Impact Factor
ISSN: 1449-1907
Int J Med Sci 2023; 20(1):142-150. doi:10.7150/ijms.77206 This issue Cite
Review
A Review of the Impact of Pharmacogenetics and Metabolomics on the Efficacy of Metformin in Type 2 Diabetes
1. Pharmacology Department, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia.
2. Department of Veterinary Medicine, University of Cambridge, Cambridge, United Kingdom.
Received 2022-7-17; Accepted 2022-12-2; Published 2023-1-1
Abstract
Metformin is the most often prescribed drug for people with type 2 diabetes (T2D). More than 120 million patients with T2D use metformin worldwide. However, monotherapy fails to achieve glycemic control in a third of the treated patients. Genetics contribute to some of the inter-individual variations in glycemic response to metformin. Numerous pharmacogenetic studies have demonstrated that variations in genes related to pharmacokinetics and pharmacodynamics of metformin's encoding transporters are mainly associated with metformin response.
The goal of this review is to evaluate the current state of metformin pharmacogenetics and metabolomics research, discuss the clinical and scientific issues that need to be resolved in order to increase our knowledge of patient response variability to metformin, and how to improve patient outcomes. Metformin's hydrophilic nature and absorption as well as its action mechanism and effectiveness on T2D initiation are discussed. The impacts of variations associated with various genes are analysed to identify and evaluate the effect of genetic polymorphisms on the therapeutic activity of metformin. The metabolic pattern of T2D and metformin is also indicated. This is to emphasise that studies of pharmacogenetics and metabolomics could expand our knowledge of metformin response in T2D.
Keywords: Type 2 diabetes, Metformin, Pharmacogenetics, Metabolomics, Metformin Transporters.
Introduction
Diabetes mellitus (DM) commonly known as diabetes is on the rise worldwide. According to the International Diabetes Federation's most recent estimate, there were roughly 537 million diabetics worldwide in 2021, and that number is projected to increase to 643 million by the year 2030 and 783 million by the year 2045 (1, 2). This equates to 1 in 10 adults aged between 20 and 79 years in the world suffering from diabetes. In addition, 541 million individuals were anticipated to be at risk of becoming diabetic in 2021 because of their impaired glucose tolerance. This results in extreme healthcare costs in addition to the enormous personal and social costs. Indeed, total diabetes healthcare costs were estimated to be nearly one trillion USD and will exceed this amount by 2030 (2). According to the World Health Organization (WHO), diabetes was the ninth leading cause of death worldwide in 2019 (3). Moreover, recent studies have revealed that patients with diabetes had a 2.3-fold higher risk of death and a 3.6-fold higher likelihood of being hospitalised with COVID-19 than those without diabetes (4-6). All this adds up to demonstrate the magnitude of the issue we face in the form of a global diabetes epidemic.
The prevalence of diabetes is growing at a fast rate in Saudi Arabia. In total, one-fourth of the adult population is affected by diabetes, and the number is anticipated to double by 2030. The most concerning trend is the rise in diabetes prevalence in recent years, with a nearly ten-fold increase in Saudi Arabia during the previous thirty years (7). Recently, the findings of a multi-center, retrospective, cross-sectional study conducted nationwide found that a significant number of COVID-19 patients (7.6 to 68.3%) had diabetes (8). Diabetes accounts for 24 percent of the nation's total healthcare spending, making it the highest per capita spending state in the Middle East and North Africa (2).
Ninety to ninety five percent of all diabetes cases are caused by type 2 diabetes (T2D). Despite at least eleven other classes of drugs being used for the same condition, metformin is still the most recommended treatment (80%-85%) for people with T2D. Most medical associations, including the International Diabetes Foundation and the American Diabetes Association, recommend metformin as the primary oral glucose-lowering medication in addition to lifestyle control to treat T2D (1, 9, 10). Previous studies have shown that metformin was prescribed as first-line monotherapy to over 120 million T2D patients worldwide (11), to 73 percent of T2D patients in the UK between 2000 and 2017, and to 89 percent of T2D patients in 2017 (12). Metformin is the drug of choice for T2D patients who are overweight because it reduces weight gain and the risk of microvascular problems (13). Furthermore, metformin use was associated with a continued decrease in microvascular risk, as well as a decrease in the risk of myocardial infarction in prediabetic patients (14). It is also a popular treatment for pre-diabetes, gestational diabetes, and polycystic ovary syndrome (15). Some studies have shown that in patients with chronic kidney disease (stage 3), metformin is contraindicated and users with kidney damage are more likely to develop lactic acidosis (16). The risk of most causes of death and incidence of terminal renal disease was, however, shown to be reduced by metformin in individuals with chronic kidney disease and renal fibrosis, according to recent research (17, 18). The disparity between the various studies is due to the complex renal effects of metformin, which depend on the kind of disease and also the type and onset of the injury.
The pharmacological response to metformin varies significantly across individuals. Earlier studies showed that metformin did not achieve the optimal glycemic regulation in 35% of patients, necessitating dose escalation or the use of a combined hypoglycemic medication (19, 20). In children and adolescents with newly diagnosed T2D, the failure rate was as high as 50% (21). Other studies indicated that African Americans had better glycemic regulation with metformin than European Americans (22).
The study of pharmacogenetics plays a constructive role in identifying the crucial genetic factors that significantly influence typical drug responses in the body. The high polymorphisms rate associated with certain genes generally accounts for subsequent drug responses in certain individuals. Certain variations can either enhance or reduce the overall exposure and activity of a drug in the body; therefore, study of these factors should be necessarily taken into account to achieve the required responses in patients (23). Moreover, studies examining links between metabolites and T2D risk and symptoms may also shed light on the disease's pathophysiological processes and lead to the discovery of new, reliable biomarkers that will help T2D be detected and treated more effectively (24, 25).
The primary goals of this review are to advance mechanistic knowledge of metformin's pharmacokinetics and pharmacodynamics in T2D and to examine the effects of its pharmacogenetics and metabolomics.
Pharmacological aspects of metformin
Metformin transporters involved in its pharmacokinetics
Metformin is excreted in its whole form in urine. The drug's hydrophilic nature prevents it from diffusing across cell membranes, so it must rely on organic cation transporters for active transport into and out of enterocytes, hepatocytes, and renal epithelial cells (26).
