Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(5):607-615. doi:10.7150/ijms.82331 This issue Cite
Research Paper
Sex-specific genetic variants associated with adult-onset inguinal hernia in a Taiwanese population
1. Division of Pediatric Genetics and Metabolism, Children's Medical Center, Taichung Veterans General Hospital, Taichung, Taiwan
2. Department of Medical Research, Taichung Veterans General Hospital, Taichung, Taiwan
3. School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan
4. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan
5. Children's Medical Center, Taichung Veterans General Hospital, Taichung, Taiwan
6. Department of Food and Nutrition, Providence University, Taichung, Taiwan
7. School of Medicine, Chung Shan Medical University, Taichung, Taiwan
8. Division of Allergy, Immunology, and Rheumatology, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan
9. Institute of Biomedical Science and Rong Hsing Research Center for Translational Medicine, National Chung Hsing University, Taiwan
Received 2023-1-3; Accepted 2023-3-16; Published 2023-3-27
Abstract
Introduction: Inguinal hernia repair is one of the most common surgeries worldwide. However, there is limited information on its underlying genetic mechanism. Studies on the genetic factors related to inguinal hernia in Han Chinese are lacking. Therefore, we aimed to conduct a hospital-based study to assess the genetic factors and comorbidities underlying inguinal hernia in Taiwan.
Materials and Methods: This was a retrospective case-control study. Utilizing data from the Taiwan Precision Medicine Initiative, we identified 1000 patients with inguinal hernia and 10,021 matched controls without inguinal hernia between June 2019 and June 2020. Four susceptibility loci (rs2009262, rs13091322, rs6991952, and rs3809060) associated with inguinal hernia were genotyped by the Taiwan Biobank version 2 (TWBv2) array. Inguinal hernia, surgery types, and comorbidities were obtained from the electronic health records of Taichung Veterans General Hospital.
Results: Adult-onset inguinal hernia was associated with WT1 rs3809060 GT/TT genotype in males and EFEMP1 rs2009262 TC/CC genotype in females. In addition, we identified sex-specific risk factors associated with inguinal hernia; benign prostatic hyperplasia in males (OR: 3.19, 95% CI: 2.73 - 3.73, p< 0.001), chronic obstructive pulmonary disease in females (OR: 2.34, 95% CI: 1.33 - 4.11, p = 0.003) and overweight, defined by body mass index ≧24 kg/m2 (OR: 0.75, 95% CI: 0.65 - 0.86, p<0.001 in males, and OR: 0.60, 95% CI:0.37 - 0.98, p = 0.042 in females), were inversely associated with inguinal hernia.
After stratifying BMI, overweight males with EFEMP1 rs2009262 TC/CC genotype exhibited a higher risk of inguinal hernia (OR: 1.31, 95% CI: 1.07 - 1.61, p = 0.01). Additionally, rs3809060 was specifically associated with male patients with direct-type inguinal hernia (OR: 1.62, 95% CI: 1.19 - 2.22, p = 0.002).
Conclusion: Genetic susceptibility appears to participate in the pathogenesis of inguinal hernia in the Taiwanese population in a sex-specific manner. Future studies are needed to illuminate the complex interplay between heredity and comorbidities.
Keywords: Inguinal hernia, WT1 rs3809060, EFEMP1 rs2009262, BPH, COPD, BMI
Introduction
Inguinal hernia is the consequence of protrusion of abdominal viscus out of the lining of the abdominal cavity through a weak spot in the transversalis and oblique tissues of the low abdominal wall.[1] An epidemiological investigation revealed an estimated prevalence of 4.36% in the Americas, 4.06% in Europe, and 4.88% in Southeast Asia.[2] Men are reported to have a much higher lifetime prevalence of inguinal hernias (20-27%) than women (3-6%).[3] In terms of anatomical location, inguinal hernias are divided into either direct or indirect types. Direct inguinal hernias bulge medially toward the inferior epigastric vessels, due to an acquired defect in the fibromuscular tissues of the inguinal canal, and indirect inguinal hernia sacs protrude laterally toward the inferior epigastric artery, due to late closure of the processus vaginalis.
Inguinal hernias have a variety of symptoms ranging from a painless lump in the groin area to serious complications such as strangulation and bowel incarceration, and surgical repair is currently the only definitive treatment. The rate of recurrent inguinal hernia was reported to range from 0.5 to 15 percent, depending upon the type of repair, hernia site, and the clinical circumstances.[4] Multiple risk factors for the formation of inguinal hernia have been recognized, including older age, male sex,[5] lower body mass index,[6] patent processus vaginalis,[7] matrix metalloproteinase (MMP) abnormalities,[1] chronic obstructive pulmonary disease,[8] and history of prostatectomy.[9] Greater body height and rural residence were also reported to be associated with direct inguinal hernia in females.[3] Although male sex is a well-known predictor for inguinal hernia, it remains unclear whether a sex-specific risk factor exists, based on prior studies.
Family history is a strong risk factor for inguinal hernia.[10, 11] Susceptible genes associated with inguinal hernia have been reported,[12-15] suggesting heredity may be conducive to the pathogenesis of inguinal hernias. Jorgenson et al. conducted the first genome-wide association study and recognized four significant inguinal hernia risk loci (EBF2, ADAMTS6, EFEMP1 and WT1).[16] It has been demonstrated that these four genes contribute to elastin maintenance and collagen equilibrium and are involved in the development of inguinal hernia. Genetic heterogeneity may exist among different ethnicities, but the majority of previous genetic studies were conducted in Caucasian populations. The risk loci associated with inguinal hernia in Asians remain unclear.
