journal of cellular and molecular medicine. 2021;25:10761-9
23. Chou YE, Yang PJ, Lin CY, Chen YY, Chiang WL, Lin PX. et al. The Impact of HMGB1 Polymorphisms on Prostate Cancer Progression and Clinicopathological Characteristics. International journal of environmental research and public health. 2020 17
24. Hu JC, Lin CY, Wang SS, Chiu KY, Li JR, Chen CS. et al. Impact of H19 Polymorphisms on Prostate Cancer Clinicopathologic Characteristics. Diagnostics (Basel, Switzerland). 2020 10
25. Hu JC, Wang SS, Chou YE, Chiu KY, Li JR, Chen CS. et al. Associations between LncRNA MALAT1 Polymorphisms and Lymph Node Metastasis in Prostate Cancer. Diagnostics (Basel, Switzerland). 2021 11
26. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC. et al. Impact of GAS5 genetic polymorphism on prostate cancer susceptibility and clinicopathologic characteristics. International journal of medical sciences. 2019;16:1424-9
27. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC. et al. Genetic polymorphism and carbonic anhydrase 9 expression can predict nodal metastatic prostate cancer risk in patients with prostate-specific antigen levels ≤10 ng/ml at initial biopsy. Urologic oncology. 2019;37:814 e9- e16
28. Cheng HL, Liu YF, Su CW, Su SC, Chen MK, Yang SF. et al. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7:69384-96
29. Hung SC, Chou YE, Li JR, Chen CS, Lin CY, Chang LW. et al. Functional genetic variant of WW domain containing oxidoreductase gene associated with urothelial cell carcinoma clinicopathologic characteristics and long-term survival. Urologic oncology. 2020;38:41 e1- e9
30. Lee HL, Cheng HL, Liu YF, Chou MC, Yang SF, Chou YE. Functional genetic variant of WW domain-containing oxidoreductase (WWOX) gene is associated with hepatocellular carcinoma risk. PloS one. 2017;12:e0176141
31. Li JP, Chang JT, Ju PC, Hsieh MH, Chao YH, Tsao TC. et al. Effect of WW Domain-Containing Oxidoreductase Gene Polymorphism on Clinicopathological Characteristics of Patients with EGFR Mutant Lung Adenocarcinoma in Taiwan. International journal of environmental research and public health. 2021 18
32. Lin YH, Hsiao YH, Wu WJ, Yang SF, Hsu CF, Kang YT. et al. Relationship of genetic variant distributions of WW domain-containing oxidoreductase gene with uterine cervical cancer. International journal of medical sciences. 2018;15:1005-13
33. Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. The American journal of surgical pathology. 2016;40:244-52
34. D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA. et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. Jama. 1998;280:969-74
35. Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D. et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11417-22
36. Cancemi L, Romei C, Bertocchi S, Tarrini G, Spitaleri I, Cipollini M. et al. Evidences that the polymorphism Pro-282-Ala within the tumor suppressor gene WWOX is a new risk factor for differentiated thyroid carcinoma. Int J Cancer. 2011;129:2816-24
37. Hung SC, Chou YE, Li JR, Chen CS, Lin CY, Chang LW. et al. Functional genetic variant of WW domain containing oxidoreductase gene associated with urothelial cell carcinoma clinicopathologic characteristics and long-term survival. Urologic oncology. 2020;38:41.e1-e9
38. Schirmer MA, Lüske CM, Roppel S, Schaudinn A, Zimmer C, Pflüger R. et al. Relevance of Sp Binding Site Polymorphism in WWOX for Treatment Outcome in Pancreatic Cancer. J Natl Cancer Inst. 2016 108
39. Guo W, Wang G, Dong Y, Guo Y, Kuang G, Dong Z. Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog. 2013;52:265-74
40. Huang D, Qiu F, Yang L, Li Y, Cheng M, Wang H. et al. The polymorphisms and haplotypes of WWOX gene are associated with the risk of lung cancer in southern and eastern Chinese populations. Mol Carcinog. 2013;52(Suppl 1):E19-27