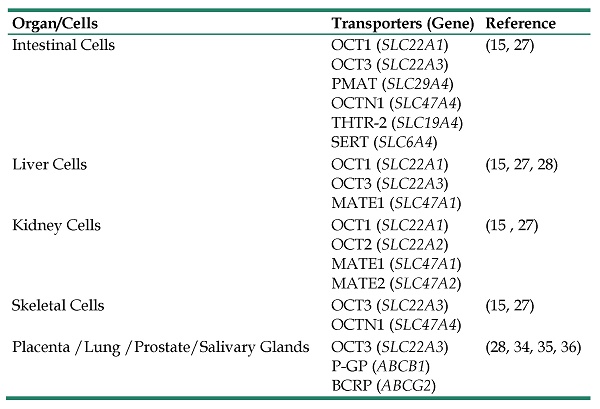
After oral intake, metformin is absorbed by enterocytes from the gut endothelium through Solute Carrier Family 29 Member 4 (SLC29A4), plasma monoamine transporter (PMAT), organic cation transporter 1 (OCT1; SLC22A1), OCT3 (SLC22A3) and OCTN1 (SLC22A4) located on the inner surface of the gut epithelium. Additionally, SERT (SLC6A4) and THTR-2 (SLC19A4) may contribute to the intestinal absorption of metformin, whereas OCT1 transports it into the bloodstream. OCT1 and OCT3 both take metformin from the bloodstream and store it in hepatocytes (27, 28). Polyspecificity is one of the OCT1's most distinguishing characteristics (29). Currently, it has been determined that 150 cationic organic molecules of various chemical compositions, including well used medications like metformin, are substrates for the OCT1 (30-32).
Metformin is transported into kidney epithelial cells by OCT1 (SLC22A1) and OCT2 (SLC22A2). Additionally, the disposal of metformin from proximal tubule cells into the urine is carried out by the multidrug and toxin extrusion 1 protein (MATE1) encoded by the gene (SLC47A1) and MATE2 encoded by the gene (SLC47A2), while MATE1 (SLC47A1) is also in charge of the disposal of metformin from hepatocytes into the bile (27, 33). Furthermore, OCT3 is expressed in many organs such as the lungs, kidney, placenta, prostate and salivary glands, where it may play a role in metformin's transportation (34, 35). ATP-binding cassette (ABC) transporters P-glycoprotein (P-GP) and the breast cancer resistance protein (BCRP) transporters have been shown to be mainly responsible for foetal-to-maternal efflux of metformin in gestational diabetes subjects (36). A summary of various organ transporters involved in the pharmacokinetics of metformin is presented in Table 1. When metformin is given orally, half of it is lost in the systemic circulation and has a plasma half-life of 4 to 8 hours in individuals with normal renal function (37, 38).
A Summary of Various Transport Proteins with Tissue-Specific Expression Pattern Involved in Pharmacokinetics of Metformin.
| Organ/Cells | Transporters (Gene) | Reference |
|---|---|---|
| Intestinal Cells | OCT1 (SLC22A1) OCT3 (SLC22A3) PMAT (SLC29A4) OCTN1 (SLC47A4) THTR-2 (SLC19A4) SERT (SLC6A4) | (15, 27) |
| Liver Cells | OCT1 (SLC22A1) OCT3 (SLC22A3) MATE1 (SLC47A1) | (15, 27, 28) |
| Kidney Cells | OCT1 (SLC22A1) OCT2 (SLC22A2) MATE1 (SLC47A1) MATE2 (SLC47A2) | (15 , 27) |
| Skeletal Cells | OCT3 (SLC22A3) OCTN1 (SLC47A4) | (15, 27) |
| Placenta /Lung /Prostate/Salivary Glands | OCT3 (SLC22A3) P-GP (ABCB1) BCRP (ABCG2) | (28, 34, 35, 36) |
Metformin pharmacodynamics
Although metformin has been the most successful treatment for T2D for more than 60 years, its specific action is still uncertain. Metformin is thought to regulate blood sugar levels through pleiotropic processes involving multiple pathways (38, 39). The drug works by decreasing glucose production from the liver by inhibiting hepatic gluconeogenesis. Metformin has been shown to act through the activation of AMP-activated protein kinase (AMPK), an enzyme involved in cellular regulation of energy homeostasis, as well as lipid and glucose metabolism, however, AMPK independent mechanisms have also gained interest recently (39). Earlier studies had shown that metformin inhibited mitochondrial Complex I (40, 41). This was believed, at least in part to contribute to prevent hepatic glucose production and increase glucose utilization in peripheral tissues. A recent study confirmed that metformin inhibits mitochondrial Complex 1; prevent mitochondrial ATP production, thereby, increase cytoplasmic AMP:ATP and ADP:ATP ratios and thus activate AMPK dependent-pathways (42). Higher AMP:ATP ratio or compromised cellular energy balance has been shown to inhibit fructose-1,6-bisphosphatase and thereby causing acute inhibition of gluconeogenesis (43). Activated AMPK regulates energy homeostasis by phosphorylation of acetyl-CoA carboxylase 1 and 2, thereby inhibiting fat synthesis, accelerating fat oxidation and depletion of fat stores, ultimately improving insulin sensitivity (44). Gluconeogenesis is an energy intensive process that requires 6 molecules of ATP per molecule of glucose produced. Therefore, metformin inhibition of ATP production explains its inhibition of gluconeogenesis. Moreover, changes in the NAD+:NADH ratio resulting from respiratory chain inhibition by metformin are also believed to contribute towards inhibition of gluconeogenesis. Mitochondrial glycerophosphate dehydrogenase, a redox shuttle enzyme, has also been proposed as a mitochondrial target of metformin (45), however, further studies are warranted to establish its role in inhibition of gluconeogenesis. Multiple mechanisms of metformin action especially its effects on energy metabolism are reviewed in (39).
Metformin also affects the function of AMPK through the serine/threonine kinases Ataxia-telangiectasia (ATM) and serine threonine kinase 11 (STK11). In liver cells, activated AMPK reduces the chances of fat diseases by directly repressing lipogenesis and cholesterol biosynthesis, which is mainly achieved by enhancing the fatty acid beta-oxidation and mitochondrial biogenesis in the body. Metformin's pleiotropic effects can help to reduce diabetes complications, especially cardiovascular diseases (26). In the skeletal muscle cells, metformin activates AMPK to promote glucose uptake by increasing SLC2A4 encoded glucose transporter type 4 (GLUT-4) translocation (46, 47).
In gut, metformin increases glucose uptake, glycolytic lactate production, glucagon-like peptide-1 (GLP-1) secretion and bile acid pool in addition to microbiome alterations thus having pleiotropic effects on blood glucose homeostasis (48, 39). Recent studies have shown that metformin treatment leads to changes in gut microbiota and relative abundance of microbial metabolites (49,50). Cumulative evidence from those reports confirms that hypoglycemic effects of metformin were associated with AMPK activation and microbial metabolites involved in energy metabolism, gluconeogenesis and branched-chain amino acid metabolism. To fully understand how metformin works in the gut, however, further research is required.