The primary aim of this study was to verify whether susceptibility loci (rs2009262, rs13091322, rs6991952, and rs3809060) previously reported in Europeans[16, 17] contribute to inguinal hernia and comorbidities in the Taiwanese population. The secondary aim was to distinguish sex-specific risk factors for the development of inguinal hernia in a hospital-based cohort.
Material and Methods
Study Design and Data Source
This was a retrospective case-control study. The data source was the Taiwan Precision Medicine Initiative (TPMI), a countrywide genetic scheme overseen by Academia Sinica and partner hospitals. The TPMI cohort consisted of Taiwanese participants in 16 hospitals across the country, from whom specimens and information were collected, as well as patients at Taichung Veterans General Hospital (TCVGH), a tertiary medical center, who comprised the greater part of the cohort. A total of 58,091 patients aged >18 years who visited 28 surgical and medical outpatient clinics in TCVGH were selected for enrollment in the TPMI study between June 2019 and June 2020. Our study cohort comprised 11021 patients, whose genetic profiles were connected to medical claims data in TCVGH, including demographic characteristics, procedures, examinations, diagnoses, medication prescriptions, surgeries, outpatient services, and inpatient services. Diagnoses are coded employing the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) format. This study protocol was approved by the institutional review board of Taichung Veterans General Hospital (IRB no. SF19153A). All of the participants provided informed consent.
Case identification
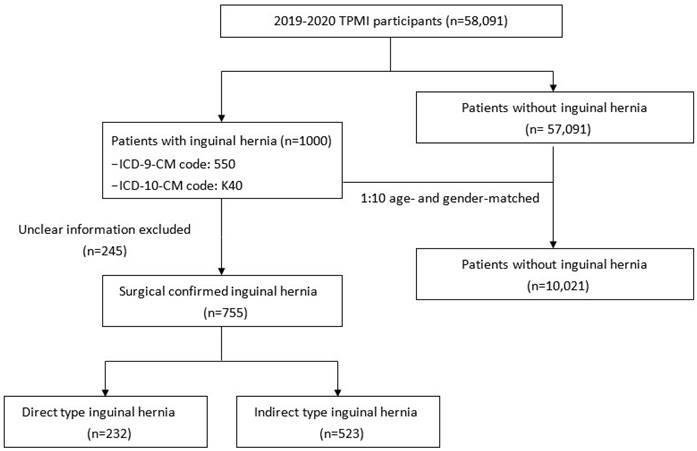
A total of 1,000 patients who had a clinical diagnosis of inguinal hernia (ICD-9-CM code 550, ICD-10-CM code K40) during hospitalization or in an outpatient clinic were selected as the case group (Figure 1). Among them, 755 had available surgery records in TCVGH with postoperative diagnosis of direct or indirect inguinal hernia. A total of 10,021 patients without a medical history of inguinal hernia were matched to the case group by gender and age at a ratio of 1:10 to serve as the control group. We obtained comorbidity information from the electronic health records of TCVGH based on ICD-9 diagnostic codes. Comorbidities of hyperlipidemia (ICD-9-CM code 272, ICD-10-CM code E78.1-E78.5), hypertension (ICD-9-CM code 401-405, ICD-10-CM code I10-I15), diabetes mellitus (ICD-9-CM code 250, ICD-10-CM code E10.9 and E11.9), benign prostatic hyperplasia (BPH) (ICD-9-CM code 600.2, ICD-10-CM code N40.0), chronic kidney disease (CKD) (ICD-9-CM code 585 and ICD-10 code N18.1-N18.9), and chronic obstructive pulmonary disease (COPD) (ICD-9 code 491, ICD-10-CM code J44) were identified if the diagnostic code was used once during admission or at least twice in the outpatient service.
Flow chart of study design.
Genotyping
In our study, genomic DNA was extracted from 58,091 TPMI participants using DNA isolation kits (TIANGEN Biotech, Beijing, China) and quantified using a NanoDrop 2000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Then, the Taiwan Biobank version 2 (TWBv2) array was used to perform next generation sequencing (NGS) for the participants, including 114,000 risk variants in 2,831 unusual disease genes selected from ClinVar, ACMG, GWAS Catalog, HGMD, locus-specific databases, and the published literature.[18] After literature review, we chose four available single nucleotide polymorphisms (SNP), namely, rs2009262, rs13091322, rs6991952, and rs3809060, for further analyses, which were previously reported by Jorgenson et al.[16] and Hikino et al.[17] Quality control of genotyping included an exclusion of SNPs if only one allele appeared in the study cohort; the total call rate < 95% or the total MAF < 0.01. In addition, SNPs significantly departing from the Hardy-Weinberg equilibrium (P < 1 × 10-4) were also excluded. PLINK 1.9 was utilized for statistical genetic analyses and quality control of genetic data.
Statistics
We analyzed all data using the Statistical Package for the Social Sciences (SPSS, IBM Corp., Armonk, NY, USA) version 22.0. The baseline demographics are expressed as number (percent) for categorical variables and the mean ± standard deviation (SD) for continuous variables. Chi-square test for categorical variables and Student's t-test for continuous variables were utilized to compare variables between inguinal hernia cases and controls. Univariate and multivariate logistic regression analyses were applied to estimate the associations of genotypes with the risks of hernia. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. Significant covariates were contained in the final model and a 2-sided p value of < 0.05 was interpreted as statistically significant in this study.