41. Chen W, Zhou C, Zhang W, Atyah M, Yin Y, Guo L. et al. Association of WWOX rs9926344 polymorphism with poor prognosis of hepatocellular carcinoma. Journal of Cancer. 2018;9:1239-47
42. Morote J, Del Amo J, Borque A, Ars E, Hernández C, Herranz F. et al. Improved prediction of biochemical recurrence after radical prostatectomy by genetic polymorphisms. The Journal of urology. 2010;184:506-11
43. Walz J, Chun FK, Klein EA, Reuther A, Saad F, Graefen M. et al. Nomogram predicting the probability of early recurrence after radical prostatectomy for prostate cancer. The Journal of urology. 2009;181:601-7 discussion 7-8
44. Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ Jr, Dotan ZA, DiBlasio CJ. et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:7005-12
45. Salomon L, Saint F, Anastasiadis AG, Sebe P, Chopin D, Abbou CC. Combined reporting of cancer control and functional results of radical prostatectomy. European urology. 2003;44:656-60
46. Bolla M, van Poppel H, Tombal B, Vekemans K, Da Pozzo L, de Reijke TM. et al. Postoperative radiotherapy after radical prostatectomy for high-risk prostate cancer: long-term results of a randomised controlled trial (EORTC trial 22911). Lancet. 2012;380:2018-27
47. Huelster HL, Laviana AA, Joyce DD, Huang LC, Zhao Z, Koyama T. et al. Radiotherapy after radical prostatectomy: Effect of timing of postprostatectomy radiation on functional outcomes. Urologic oncology. 2020;38:930.e23-e32
48. Zaffuto E, Gandaglia G, Fossati N, Dell'Oglio P, Moschini M, Cucchiara V. et al. Early Postoperative Radiotherapy is Associated with Worse Functional Outcomes in Patients with Prostate Cancer. The Journal of urology. 2017;197:669-75
49. Adam M, Tennstedt P, Lanwehr D, Tilki D, Steuber T, Beyer B. et al. Functional Outcomes and Quality of Life After Radical Prostatectomy Only Versus a Combination of Prostatectomy with Radiation and Hormonal Therapy. European urology. 2017;71:330-6
50. Oh JJ, Park S, Lee SE, Hong SK, Lee S, Kim TJ. et al. Genetic risk score to predict biochemical recurrence after radical prostatectomy in prostate cancer: prospective cohort study. Oncotarget. 2017;8:75979-88
51. Luo C, He S, Zhang H, He S, Qi H, Wei A. Clinical and Biological Significance of DNA Methylation-Driven Differentially Expressed Genes in Biochemical Recurrence After Radical Prostatectomy. Front Genet. 2022;13:727307
52. Zhao Y, Tao Z, Li L, Zheng J, Chen X. Predicting biochemical-recurrence-free survival using a three-metabolic-gene risk score model in prostate cancer patients. BMC cancer. 2022;22:239
53. Boström PJ, Bjartell AS, Catto JW, Eggener SE, Lilja H, Loeb S. et al. Genomic Predictors of Outcome in Prostate Cancer. European urology. 2015;68:1033-44
54. Abou-Ouf H, Alshalalfa M, Takhar M, Erho N, Donnelly B, Davicioni E. et al. Validation of a 10-gene molecular signature for predicting biochemical recurrence and clinical metastasis in localized prostate cancer. Journal of cancer research and clinical oncology. 2018;144:883-91
55. Shi R, Bao X, Weischenfeldt J, Schaefer C, Rogowski P, Schmidt-Hegemann NS. et al. A Novel Gene Signature-Based Model Predicts Biochemical Recurrence-Free Survival in Prostate Cancer Patients after Radical Prostatectomy. Cancers (Basel). 2019 12
56. Meller S, Zipfel L, Gevensleben H, Dietrich J, Ellinger J, Majores M. et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016;11:871-80
57. Lee H, Lee M, Hong SK. CRTC2 as a novel prognostic biomarker for worse pathologic outcomes and biochemical recurrence after radical prostatectomy in patients with prostate cancer. Investig Clin Urol. 2019;60:84-90
Author contact
![]() Corresponding author: Shun-Fa Yang, PhD. Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: ysfedu.tw.
Corresponding author: Shun-Fa Yang, PhD. Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: ysfedu.tw.

 Global reach, higher impact
Global reach, higher impact