Metformin pharmacogenetics
Previous studies have greatly focused on drawing associations among various genomic factors and their impacts on drug responses. In the case of metformin, OCT1 is highly polymorphic and is preferentially encoded by the gene SLC22A1. This transporter protein is greatly expressed in the liver and few other body cells. Interindividual differences in metformin responses among T2D patients can be attributed to the high polymorphisms associated with OCT1. A loss of functional variant associated with OCT1 has been linked to lower metformin response in some patients (24, 51). Four functional variations (rs12208357/R61C, rs72552763/M420del, rs34059508/G465R, and/or rs34130495/G401S) were connected to a decreased metformin's ability to lower blood sugar in healthy human volunteers following an oral glucose tolerance test (OGTT), according to research by Shu and colleagues (52). Reduced functional variants of OCT1 (rs72552763/M420del, rs12208357/R61C, rs34059508/G465R, and rs34130495/G401S) were also associated with trough concentrations of metformin and improvement in HbA1c after six months of metformin treatment in a cohort of 151 T2D patients in a prospective multicentre South Danish Diabetes Study (53). In a study of 1,915 completely metformin-tolerant and 251 intolerant T2D patients, allelic variants of OCT1 rs12208357/R61C, rs72552763/M420del, rs34130495/G401S, rs55918055/C88R, and rs34059508/G465R were found to be related to metformin intolerance due to increased metformin accumulation in enterocytes (54). Later studies reported three variants of OCT1 (rs622342, rs628031, and rs594709) that showed overall reduction in the efficacy of metformin among various populations (55, 56). Metformin hepatic accumulation was also found to be decreased in humans carrier of rs12208357/R61C and rs72552763/M420del SLC22A1 forms, although the blood levels of metformin were unaltered (24). OCT1 gene polymorphisms, however, may not have a significant impact on the clinical effectiveness of metformin, according to a prior study by Shikata et al. (57). Similar to this, the GoDARTS research revealed no association between the two most prevalent OCT1 variations in terms of allelic frequency in persons of European descent—rs12208357/R61C and rs72552763/M420del—with glycaemic response to metformin in a sample of 1,531 patients with T2D (58). Therefore, it is imperative to say that some OCT1 variants have effects on metformin response but the small number of patients and the co-medication of other anti-hyperglycaemic drugs in some study groups represent a drawback and explain the discrepancies between the various studies.
Several studies have found no evidence of a connection between OCT2 variants (encoded by the gene SLC22A2) and metformin response (24, 51, 59, 60). However, it has been demonstrated that the particular C allele of the variant rs8192675 in the gene SLC2A2, which codes for the glucose transporter (GLUT2), is crucial for controlling the metformin action (61). Reduced metformin uptake and altered substrate selectivity were present in OCT3 variants T44M (c.131C>T), V423F (c.1267G>T), and T400I (c.1199C>T), particularly in the latter two (20).
Inconsistent findings have been reported for metformin response by variants of SLC47A1 gene encoding MATE1 (53, 60, 62, 63). According to studies, homozygous carriers of the SLC47A1 rs2289669 A-allele have a much greater decrease in HbA1c following a six-month therapy with metformin than do carriers of the more prevalent G-allele. Therefore, approximately 20% of T2D patients showed a higher two-fold (0.55%) reduction in HbA1c levels than the rest of the patients (63). However, meta-analysis of Metformin Genetics Consortium studies involving nearly 8,000 T2D patients found no substantial association between glycaemic response to metformin and MATE1 transporter genes besides other genes encoding OCT1, OCT2, MATE2-K, and OCTN1 transporters (60). A recent pharmacogenetics study found no association of metformin transporters OCT1, OCT2, OCT3 and P-GP with therapeutic inefficacy among Mexican T2D patients (36).
The transcription factor 7-like 2 (TCF7L2) gene has been demonstrated to be closely related with T2D through poor glucose control and insulin production, in addition to genes that encode transporter proteins and influence responses in patients with T2D (64). Physiological investigations have also indicated that rs290487 variant played a role in insulin resistance, implying that Wnt signalling is involved in insulin resistance (65). Furthermore, TCF7L2 variants were also linked to a faster decline in pancreatic ß-cell function, higher glycemic levels, and lower insulin production, all of which are linked to familial history of T2D (66). Other studies have shown that the TCF7L2 rs 7903146T genotype is associated with both T2D and obesity, and that carriers may have changes in their plasma metabolic profiles due to phospholipids, suggesting that phospholipids may cause metabolic abnormalities prior to the onset of glucose intolerance (67, 68). These effects were recently brought to light by research demonstrating that the TCF7L2 gene controls a number of processes, including adipogenesis and disorders like T2D, as a downstream effector of the Wnt/ß-catenin signaling system (69).
From these various studies there seems to be lack of consistency in molecular pathways, and the need for more functional studies on metformin pharmacodynamics. Known metformin pharmacokinetic and pharmacodynamic gene variants significantly linked to metformin clinical response are presented in Table 2.
Known metformin pharmacokinetic and pharmacodynamic gene variants (SNPs) significantly linked to metformin clinical response.