Results
Baseline characteristics and genetic variations associated with inguinal hernia
The baseline demographics are illustrated in Table 1. Inguinal hernia developed mostly at the age of > 65 years (67.9%), with significant male dominance (90.5%). Participants with inguinal hernia exhibited higher percentages of BPH (60% vs. 36.8%, p < 0.001), and COPD (23.8% vs. 19.4%, p = 0.001). Overweight (BMI ≧ 24 kg/m2) (45.5% vs. 54.1%, p < 0.001), hyperlipidemia (40.2% vs. 45.5%, p = 0.001), diabetes mellitus (30.9% vs. 43.9%, p < 0.001), and CKD (31.0% vs. 34.4%, p = 0.04) were inversely associated with inguinal hernia.
Baseline characteristics of the participants.
| Characteristics | Non-inguinal hernia group (n=10,021) | Inguinal hernia group (n=1,000) | p value |
|---|---|---|---|
| n (%) | n (%) | ||
| Age (years) | 0.93 | ||
| < 45 | 552 (5.5) | 57 (5.7) | |
| 45-65 | 2694 (26.9) | 264 (26.4) | |
| > 65 | 6775 (67.7) | 679 (67.9) | |
| Gender | 1.00 | ||
| Female | 957 (9.5) | 95 (9.5) | |
| Male | 9064 (90.5) | 905 (90.5) | |
| Overweight (BMI≧24 kg/m2) | < 0.001 | ||
| No | 4603 (45.9) | 545 (54.5) | |
| Yes | 5418 (54.1) | 455 (45.5) | |
| Hyperlipidemia | 0.001 | ||
| No | 5460 (54.5) | 598 (59.8) | |
| Yes | 4561 (45.5) | 402 (40.2) | |
| Hypertension | 0.295 | ||
| No | 4801 (47.9) | 496 (49.6) | |
| Yes | 5220 (52.1) | 504 (50.4) | |
| Diabetes mellitus | < 0.001 | ||
| No | 5623 (56.1) | 691 (69.1) | |
| Yes | 4398 (43.9) | 309 (30.9) | |
| Chronic kidney disease | 0.04 | ||
| No | 6577 (65.6) | 690 (69.0) | |
| Yes | 3444 (34.4) | 310 (31.0) | |
| BPH | < 0.001 | ||
| No | 6336 (63.2) | 400 (40.0) | |
| Yes | 3685 (36.8) | 600 (60.0) | |
| COPD | 0.001 | ||
| No | 8072 (80.6) | 762 (76.2) | |
| Yes | 1949 (19.4) | 238 (23.8) |
Abbreviations: BMI: Body mass index; BPH: benign prostatic hyperplasia; COPD: chronic obstructive pulmonary disease
The genotype frequencies of the four susceptibility loci (rs2009262, rs13091322, rs6991952, and rs3809060) are shown in Table 2. A greater prevalence of the EFEMP1 rs2009262 TC/CC genotype (36.6% vs. 32.6%, p = 0.01) and the WT1 rs3809060 GT/TT genotype (48.1% vs. 44.1%, p = 0.02) was found in the inguinal hernia group than in the control group.
Allele frequencies and genetic association with inguinal hernia
| Gene / SNP | Non- inguinal hernia group (n=10,021) | Inguinal hernia group (n=1,000) | p value |
|---|---|---|---|
| n (%) | n (%) | ||
| EFEMP1 rs2009262 | 0.01 | ||
| TT | 6756 (67.4) | 634 (63.4) | |
| TC/CC | 3265 (32.6) | 366 (36.6) | |
| WT1 rs3809060 | 0.02 | ||
| GG | 5598 (55.9) | 519 (51.9) | |
| GT/TT | 4423 (44.1) | 481 (48.1) | |
| ERC2 rs13091322 | 0.72 | ||
| AA | 6416 (64.0) | 634 (63.4) | |
| AG/GG | 3605 (36.0) | 366 (36.6) | |
| EBF2 rs6991952 | 0.38 | ||
| AA | 6101 (60.9) | 594 (59.4) | |
| AG/GG | 3920 (39.1) | 406 (40.6) |
Abbreviations: SNP: Single Nucleotide Polymorphism
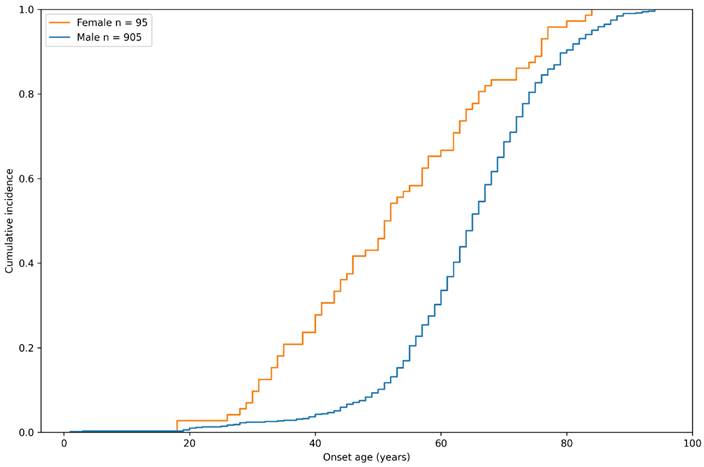
Sex-specific cumulative incidence curve of inguinal hernia in the study group by onset age.
Sex-stratified risk of inguinal hernia with EFEMP1 rs2009262 and WT1 rs3809060
| Male | |||||
|---|---|---|---|---|---|
| Variables | Non- inguinal hernia group (n=9,064) | Inguinal hernia group (n=905) | P value | Risk of Hernia | |
| n (%) | n (%) | OR (95%CI) | p value | ||
| Age, years (%) | |||||
| <45 | 303 (3.3) | 32 (3.5) | 1 | ||
| 45-65 | 2339 (25.8) | 230 (25.4) | 0.93 (0.63-1.37) | 0.72 | |
| >65 | 6422 (70.9) | 643 (71.0) | 0.93 | 0.95 (0.65-1.38) | 0.78 |
| Overweight (BMI ≧ 24 kg/m2) | |||||
| No Yes | 4035 (44.5) 5029 (55.5) | 476 (52.6) 429 (47.4) | <0.001 | 1 0.72 (0.63-0.83) | < 0.001 |
| EFEMP1 rs2009262 | |||||
| TT | 6111 (67.4) | 582 (64.3) | 1 | ||
| TC/CC | 2953 (32.6) | 323 (35.7) | 0.06 | 1.15 (0.99-1.32) | 0.06 |
| WT1 rs3809060 | |||||
| GG | 5068 (55.9) | 470 (51.9) | 1 | ||
| GT/TT | 3996 (44.1) | 435 (48.1) | 0.02 | 1.17 (1.02-1.35) | 0.02 |
| Female | |||||
| Age, years (%) | |||||
| <45 | 249 (26.0) | 25 (26.3) | 1 | ||
| 45-65 | 355 (37.1) | 34 (35.8) | 0.95 (0.55-1.64) | 0.86 | |
| >65 | 353 (36.9) | 36 (37.9) | 0.97 | 1.01 (0.59-1.73) | 0.95 |
| Overweight (BMI ≧ 24 kg/m2) | |||||
| No Yes | 568 (59.4) 389 (40.6) | 69 (72.6) 26 (27.4) | 0.016 | 1 0.55 (0.34-0.88) | 0.01 |
| EFEMP1 rs2009262 | |||||
| TT | 645 (67.4) | 52 (54.7) | 1 | ||
| TC/CC | 312 (32.6) | 43 (45.3) | 0.02 | 1.71 (1.12-2.62) | 0.01 |
| WT1 rs3809060 | |||||
| GG | 530 (55.4) | 49(51.6) | 1 | ||
| GT/TT | 427 (44.6) | 46 (48.4) | 0.55 | 1.17 (0.76-1.78) | 0.48 |
Gender differences in the onset age of inguinal hernia
The sex-specific cumulative incidence of inguinal hernia in the case group of the study population is depicted in Figure 2. Interestingly, we found that although there was a higher prevalence of male patients with inguinal hernia, female participants seemed to develop this disease at an earlier age. By the age of 60 years, 66.7% of female participants developed inguinal hernia, compared to only 33.6% of males, indicating that the pathogenesis of hernia might be different between genders.
Sex-specific factors associated with inguinal hernia
In order to investigate sex-specific risk factors for inguinal hernia, univariate analysis was performed (Table 3). In the female group, the EFEMP1 rs2009262 TC/CC genotype was significantly associated with inguinal hernia (OR: 1.71, 95% CI: 1.12-2.62, p = 0.01). Comparatively, in males with inguinal hernia, WT1 rs3809060 GT/TT was found to be a genetic variation significantly correlated with inguinal hernia (OR: 1.17, 95% CI: 1.02-1.35, p = 0.02). Both males and females in the inguinal hernia group had a lower BMI as compared to their counterparts.
In the multivariate analysis of males (Table 4), genotype WT1 rs3809060 GT/TT and BPH (OR: 3.19, 95% CI: 2.73 - 3.73, p < 0.001) were independently associated with a higher risk of inguinal hernia, while diabetes mellitus (OR: 0.61, 95% CI: 0.52 - 0.72, p < 0.001) was negatively associated with development of inguinal hernia. In the female group, genotype EFEMP1 rs2009262 TC/CC and COPD (OR: 2.34, 95% CI: 1.33 - 4.11, p = 0.003) were associated with greater incidence of inguinal hernia. The risk of inguinal hernia was significantly inversely associated with BMI ≧ 24 kg/m2 in men (OR: 0.75, 95% CI: 0.65 - 0.86, p<0.001) and women (OR: 0.60, 95% CI:0.37 - 0.98, p=0.042), compared with their counterparts.
BMI and genetic variants in different genders
We further stratified participants based on sex and BMI levels to explored the association of the genetic variants and overweight (BMI ≧ 24 kg/m2) with inguinal hernia. We conducted a multivariate logistic regression analysis as shown in Table 5, overweight males with EFEMP1 rs2009262 variant had a higher risk of inguinal hernia development after adjusted for potential confounders (OR: 1.31, 95% CI: 1.07-1.61, p = 0.010). Conversely, among females with BMI < 24 kg/m2, carriers of EFEMP1 rs2009262 had a 1.83-fold increased risk of inguinal hernia (OR: 1.83, 95% CI: 1.10-3.05, p = 0.02).