| Gene | dbSNP ID | Cohort | Reference |
|---|---|---|---|
| SLC22A1 (OCT1) | rs1867351 rs4709400 rs628031 rs2297374 | Han Chinese 153 T2D 124 Control | (55) |
| 256 healthy brazilian | (70) | ||
| rs622342 | 125 Chinese | (71) | |
| 63 Lebanese patients | (72) | ||
| 256 Brazilian healthy | (70) | ||
| rs12208357 rs72552763 rs34059508 | 256 healthy brazilian | (70) | |
| rs34130495 rs461473 rs34104736 | 371 Danish T2D patients | (53) | |
| rs594709 | 267 T2D Chinese patients and182 healthy subjects | (56) | |
| SLC22A2(OCT2) | rs315978 rs662301 | 2,994 Participants in Diabetes Prevention Program | (62) |
| rs316019 | 125 Chinese participants | (71) | |
| 256 Brazilian adults | (70) | ||
| 91 Korean subjects | (73) | ||
| rs201919874 | 400 Pakistani patients | (74) | |
| SLC22A3 (OCT3) | rs3127602 | 258 White Europeans | (75) |
| rs520685 rs520829 | 25 Korean subjects | (76) | |
| SLC29A4 (PMAT) | rs10234709 | 258 White Europeans | (75) |
| rs2685753 | 91 Korean subjects | (73) | |
| rs3889348 rs4720572 rs4299914 rs6971788 | 91 Korean subjects | (73) | |
| SLC47A1(MATE1) | rs2289669 | 267 T2D Chinese patients and 182 healthy subjects | (56) |
| 125 Chinese subjects | (71) | ||
| 256 Brazilian subjects | (70) | ||
| 91 Korean subjects | (73) | ||
| rs8065082 rs2453583 | 2,994 DPP | (62) | |
| rs2120274 | 258 White Europeans | (75) | |
| rs2252281 | 125 Chinese subjects | (71) | |
| 256 Brazilian subjects | (70) | ||
| SLC47A2(MATE2) | rs4621031 | 258 White Europeans | (75) |
| rs34399035 | 371 Danish T2D Patients | (53) | |
| rs12943590 | 256 Brazilian subjects | (70) | |
| 91 Korean subjects | (73) | ||
| rs34834489 | 91 Korean subjects | (73) | |
| Rs138244461 | 400 Pakistani patients | (74) | |
| STK11 | rs2301759 | 258 White Europeans | (75) |
| rs2075604 | Chinese 94 T2DMpatients | (71) | |
| ATM | rs1800058 | 258 White Europeans | (75) |
| rs11212617 | 460 Russian T2DM patients | (77) | |
| TCF7L2 | rs7903146 | 525 Taiwanese subjects | (65) |
| 1023 Brazillian subjects | (68) | ||
| ABCB1(P-GP) | rs1128503 rs2032582 | 103 Mexican DMT2 patients | (78) |
Metformin and metabolomics studies in T2D
The systematic identification and measurement of all metabolic products in the human body is known as metabolomics. This field has the potential to give researchers with new diagnostic biomarkers for disease states, as well as assessing therapy response to drugs on an individual basis. A systematic review and meta-analysis of cross-sectional and prospective human metabolomics studies in prediabetes and T2D patients showed higher levels of carbohydrates (glucose and fructose), lipids (phospholipids, sphingomyelins, and triglycerides), and amino acids (branched-chain and aromatic amino acids)) in T2D patients compared to control subjects (79). Meta-analysis of prospective studies provided evidence that people with higher levels of isoleucine, leucine, valine, tyrosine and phenylalanine had higher risk of developing T2D while increased glycine and glutamine levels were found to be associated with lower risk of developing T2D (79). The authors reported that multiple (more than 10) studies reported positive correlation of sugar metabolites dihexose, glucose, mannose, fructose, and arabinose with T2D. Another study reported that 1,5-anhydroglucitol plasma levels were about 37.8% lower in T2D patients while mannose, glucose, deoxyhexose, and di-hexose levels were significantly higher in diabetic patients than in controls (80). One of the endogenous substrates for OCT2 has been discovered as tryptophan, and it has the potential to serve as a biomarker candidate for the variability of OCT2's transport activity (81). Along with a number of organic substances, such as purines and metabolites of the urea cycle, such as ornithine, arginine, and citrulline, several organic acids, such as maleic acid, acetic acid, and dimethyl ester, have also been linked to T2D. (79, 82). Studies have also investigated the relationship between T2D and branched chain amino acids leucine, isoleucine, and valine, and discovered that high levels of these amino acids are linked to insulin resistance in overweight children (83). Other studies have reported that an increase in the levels of branched-chain amino acids is a specific and reliable indicator of future insulin resistance in T2D patients (79, 84). Moreover, Yoon, (2016) have found that patients with T2D have greater plasma concentrations of aromatic and branched-chain amino acids, as well as a higher glutamate to glutamine ratio, than healthy people (82). Amino acids (including hydroxy acids and hydroxybutyrate) have been associated to increased insulin resistance and decreased glucose tolerance in diabetic individuals, whereas 3,hydroxybutyrate and hydroxybutyrate have been connected to a greater risk of prediabetes (85). Metformin treatment of obese T2D patients for 6 months resulted in 30 dysregulated (21 were up-regulated and 9 were down-regulated) metabolites mainly related to amino acids metabolism to change to obese control levels. Metformin treatment may have adjusted the dysregulated pathways in T2D or those dysregulated pathways may be common for both T2D and metformin. However, 71 dysregulated (30 up-regulated and 41 down-regulated) metabolites remained unchanged after metformin treatment (86). The study was underpowered due to smaller sample size. Similar large-scale targeted and untargeted metabolomics studies using a multi-platform approach could unravel novel molecular targets of metformin in T2D patients. Numerous glutamine-containing dipeptides, such as glutathionyl-L-cysteine and creatine, were found to be dramatically elevated in T2D. Those included glutaminyl-glutamine, glutamyl-glutamine, glutaminyl-glutamic acid, tyrosyl-glutamine, and glutamyl-tyrosine and then returned to obese comparable levels after metformin treatment (86). Given previously unestablished effect of metformin on glutamine metabolism, further studies are required to validate this finding. The 10 dysregulated metabolites only observed after metformin treatment may be involved in pharmacodynamics of metformin and shows the power of metabolomics studies in understanding drug action. The glycolytic pathway intermediate 3,phosphoglycerate (3PGA), which is a source of the metabolite serine, was downregulated in T2D, suggesting a potential change in the disease's metabolic route.
Adam and colleagues in a well-defined German Kooperative Gesundheitsforschung in der Region Augsburg (KORA) cohort looked at the metabolite profile of T2D patients undergoing metformin treatment (87). They discovered citrulline and an unknown metabolite associated with metformin treatment in T2D patients using a non-targeted mass spectrometry approach. This finding was repeated in mice and confirmed in a follow-up cohort. The same group had discovered that T2D patients taking metformin had slightly lower ornithine levels (88). The authors speculated that metformin/AMPK/eNOS/NO could mediate increased citrulline excretion in urine, resulting in lower citrulline levels in serum. Recent research has demonstrated that the metformin-treated diabetic individuals in both the lean and obese groups had lower levels of taurine, citrulline, and 5-hydroxymethyluracil (HMU), although levels of salicylic acid, L-proline, and L-alanine were increased (86). According to the same study, guanido-acetic acid and L-arginine were among the down-regulated metabolites in the metformin-treated patients with T2D in the lean and obese groups. Contrarily, the metabolites of the urea cycle, such as homoarginine and citrulline, the methylene homologue of arginine, are down-regulated in the obese T2D group receiving metformin. Some metabolic patterns as an effect of prediabetes, diabetes and also as metformin therapy in T2D are shown in Table 3.
Taken together, above studies have shown the value of using metabolomics methods to identify potential targets and molecular mechanism in metformin pharmacodynamics in T2D patients.