Multivariate logistic regression analysis of risks associated with inguinal hernia by sexa
| Variables | Male | Female | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p value | OR | 95% CI | p value | |
| EFEMP1 rs2009262 TC/CC | 1.14 | 0.99-1.32 | 0.072 | 1.74 | 1.13-2.68 | 0.013 |
| WT1 rs3809060 GT/TT | 1.15 | 1.00-1.33 | 0.044 | 1.23 | 0.79-1.89 | 0.347 |
| Overweight (BMI≧24 kg/m2) | 0.75 | 0.65-0.86 | < 0.001 | 0.60 | 0.37-0.98 | 0.042 |
| Hyperlipidemia | 0.94 | 0.80-1.10 | 0.409 | 0.63 | 0.36-1.10 | 0.107 |
| Hypertension | 1.12 | 0.96-1.31 | 0.165 | 0.81 | 0.45-1.46 | 0.479 |
| Diabetes mellitus | 0.61 | 0.52-0.72 | < 0.001 | 0.73 | 0.40-1.31 | 0.287 |
| Chronic kidney disease | 0.87 | 0.74-1.02 | 0.086 | 0.75 | 0.42-1.35 | 0.340 |
| COPD | 1.13 | 0.96-1.34 | 0.147 | 2.34 | 1.33-4.11 | 0.003 |
| BPH | 3.19 | 2.73-3.73 | < 0.001 | - | - | - |
aModel adjusted for genotypes, age, Body mass index, hyperlipidemia, hypertension, diabetes mellitus, chronic kidney disease, chronic obstructive pulmonary disease, and benign prostate hyperplasia.
Abbreviations: CI: confidence interval; OR: odds ratio; BMI: Body mass index; BPH: benign prostatic hyperplasia; COPD: chronic obstructive pulmonary disease.
Sex-specific associations of genetic variants and BMI levels with inguinal herniab
| Variables | Male | |||||
|---|---|---|---|---|---|---|
| BMI < 24 kg/m2 | BMI ≧ 24 kg/m2 | |||||
| OR | 95% CI | p value | OR | 95% CI | p value | |
| Age, years | 0.98 | 0.98-0.99 | < 0.001 | 0.99 | 0.98-0.10 | 0.002 |
| EFEMP1 rs2009262 TC/CC | 1.00 | 0.81-1.23 | 0.997 | 1.31 | 1.07-1.61 | 0.010 |
| WT1 rs3809060 GT/TT | 1.21 | 0.99-1.46 | 0.056 | 1.11 | 0.91-1.36 | 0.309 |
| Female | ||||||
| Age, years | 1.01 | 0.99-1.02 | 0.430 | 1.03 | 1.00-1.06 | 0.097 |
| EFEMP1 rs2009262 TC/CC | 1.83 | 1.10-3.05 | 0.020 | 1.44 | 0.62-3.36 | 0.395 |
| WT1 rs3809060 GT/TT | 1.32 | 0.80-2.20 | 0.281 | 0.99 | 0.44-2.27 | 0.989 |
bModel adjusted for age, genotypes, hyperlipidemia, hypertension, diabetes mellitus, chronic kidney disease, chronic obstructive pulmonary disease, and benign prostate hyperplasia.
Abbreviations: CI: confidence interval; OR: odds ratio; BMI: Body mass index; BPH: benign prostatic hyperplasia; COPD: chronic obstructive pulmonary disease.
Sex-specific genetic variations associated with different inguinal hernia subtypesc
| SNP | Type | Male | Female | ||
|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | ||
| rs2009262 | |||||
| Direct | 1.13 (0.82-1.55) | 0.46 | 0.68 (0.17-2.77) | 0.59 | |
| Indirect | 0.89 (0.68-1.17) | 0.41 | 0.75 (0.30-1.92) | 0.55 | |
| rs3809060 | |||||
| Direct | 1.62 (1.19-2.22) | 0.002 | 3.35 (0.71-15.69) | 0.13 | |
| Indirect | 0.78 (0.60-1.02) | 0.07 | 0.55 (0.21-1.43) | 0.22 | |
cModel adjusted for genotypes, hyperlipidemia, hypertension, chronic kidney disease, diabetes mellitus, chronic obstructive pulmonary disease, benign prostate hyperplasia.
Abbreviations: CI: confidence interval; OR: odds ratio.
Genotypes and inguinal hernia types
To further delineate genetic variation associated with hernia types, a multivariate regression analysis was conducted (Table 6). We identified that rs3809060 was significantly correlated with direct type inguinal hernia in males (OR: 1.62, 95% CI: 1.19 - 2.22, p = 0.002).
Discussion
This hospital-based case-control study identified preferential SNPs with increased susceptibility to adult-onset inguinal hernia; EFEMP1 rs2009262 in females and WT1 rs3809060 in males, specifically in association with direct-type inguinal hernia. We also demonstrated sex-specific risk factors associated with inguinal hernia, with COPD in women, BPH in men, and BMI in both genders. The WT1 rs3809060 locus exhibited an association with hernia risk among male participants, while overweight (BMI ≧ 24 kg/m2) males carrying the TC/CC genotype of the EFEMP1 rs2009262 variant were found to be at an increased risk of developing inguinal hernia. This study is the first to demonstrate genetic susceptibility loci of inguinal hernia in a Han Chinese population. Our result indicates that heredity plays a compelling role in the pathogenesis of inguinal hernia in a sex-specific manner.
Genetic defects in any components of the inguinal canal, including collagen, microfibrils, elastin, abdominal wall muscles, and glycosaminoglycan elements of the extracellular matrix, could result in inguinal hernia formation.[19] A number of genes or risk loci associated with inguinal hernia were identified by candidate gene approach[12] and genome-wide association studies;[16, 17, 20] most of them were correlated with connective tissue homoeostasis, such as elastin (ELN).[17] Decreased tissue concentration of ELN was observed in inguinal hernia patients,[21] and it has been reported that variants in ELN gene are associated with inguinal hernia development.[20] Epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1) belongs to the fibulin gene family, and it is crucial for ELN production and elastic fiber formation by regulating incorporation of elastin to the microfibril architecture.[22] EFEMP1 knockout mice present diminished elastic fibers in fascia and appear to have inguinal hernias or other large hernias.[23] EFEMP1 variants are also associated with various functional alterations, including body height,[24] waist circumference,[25] and forced vital capacity.[26] These collective correlations indicate that abnormal elastin tissue may result in the development of both inguinal hernia and COPD, which is compatible with our finding that COPD was associated with greater incidence of inguinal hernia in the female group.
Additionally, the altered activity of tissue inhibitor of metalloproteinases (TIMPs) and collagen degrading matrix metalloproteinases (MMPs) led to distorted collagen metabolism and diminished ratio of type I to type III collagen, which could affect the development of inguinal hernia.[27, 28] Previous studies have reported increased tissue expression of MMP2 and MMP9, while TIMP1 and TIMP3 were decreased in inguinal hernia patients.[28-31] WT1 has been revealed to activate TIMP3 and downregulate MMP-2 and MMP-9,[32] which could further correlate with dysregulation of the proportion of type I/III collagen and inguinal hernia formation. In men, there are several reasons that could explain the association of BPH and higher risk of inguinal hernia. Significant relations were reported among BPH and prostate cancer as BPH could augment risk of prostate cancer.[33] WT1 also regulates the levels of E-cadherin, contributing to the migratory potential of prostate cancer cells.[34] In addition, MMP-9, which is downregulated by WT1,[32] has been identified as a potential prostate tumor marker, while overexpression of MMP-9 was observed in prostate cancers.[35] Our result resembles the finding of a previous epidemiological study that showed BPH was associated with later inguinal hernia formation.[36]
In our study, indirect inguinal hernia accounted for 65 percent of repairs in men, which was similar to a previous study in Taiwan.[37] The OR exhibited for direct-type inguinal hernia was stronger for males with the rs3809060 GT/TT genotype than for the indirect-type inguinal hernia, indicating different underlying mechanisms and genetic susceptibility among the subtypes of inguinal hernia.[38] In females, as there were few cases with surgically identified inguinal hernia (N = 52), no genotype association was nominally observed. We also demonstrated that normal body mass index (BMI<24 kg/m2) was inversely related to the formation of inguinal hernia, which has been reported in many previous studies.[39-41] While EFEMP1 locus has been associated with anthropometric measures of body height and abdominal circumference, shared genetic effects may explain the plausible causality between BMI and inguinal hernia, as previously noted by Hélène Choquet et al.[20] However, further investigation of different pathophysiological associations of EFEMP1 in sex-specific manner may be considered as opposite correlations between EFEMP1 rs2009262 TC/CC and inguinal hernia were observed in different genders according to BMI levels.
There were some limitations in this study. First, we utilized the SNP array for genotyping. Not every variant associated with inguinal hernia was included in the probe design[18]. Hence, it is highly likely that coexistent genetic variants in the study group were not identified. Further study with next-generation sequencing is required to decode genetic contributions to inguinal hernia. Second, our study enrolled only cases with adult-onset inguinal hernia. As the age of inguinal hernia occurrence also peaks in the newborn period due to late closure of the processus vaginalis, it remains unknown whether rs2009262 and rs3809060 could also contribute to the development of pediatric inguinal hernia. Third, our participants were all Taiwanese, and therefore it might not be possible to extrapolate these results to other ethnic groups. Lastly, as our study is hospital-based, only patients who visited outpatient clinics with blood tests participated. The selection bias is likely. Therefore, we may underestimate the effect of these sex-specific risk factors associated with inguinal hernia.
Conclusion
In summary, this hospital-based study demonstrated a gender difference in the genetic susceptibility of adult-onset inguinal hernia. Further studies are needed to decipher the mechanisms underlying the complex interaction among sex, genetic background, and comorbidities.
Acknowledgements
We thank all of the participants and investigators from the Taiwan Precision Medicine Initiative.
Funding
This study was funded by Academia Sinica 40-05-GMM, AS-GC-110-MD02, and VTA111-V2-1-1, and National Science and Technology Council, Taiwan (NSTC -111-2634-F-A49-014, NSTC-111-2218-E-039-001, and NSTC-111-2314-B-075A-003-MY3), and Taichung Veterans General Hospital, Taiwan (TCVGH-1117001B, TCVGH-1127301C, TCVGH-1127302D, TCVGH-YM1120110, TCVGH-1127002B and TCVGH-1127304B). The funding organizations had no role in the design and conduct of the study.