Some metabolic patterns as an effect of prediabetes, diabetes and metformin therapy in T2D (↑: up-regulated, ↓:down-regulated).
| Prediabetes (25) | Diabetes (25) | Metformin in T2D (86) |
|---|---|---|
| Branch-chain amino acids ↑ | Branch-chain amino acids | L-arginine ↓ |
| Aromatic amino acids | Aromatic amino acids | guanidoacetic acid ↓ |
| Glutamine/glutamate ratio | Glutamine/glutamate ratio | L-proline ↑ |
| B-Hydroxybutyrate ↑ | Mannose ↑ | taurine ↓ |
| glucose ↑ | L-alanine ↑ | |
| sugar metabolites ↑ | citruline ↓ | |
| organic acid ↑ | 5-hydroxymethyluracil ↓ | |
| 1,5,Anhydroglucitol ↓ | Salicylic acid ↑ |
Conclusion
This article describes the importance of determining the pharmacogenetics and metabolomics for interindividual differences in metformin response. Exploration of metformin's clinical omics will not only lead to better prescribing, but it will also help to explain the pleiotropic mechanisms by which metformin works. Various pathways implicated in the effects of T2D drugs are possibly multifactorial. However, studies have demonstrated the value of metabolomics in exploring the pharmacodynamics of metformin in patients with T2D. Genes with metformin response polymorphisms suggest that genetic factors may play a significant role in the therapeutic response to T2D treatment. Moreover, this review provides ample evidence about the association between various transporter genes polymorphism and its significant impact on altered metformin response in T2D patients. There is a need for more research in sizable cohorts of previously examined patients from various ethnic/genetic and cultural backgrounds. Future development of specialized tools for improved therapy will be made possible by a greater knowledge of metformin metabolomics, T2D, pharmacogenetics, and frequent and unusual gene mutations in T2D patients.
Acknowledgements
The authors are grateful to the support of the Deputyship for Research and Innovation in the Ministry of Education in Saudi Arabia for funding the research work under the project number (988).
Competing Interests
H.M. Alkreathy, M.S. Ahmad, H.M. Abualhamail, F.A. Alharbi, and Z.A. Damanhouri affirm that they do not have any competing interests with regard to the current study.
References
1. IDF. 2006. Clinical guidelines task force. Global guideline for Type 2 Diabetes: recommendations for standard, comprehensive, and minimal care. doi: 10.1111/j.1464-5491. 2006 01918.x
2. IDF Diabetes Atlas. International Diabetes Federation. Tenth edition. 2021 Facts & Figures https://idf.org/aboutdiabetes/what-is-diabetes/facts-figures.html)
3. WHO Diabetes Report. 2022. https://www.who.int/news-room/fact-sheets/detail/diabetes. 2022
4. Fox T, Ruddiman K, Lo KB, Peterson E, DeJoy R, Salacup G. The relationship between diabetes and clinical outcomes in COVID-19: a single-center retrospective analysis. Acta Diabetol. 2020:1-6 DOI: 10.1007/s00592-020-01592-8
5. Sourij H, Aziz F, Bruer A, Ciardi C, Clodi M, Fasching P. COVID-19 fatality prediction in people with diabetes and prediabetes using a simple score upon hospital admission. Diabetes Obes Metab. 2021 23(2). DOI: 10.1111/dom.14256
6. Schlesinger S, Neuenschwander M, Lang A, Pafili K, Kuss O, Herder C. Risk phenotypes of diabetes and association with COVID-19 severity and death: a living systematic review and meta-analysis. Diabetologia. 2021;64(7):1480-1491 DOI: 10.1007/s00125-021-05458-8
7. Robert A A, Al Dawish A M. The Worrying Trend of Diabetes Mellitus in Saudi Arabia: An Urgent Call to Action. Current Diabetes Reviews. 2020;16(3):204-210 doi:10.2174/1573399815666190531093735
8. Robert A A, Al SaeedA, Al Dawish M A. COVID-19 among people with diabetes millitus in Saudi Arabia: current situation and new perspectives. Diabetes Metab Syndr. 2021;15(5):102231. doi:10.1016/j.dsx.2021.102231
9. American Diabetes Association. Standards of Medical Care in Diabetes-2019 Abridged for Primary Care Providers. Clinical diabetes: a publication of the American Diabetes Association. 2019;37(1):11-34 doi:10.2337/cd18-0105
10. Baker C, Retzik-Stahr C, Singh V, Plomondon R, Anderson V, Rasouli N. Should metformin remain the first-line therapy for treatment of type 2 diabetes? Ther Adv Endocrinol Metab. 2021;12:1-13 doi:10.1177/2042018820980225
11. Viollet B, Guigas B, Garcia NS, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci. 2012;122(6):253-70
12. Wilkinson S, Douglas I, Stirnadel-Farrant H, Fogarty D, Pokrajac A, Smeeth L, Tomlinson L. Changing use of antidiabetic drugs in the UK: trends in prescribing 2000-2017. BMJ open. 2018;8(7):e022768
13. Lentferink YE, Knibbe CA, Van der Vorst MM. Efficacy of Metformin Treatment with Respect to Weight Reduction in Children and Adults with Obesity: A Systematic Review. Drugs. 2018;78(18):1887-1901 DOI: 10.1007/s40265-018-1025-0. PMID: 30511324
14. Sardu C, D'Onofrio N, Torella M. et al. Pericoronary fat inflammation and Major Adverse Cardiac Events (MACE) in prediabetic patients with acute myocardial infarction: effects of metformin. Cardiovasc Diabetol. 2019;18:126. doi: 10.1186/s12933-019-0931-0
15. Sam S, Ehrmann DA. Metformin therapy for the reproductive and metabolic consequences of polycystic ovary syndrome. Diabetologia. 2017;60(9):1656-61
16. Connelly PJ, Lonergan M, Soto-Pedre E, Donnell L, Zhou K, Pearson ER. Acute kidney injury, plasma lactate concentrations and lactic acidosis in metformin users: A GoDarts study. Diabetes Obes Metab. 2017;19(11):1579-86
17. Kwon S, Kim YC, Park JY, Lee J, An JN, Kim CT, Oh S, Park S, Kim DK, Oh YK, Kim YS, Lim CS, Lee JP. The Long-term Effects of Metformin on Patients with Type 2 Diabetic Kidney Disease. Diabetes Care. 2020;43(5):948-955 doi: 10.2337/dc19-0936
18. Pan Q, Lu X, Zhao C, Pan Q, Lu X, Zhao C, Liao S, Chen X, Guo F, Yang C, Liu H F. Metformin: the updated protective property in kidney disease. Aging. 2020;12(9):8742-8759 doi: 10.18632/aging.103095
19. Cook MN, Girman CJ, Stein PP, Alexander CM. Initial monotherapy with either metformin or sulphonylureas often fails to achieve or maintain current glycaemic goals in patients with Type 2 diabetes in UK primary care. Diabet Med. 2007;24(4):350-8
20. Chen LB, Pawlikowski B, Schlessinger A, More SS, Stryke D, Johns SJ, Portman MA, Chen E, Ferrin TE, Sali A, Giacomini KM. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics. 2010;20(11):687-699
21. Hermann LS. 1990. Biguanides and sulfonylureas as combination therapy in NIDDM, Diabetes Care. 1990;1(13):37-41
22. Williams LK, Padhukasahasram B, Ahmedani BK, Peterson EL, Wells KE, González Burchard E, Lanfear DE. Differing effects of metformin on glycemic control by race-ethnicity. J Clin Endocrinol Metab. 2014;99(9):3160-8
23. Weinshilboum RM, Liewei W. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. 2017;92(11):1711-1722
24. Sundelin EI, Gormsen LC, Jensen JB, Vendelbo MH, Jakobsen S, Munk OL, Christensen MM, Brøsen K, Frøkiaer J, Jessen N. Genetic polymorphisms in organic cation transporter 1 attenuates hepatic metformin exposure in humans. Clin Pharmacol Ther. 2017;102(5):841-8
25. Arneth B, Arneth R, Shams M. Metabolomics of Type 1 and Type 2 Diabetes. Int J Mol Sci. 2017;20:2467. doi:10.3390/ijms20102467
26. Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Bio. 2012;19(10):1222-36
27. Liang X, Giacomini KM.Transporters Involved in Metformin Pharmacokinetics, Treatment Response. J Pharm Sci. 2017; 106: 2245-2250. http://dx.doi.org/10.1016/j.xphs. 2017 04.078
28. Yang Y, Zhang Z, Li P, Kong W, Liu X, Liu L. A Whole-Body Physiologically Based Pharmacokinetic Model Characterizing Interplay of OCTs and MATEs in Intestine, Liver and Kidney to Predict Drug-Drug Interactions of Metformin with Perpetrators. Pharmaceutics. 2021;13:698. doi: 10.3390/ pharmaceutics13050698
29. Meyer MJ, Tzvetkov MV. OCT1 Polyspecificity—Friend or Foe? Front Pharmacol. 2021;12:698153. doi: 10.3389/fphar.2021.698153
30. Shen C-H, Zhang Y-X, Lu R-Y, Jin B, Wang S, Liu Z-R. et al. Specific OCT1 and ABCG2 Polymorphisms Are Associated with Lamotrigine Concentrations in Chinese Patients with Epilepsy. Epilepsy Res. 2016;127:186-190 doi:10.1016/j.eplepsyres.2016.09.004
31. Tzvetkov MV, Matthaei J, Pojar S, Faltraco F, Vogler S, Prukop T. et al. Increased Systemic Exposure and Stronger Cardiovascular and Metabolic Adverse Reactions to Fenoterol in Individuals with Heritable OCT1 Deficiency. Clin Pharmacol Ther. 2018;103:868-878 doi:10.1002/cpt.812
32. Haberkorn B, Fromm M F, König J. Transport of Drugs and Endogenous Compounds Mediated by Human OCT1: Studies in Single- and Double-Transfected Cell Models. Front Pharmacol. 2021; 12. http://doi:10.3389/fphar. 2021 662535
33. Han T, Proctor WR, Costales CL, Cai H, Everett RS, Thakker DR. Four cationselective transporters contribute to apical uptake and accumulation of metformin in Caco-2 cell monolayers. J Pharmacol Exp Ther. 2015;352(3):519-528
34. Chen EC, Liang X, Lee SW, Geier EG, Stocker SL, Chen L, Giacomini KM. Targeted Disruption of Organic Cation Transporter 3 Attenuates the Pharmacologic Response to Metformin. Mol Pharmacol. 2015;88(1):75-83 DOI: doi: 10.1124/mol.114.096776
35. Zazuli Z, Duin NJ, Jansen K, Vijverberg SJ, Maitland-van der Zee A, Masereeuw R. The Impact of Genetic Polymorphisms in Organic Cation Transporters on Renal Drug Disposition. Int J Mol Sci. 2020;21:6627. doi: 10.3390/ijms21186627
36. Hemauer SJ, Patrikeeva SL, Nanovskaya TN, Hankins GDV, Ahmed MS. Role of human placental apical membrane transporters in the efflux of glyburide, rosiglitazone, and metformin. Am J Obstet Gynecol. 2010;202(4):383.e1-7 doi: 10.1016/j.ajog.2010.01.035
37. Pawlyk AC, Giacomini KM, McKeon C, Shuldiner AR, Florez JC. Metformin pharmacogenomics: current status and future directions. Diabetes. 2014;63(8):2590-9
38. Alemón-Medina R, Altamirano-Bustamante N, Lugo-Goytia G, García-Álvarez R, Rivera-Espinosa L, Torres-Espíndola LM, Chávez-Pacheco JL, Juárez-Olguín H, Gómez-Garduño J, Flores-Pérez C, Fernández-Pérez PG. Comparative Bioavailability and Pharmacokinetics Between the Solid Form of Metformin vs a Novel Liquid Extemporaneous Formulation in Children. Dose Response. 2021;19(3):15593258211033140. doi: 10.1177/15593258211033140
39. Rena G, Hardie DG, Pearson ER. The mechanisms of action of metformin. Diabetologia. 2017;60:1577-1585 doi: 10.1007/s00125-017-4342-z
40. Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(3):607-14 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1221104/
41. El-Mir MY, Nogueira V, Fontaine E, Avéret N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. Biol Chem. 2000;275(1):223-8 doi: 10.1074/jbc.275.1.223
42. Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, Towler MC, Brown LJ, Ogunbayo OA, Evans AM, Hardie DG. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010;11(6):554-65 doi: 10.1016/j.cmet.2010.04.001
43. Vincent MF, Marangos PJ, Gruber HE, Van den Berghe G. Inhibition by AICA riboside of gluconeogenesis in isolated rat hepatocytes. Diabetes. 1991;40(10):1259-66 doi: 10.2337/diab.40.10.1259
44. Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen Z, O'Neill HM, Ford RJ, Palanivel R, O'Brien M, Hardie DG, Macaulay, Schertzer SJ, Dyck JRB, van Denderen BJ, Kemp BE, Steinberg GR. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med. 2013;19(12):1649-54 doi: 10.1038/nm.3372. Epub 2013 Nov 3
45. Madiraju AK, Erion DM, Rahimi Y, Zhang X, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, Jurczak MJ, Camporez J, Lee H, Cline GW, Samuel VT, Kibbey RG, Shulman GI. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510(7506):542-6 doi: 10.1038/nature13270. Epub 2014 May 21
46. Griffin SJ, Leaver JK, rving GJ. Impact of metformin on cardiovascular disease: a meta-analysis of randomised trials among people with type 2 diabetes. Diabetologia. 2017;60(9):1620-9
47. Polianskyte-Prause Z, Tolvanen TA, Lindfors S, Dumont V, Van M, Wang H. et al. Metformin increases glucose uptake and acts renoprotectively by reducing SHIP2 activity. The FASEB Journal. 2019;33(2):2858-2869 doi: 10.1096/fj.201800529RR
48. Tongzhi Wu, Horowitz M, Rayner CK. New insights into the anti-diabetic actions of metformin: from the liver to the gut. Expert Rev Gastroenterol Hepatol. 2017;11(2):157-166 doi:10.1080/17474124.2017.1273769
49. Lee Y, Kim AH, Kim E, Lee S, Yu K-S, Jang I-J, Chung J-Y, Cho J-Y. Changes in the gut microbiome influence the hypoglycemic effect of metformin through the altered metabolism of branched-chain and nonessential amino acids. Diabetes Res Clin Pract. 2021;178:108985. doi: 10.1016/j.diabres.2021.108985 0168-8227
50. Wang D, Liu J, Zhou L, Zhang Q, Li M, Xiao X. Effects of Oral Glucose-Lowering Agents on Gut Microbiota and Microbial Metabolites. Front Endocrinol (Lausanne). 2022;13:905171. doi: 10.3389/fendo.2022.905171
51. Kerb R, Brinkmann U, Chatskaia N, Gorbunov D, Gorboulev V, Mornhinweg E, Keil A, Eichelbaum M, Koepsell H. Identification of genetic variations of the human organic cation transporter hOCT1 and their functional consequences, Pharmacogenet. Genomics. 2002;12(8):591-5
52. Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett C.M. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Investig. 2007;117(5):1422-31
53. Christensen MM, Brasch-Andersen C, Green H, Nielsen F, Damkier P, Beck-Nielsen H, Brosen K. The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c, Pharmacogenet. Genomics. 2011;21(12):837-50
54. Dujic T, Zhou K, Donnelly LA, Tavendale R, Palmer CN, Pearson ER. Association of organic cation transporter 1 with intolerance to metformin in type 2 diabetes: a GoDARTS study. Diabetes. 2015;64(5):1786-93
55. Zhou Y, Ye W, Wang Y, Jian Z, Meng X, Xiao Q, Zhao Q, Yan J. Genetic variants of OCT1 influence glycemic response to metformin in Han Chinese patients with type-2 diabetes mellitus in Shanghai. Int J clin Exp Pathol. 2015;8(8):9533-9542
56. Xiao D, Guo Y, Li X, Yin JY, Zheng W, Qiu XY, Xiao L, Liu RR, Wang SY, Gong WJ, Zhou HH. The impacts of SLC22A1 rs594709 and SLC47A1 rs2289669 polymorphisms on metformin therapeutic efficacy in Chinese type 2 diabetes patients. Int J Endocrinol. 2016 ArticleID 4350712, 7 pages. doi: 10.1155/2016/4350712
57. Shikata E, Yamamoto R, Takane H, Shigemasa C, Ikeda T, Otsubo K, Ieiri. Human organic cation transporter (OCT1 and OCT2) gene polymorphisms and therapeutic effects of metformin. J Hum Genet. 2007;52(2):117-22
58. Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney AS, Leese G, Hattersley AT, McCarthy MI, Morris AD, Palmer CN, Pearson ER. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes. 2009;58(6):1434-9
59. Rena G, Pearson ER, Sakamoto K. Molecular mechanism of action of metformin: old or new insights? Diabetologia. 2013;56(9):1898-906
60. Dujic T, Zhou K, Yee S, van Leeuwen N, de Keyser C, Javorský M, Goswami S, Zaharenko L, Hougaard Christensen M, Out M, Tavendale R, Kubo M, Hedderson M, van der Heijden A, Klimčáková L, Pirags V, Kooy A, Brøsen K, Klovins J, Semiz S, Tkáč I, Stricker B, Palmer C, 't Hart L, Giacomini K, Pearson E. Variants in Pharmacokinetic Transporters and Glycemic Response to Metformin: A Metgen Meta-Analysis. Clin Pharmacol Ther. 2017;101:763-772
61. Zhou K, Yee SW, Seiser EL, Van Leeuwen N, Tavendale R, Bennett AJ, Groves CJ, Coleman RL, Van Der Heijden A, Beulens JW, De Keyser CE. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet. 2016;48(9):1055-9
62. Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, Saxena R, Fowler S, Shuldiner A, Knowler W, Altshuler D. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59(10):2672-81
63. Tkáč I, Klimčáková L, Javorský M, Fabianová M, Schroner Z, Hermanová H, Babjaková E, Tkáčová R. Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes Metab. 2013;15(2):189-91
64. Wang J, Kuusisto J, Vanttinen M. et al. Variants of transcription factor 7- like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucoseregulation and impaired insulin secretion. Diabetologia. 2007;50:1192-1200
65. Liu P, Chang YC, Jiang YD. et al. Genetic variants of TCF7L2 are associated with insulin resistance and related metabolic phenotypes in Taiwanese adolescents and Caucasian young adults. J Clin Endocrinol Metab. 2009;94:3575-3582
66. Gautier A, Roussel R, Lange C. et al. Effects of genetic susceptibility for type 2 diabetes on the evolution of glucose homeostasis traits before and after diabetes diagnosis: data from the D.E.S.I.R. study. Diabetes. 2011;60:2654-2663
67. Geoghegan G, Simcox J, Seldin MM, Parnell T, Stubben C, Just S. et al. Targeted deletion of Tcf7l2 in adipocytes promotes adipocyte hypertrophy and impaired glucose metabolism. Mol Metab. 2019;24:44-63 doi.org/10.1016/j.molmet.2019.03.003