Author Contributions
Authors' Contributions: research idea and study design: HCY and ICC; data acquisition: HCY, ICC, and GCL; data analysis/interpretation: ICC, GCL, YYK, MCL, YMC and CCH; statistical analysis: ICC and GCL; supervision or mentorship: YYK, MCL, YMC, and CCH. Each author contributed important intellectual content during manuscript drafting or revision and agrees to be personally accountable for the individual's own contributions and to ensure that questions pertaining to the accuracy or integrity of any portion of the work, even one in which the author was not directly involved, are appropriately investigated and resolved, including with documentation in the literature if appropriate.
Data Sharing Statement
All data used in this study are available in this article.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fitzgibbons RJ Jr, Forse RA. Clinical practice. Groin hernias in adults. N Engl J Med. 2015;372:756-63
2. Debas HT, Donkor P, Gawande A, Jamison DT, Kruk ME, Mock CN. et al. Essential Surgery. In: Beard JH, ed. Hernia and Hydrocele, 3rd ed(Volume 1). Washington (DC): The International Bank for Reconstruction and Development / The World Bank. 2015
3. Ruhl CE, Everhart JE. Risk factors for inguinal hernia among adults in the US population. Am J Epidemiol. 2007;165:1154-61
4. Lee CS, Kim JH, Choi BJ, Lee JI, Lee SC, Lee YS. et al. Retrospective study on prevalence of recurrent inguinal hernia: a large-scale multi-institutional study. Ann Surg Treat Res. 2020;98:51-5
5. Burcharth J, Pedersen M, Bisgaard T, Pedersen C, Rosenberg J. Nationwide prevalence of groin hernia repair. PLoS One. 2013;8:e54367
6. Melwani R, Malik SJ, Arija D, Sial I, Bajaj AK, Anwar A. et al. Body Mass Index and Inguinal Hernia: An Observational Study Focusing on the Association of Inguinal Hernia With Body Mass Index. Cureus. 2020;12:e11426
7. Watanabe T, Yokoyama S, Iwahashi M, Mori K, Yamade N, Yamaguchi K. et al. Asymptomatic patent processus vaginalis is a risk for developing external inguinal hernia in adults: A prospective cohort study. Ann Med Surg (Lond). 2021;64:102258
8. Lau H, Fang C, Yuen WK, Patil NG. Risk factors for inguinal hernia in adult males: a case-control study. Surgery. 2007;141:262-6
9. Nilsson H, Stranne J, Stattin P, Nordin P. Incidence of groin hernia repair after radical prostatectomy: a population-based nationwide study. Ann Surg. 2014;259:1223-7
10. Zöller B, Ji J, Sundquist J, Sundquist K. Shared and nonshared familial susceptibility to surgically treated inguinal hernia, femoral hernia, incisional hernia, epigastric hernia, and umbilical hernia. J Am Coll Surg. 2013;217:289-99.e1
11. Burcharth J, Pommergaard HC, Rosenberg J. The inheritance of groin hernia: a systematic review. Hernia. 2013;17:183-9
12. Zhang Y, Han Q, Li C, Li W, Fan H, Xing Q. et al. Genetic analysis of the TBX1 gene promoter in indirect inguinal hernia. Gene. 2014;535:290-3
13. Zhao Z, Tian W, Wang L, Wang H, Qin X, Xing Q. et al. Genetic and functional analysis of the TBX3 gene promoter in indirect inguinal hernia. Gene. 2015;554:101-4
14. Qiao Y, Zhang Z, Huang W, Pang S, Xing Q, Yan B. Two functional sequence variants of the GATA6 gene promoter in patients with indirect inguinal hernia. Gene. 2014;547:86-90
15. Sezer S, Şimşek N, Celik HT, Erden G, Ozturk G, Düzgün AP. et al. Association of collagen type I alpha 1 gene polymorphism with inguinal hernia. Hernia. 2014;18:507-12
16. Jorgenson E, Makki N, Shen L, Chen DC, Tian C, Eckalbar WL. et al. A genome-wide association study identifies four novel susceptibility loci underlying inguinal hernia. Nat Commun. 2015;6:10130
17. Hikino K, Koido M, Tomizuka K, Liu X, Momozawa Y, Morisaki T. et al. Susceptibility loci and polygenic architecture highlight population specific and common genetic features in inguinal hernias: genetics in inguinal hernias. EBioMedicine. 2021;70:103532
18. Wei CY, Yang JH, Yeh EC, Tsai MF, Kao HJ, Lo CZ. et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom Med. 2021;6:10
19. Barnett C, Langer JC, Hinek A, Bradley TJ, Chitayat D. Looking past the lump: genetic aspects of inguinal hernia in children. J Pediatr Surg. 2009;44:1423-31
20. Choquet H, Li W, Yin J, Bradley R, Hoffmann TJ, Nandakumar P. et al. Ancestry- and sex-specific effects underlying inguinal hernia susceptibility identified in a multiethnic genome-wide association study meta-analysis. Hum Mol Genet. 2022;31:2279-93
21. Ringpfeil F. Selected disorders of connective tissue: pseudoxanthoma elasticum, cutis laxa, and lipoid proteinosis. Clin Dermatol. 2005;23:41-6
22. Yanagisawa H, Davis EC. Unraveling the mechanism of elastic fiber assembly: The roles of short fibulins. Int J Biochem Cell Biol. 2010;42:1084-93