68. Bride L, Naslavsky M, Lopes Yamamoto G, Scliar M, Pimassoni LH, Sossai Aguiar P, de Paula F, Wang J, Duarte Y, Passos-Bueno M, Zatz M, Imbroisi Valle Errera F. TCF7L2 rs7903146 polymorphism association with diabetes and obesity in an elderly cohort from Brazil. Peer J. 2021 5(9). doi: 10.7717/peerj.11349. PMID: 33996288; PMCID: PMC8106398
69. Del Bosque-Plata L, Martineze-Martinez E, Espinoza-Camacho M, Gragnoli C. The Role of TCF7L2 in Type 2 Diabetes. Diabetes. 2021;70:1220-1228 | doi: 10.2337/db20-0573
70. Santoro A, Botton M, Struchiner C, Suarez-Kurtz G. Influence of pharmacogenetic polymorphisms and demographic variables on metformin pharmacokinetics in an admixed Brazilian cohort. Br J Clin Pharmacol. 2018;84:987-998 doi: 10.1111/bcp.13522
71. Li L, Guan Z, Li R, Zhao W, Hao G, Yan Y, Xu Y, Liao L, Wang H, Gao Wu K, Gao Y, Li Y. Population pharmacokinetics and dosing optimization of metformin in Chinese patients with type 2 diabetes mellitus. Medicine. 2020 99 (46): (e23212) http://dx.doi.org/10.1097/MD.0000000000023212
72. Naja K, El Shamieh S, Fakhoury R. rs622342A>C in SLC22A1 is associated with metformin pharmacokinetics and glycemic response. Drug Metab Pharmacokinet. 2020;35:160-164 doi: 10.1016/j.dmpk.2019.10.007
73. Moon S, Jaeseong O, Seung H, Yewon Kyung-Sang Y, Jae-Yong C. Effect of plasma membrane monoamine transporter genetic variants on pharmacokinetics of metformin in humans. Transl Clin Pharmacol. 2018;26(2):79-85 doi: 10.12793/tcp.2018.26.2.79
74. Sadaf M, Zoya K, Fazal J, Muhammad I, Mohammad A, Rauf N, Sumbul K. Effects of SLC22A2 (rs201919874) and SLC47A2 (rs138244461) genetic variants on Metformin Pharmacokinetics in Pakistani T2DM patients. J Pak Med Assoc. 2019;69:155-163
75. Breitenstein M, Simon G, Ryu E, Armasu S, Weinshilboum R, Wang L, Pathak J. Using EHR-linked biobank data to study metformin pharmacogenomics. Stud Health Techn Inform. 2015;210:210-914
76. Kwon E, Chung J, Park H. et al. OCT3 promoter haplotype is associated with metformin pharmacokinetics in Koreans. Sci Rep. 2018;8:16965. doi: 10.1038/s41598-018-35322-6
77. Bondar I, Yu O, Shabel'nikova O, Sokolova E, Filipenko M.L. Results of studying the association between the rs11212617 polymorphism in the ATM gene and response to metformin therapy in patients with type 2 diabetes mellitus. Probl Endocrinol (Mosk). 2017;63(1):9-16 doi: 10.14341/probl20176319-16
78. Ortega-Ayala A, Rodríguez-Rivera NS, Andrés F, LLerena A, Eliseo Pérez-Silva E, Adriana Guadalupe Espinosa-Sánchez AG, Molina-Guarneros JA. Pharmacogenetics of Metformin Transporters Suggests No Association with Therapeutic Inefficacy among Diabetes Type 2 Mexican Patients. Pharmaceuticals (Basel). 2022;15(7):774. doi: 10.3390/ph15070774
79. Guasch-Ferré M, Hruby A, Toledo E, Clish C, Martínez-González M, Salas-Salvadó J, Hu F. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care. 2016;93:833-846 doi: 10.2337/dc15-2251
80. Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes H, Hrabé de Angelis M, Wichmann H, Kronenber Fg, Adamski, Illig JT. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5(11):e13953. doi: 10.1371/journal.pone.0013953
81. Song I, Lee D, Shin M, Kim H, Ahn Y, Park I, Kim K, Kind T, Shin J, Fiehn O, Liu K. Pharmacogenetics meets metabolomics: discovery of tryptophan as a new endogenous OCT2 substrate related to metformin disposition. PloS one. 2012;7(5):e36637. doi: 10.1371/journal.pone.0036637
82. Yoon M. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients. 2016;8(7):405. doi: 10.3390/nu8070405
83. Polidori N, Grasso EA, Chiarelli F, Giannini F. Amino Acid-Related Metabolic Signature in Obese Children and Adolescents. Nutrients. 2022;14(7):1454. doi: 10.3390/nu14071454
84. Savolainen O, Fagerberg B, Vendelbo B, Li B, Sandberg A, Ross A, Bergström G. Biomarkers for predicting type 2 diabetes development—Can metabolomics improve on existing biomarkers? PLoS ONE. 2017;12:e0177738. doi: 10.1371/journal.pone.0177738
85. Knebel B, Strassburger K, Szendroedi J, Kotzka J, Scheer M, Nowotny B, Mussig K, Lehr S, Pacini G, Finner H. et al. Specific metabolic profiles and their relationship to insulin resistance in recent-onset type-1and type-2 diabetes. J Clin Endocrinol Metab. 2016;101:2130-2140 doi: 10.1210/jc.2015-4133. Epub 2016 Feb 1
86. Aleidi S, Dahabiyeh L, Gu X, Al Dubayee M, Alshahrani A, Benabdelkamel H, Mujammami M, Li L, Aljada A, Abdel Rahman A. Obesity Connected Metabolic Changes in Type 2 Diabetic Patients Treated with Metformin. Front Pharmacol. 2021;11:616157. doi: 10.3389/fphar.2020.616157
87. Adam J, Brandmaier S, Leonhardt J, Scheerer MF, Mohney RP, Xu T, Bi J, Rotter M, Troll M, Chi S, Heier M. Metformin effect on nontargeted metabolite profiles in patients with type 2 diabetes and in multiple murine tissues. Diabetes. 2016;65(12):3776-85 doi: 10.2337/db16-0512. Epub 2016 Sep 12
88. Xu T, Brandmaier S, Messias AC, Herder C, Draisma HH, Demirkan A, Yu Z, Ried JS, Haller T, Heier M, Campillos M. 2015. Effects of metformin on metabolite profiles and LDL cholesterol in patients with type 2 diabetes. Diabetes care. 2015;38(10):1858-67 doi: 10.2337/dc15-0658
Author contact
![]() Corresponding author: Zoheir A. Damanhouri, Department of Pharmacology, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia, Tel/Fax +966 12 6408404 email address zdamanhouriedu.sa
Corresponding author: Zoheir A. Damanhouri, Department of Pharmacology, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia, Tel/Fax +966 12 6408404 email address zdamanhouriedu.sa

 Global reach, higher impact
Global reach, higher impact