23. McLaughlin PJ, Bakall B, Choi J, Liu Z, Sasaki T, Davis EC. et al. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum Mol Genet. 2007;16:3059-70
24. Chiou JS, Cheng CF, Liang WM, Chou CH, Wang CH, Lin WD. et al. Your height affects your health: genetic determinants and health-related outcomes in Taiwan. BMC Med. 2022;20:250
25. Justice AE, Winkler TW, Feitosa MF, Graff M, Fisher VA, Young K. et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat Commun. 2017;8:14977
26. Loth DW, Soler Artigas M, Gharib SA, Wain LV, Franceschini N, Koch B. et al. Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nat Genet. 2014;46:669-77
27. Henriksen NA. Systemic and local collagen turnover in hernia patients. Dan Med J. 2016 63
28. Antoniou GA, Tentes IK, Antoniou SA, Simopoulos C, Lazarides MK. Matrix metalloproteinase imbalance in inguinal hernia formation. J Invest Surg. 2011;24:145-50
29. Smigielski J, Brocki M, Kuzdak K, Kołomecki K. Serum MMP 2 and TIMP 2 in patients with inguinal hernias. Eur J Clin Invest. 2011;41:584-8
30. Pascual G, Rodríguez M, Gómez-Gil V, Trejo C, Buján J, Bellón JM. Active matrix metalloproteinase-2 upregulation in the abdominal skin of patients with direct inguinal hernia. Eur J Clin Invest. 2010;40:1113-21
31. Peng X, Guo Z, Zhang Y, Sun B, Zhang Q. EFEMP1 in Direct Inguinal Hernia: correlation with TIMP3 and Regulation Toward Elastin Homoeostasis as Well as Fibroblast Mobility. J Invest Surg. 2022;35:203-11
32. Marcet-Palacios M, Ulanova M, Duta F, Puttagunta L, Munoz S, Gibbings D. et al. The transcription factor Wilms tumor 1 regulates matrix metalloproteinase-9 through a nitric oxide-mediated pathway. J Immunol. 2007;179:256-65
33. Zhang L, Wang Y, Qin Z, Gao X, Xing Q, Li R. et al. Correlation between Prostatitis, Benign Prostatic Hyperplasia and Prostate Cancer: A systematic review and Meta-analysis. J Cancer. 2020;11:177-89
34. Brett A, Pandey S, Fraizer G. The Wilms' tumor gene (WT1) regulates E-cadherin expression and migration of prostate cancer cells. Mol Cancer. 2013;12:3
35. Ma F, Wang Z, Abdularab A, Dai L, Niu Y, Jiang N. Matrix metalloproteinase 9 and prostate cancer risk: a meta-analysis of 1059 participants. Minerva Urol Nefrol. 2017;69:324-9
36. Wu YH, Juan YS, Shen JT, Wang HS, Jhan JH, Lee YC. et al. Lower urinary tract symptoms-Benign prostatic hyperplasia may increase the risk of subsequent inguinal hernia in a Taiwanese population: A nationwide population-Based cohort study. PLoS One. 2020;15:e0234329
37. Lee CH, Chiu YT, Cheng CF, Wu JC, Yin WY, Chen JH. Risk factors for contralateral inguinal hernia repair after unilateral inguinal hernia repair in male adult patients: analysis from a nationwide population based cohort study. BMC Surg. 2017;17:106
38. Somuncu S, Somuncu Ö S, Ballıca B, Tabandeh B. Deficiency of epithelial-mesenchymal transition causes child indirect inguinal hernia. J Pediatr Surg. 2020;55:665-71
39. Shrestha S, Upadhyay PK. Prevalence of Obesity in Inguinal Hernia Repair Patients in a Tertiary Care Center. JNMA J Nepal Med Assoc. 2021;59:156-9
40. Lau H, Fang C, Yuen WK. et al. Risk factors for inguinal hernia in adult males: a case-control study. Surgery. 2007;141:262-6
41. Burcharth J, Pommergaard HC, Bisgaard T, Rosenberg J. Patient-related risk factors for recurrence after inguinal hernia repair: a systematic review and meta-analysis of observational studies. Surg Innov. 2015;22:303-17
Author contact
![]() Corresponding authors: Yi-Ming Chen, M.D., Ph.D. Division of Allergy, Immunology and Rheumatology, Department of Internal Medicine, Taichung Veterans General Hospital, 1650, Section 4, Taiwan Boulevard, Xitun Dist., Taichung City 40705, Taiwan. Tel.: +886-4-2359-2525 ext. 3354; Fax: +886-4-2350-3285; E-mail: ymchen1gov.tw. Chia-Chi Hsu, M.D. Division of Pediatric Genetics and Metabolism, Children's Medical Center, Taichung Veterans General Hospital, Taichung, Taiwan. Tel.: +886-4-2359-2525 ext. 5987; Fax: +886-4-2359-5046; E-mail: chiackygov.tw.
Corresponding authors: Yi-Ming Chen, M.D., Ph.D. Division of Allergy, Immunology and Rheumatology, Department of Internal Medicine, Taichung Veterans General Hospital, 1650, Section 4, Taiwan Boulevard, Xitun Dist., Taichung City 40705, Taiwan. Tel.: +886-4-2359-2525 ext. 3354; Fax: +886-4-2350-3285; E-mail: ymchen1gov.tw. Chia-Chi Hsu, M.D. Division of Pediatric Genetics and Metabolism, Children's Medical Center, Taichung Veterans General Hospital, Taichung, Taiwan. Tel.: +886-4-2359-2525 ext. 5987; Fax: +886-4-2359-5046; E-mail: chiackygov.tw.

 Global reach, higher impact
Global reach, higher impact