Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(6):1082-1092. doi:10.7150/ijms.72621 This issue Cite
Review
Crucial Roles of LncRNAs-Mediated Autophagy in Breast Cancer
1. Tai Zhou Central Hospital (Taizhou University Hospital), No.999 Donghai Road, Jiaojiang District, Taizhou, Zhejiang, 318000, China.
2. Cytotherapy Laboratory, Shenzhen People's Hospital, 1017, Dongmen North Road, Luohu, Shenzhen, 518020, China.
*These authors contributed equally to this work.
Received 2022-3-4; Accepted 2022-6-9; Published 2022-6-21
Abstract
Breast cancer remains a worldwide public health issue. LncRNA and autophagy respectively or simultaneously, get involved in cellular and molecular processes of many different cancers, including genesis, metastasis, and deterioration of breast cancer and other malignant tumors. In this review, relevant studies have been summarized, and we have found that lncRNA-mediated autophagy in luminal A breast cancer, luminal B breast cancer, HER-2 positive breast cancer, and basal-like breast cancer may play an important role in mediating drug resistance sensitivity. LncRNAs target genes and affect different signaling pathways to a complex network, which attenuates the occurrence and development of primary breast cancer by coordinating autophagy. Abnormal expression of LncRNA may lead to dysregulation of autophagy, resulting in tumor genesis, expansion, and resistance to anti-tumor therapy. Targeting specific lncRNAs for autophagy regulation may conduct as a bio-marker for reliable diagnosis and prognosis treatment of breast cancer or provide a promising therapeutic strategy.
Keywords: LncRNA, Autophagy, Molecular mechanisms, Breast cancer, Drug resistance
Introduction
Breast cancers basically include luminal A breast cancer, luminal B breast cancer, HER-2 positive breast cancer, and basal-like breast cancer. According to GLOBCAN, 2020, the morbidity and mortality of breast cancer are separately reported at 11.7% and 6.9% [1]. Breast cancer is one of the most common malignancies and the first inducement of cancer-associated death among female subjects in the world. It is reported that in 2020 breast cancer has already been the 5th most cancer worldwide [1].
As one of the most frequently diagnosed cancers among females, breast cancer represents 7%-10% of all malignant tumors. It is predicted that the incidence of estrogen receptor-positive cancers may elevate substantially amid a growing. The statistics of 2020 showed that in the United States the annual spending on the treatment of breast cancer was projected to reach $22.6 billion [2]. Although with the development of screening technology, early breast cancer can be screened through mammography technology, for early prevention and treatment, such as surgery and chemoradiotherapy [3], especially in patients who are in advanced and metastatic conditions, are still served as the leading causes of death. Moreover, because of the high resistance to chemotherapy or radiotherapy, unresectable patients with advanced-stage of tumors often have a poor prognosis.
Breast cancer is classified to determine treatment options based on the following molecular markers: The expression of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor-2 (HER-2), and Ki-67 can be divided into the following four subtypes [4]: Luminal A (ER+ and/or PR+ and HER2-, low expression of Ki67) and Luminal B (ER+ and/or PR+ and HER2-/HER2+, high expression of Ki67) [5] accounted for 40% and 30% of cases, respectively, and were the most common types of breast cancer. About 15% of breast cancers are HER2-enriched subtypes with ER-/PR-/HER2+ phenotypes [6]. HER2-enriched breast cancer shows increased expression of proliferative markers as well and has a worse prognosis than luminal subtypes [7].
Basal-like breast cancer, also named triple-negative breast cancer (TNBC) (ER-/PR-/HER2-), has the worst prognosis of all the breast cancer types [8, 9].
The molecular mechanism of breast cancer can be a prerequisite for the diagnosis, therapeutic treatment, and prognosis of this cancer. It is urgent to increase the sensitivity of chemotherapy for breast cancer. Apoptosis metabolism disorder plays a key role in the occurrence and development of various malignancies including breast tumors. More and more studies have shown that lncRNA can regulate autophagy to promote apoptosis and thus attenuate the growth of primary tumors [10]. In this review, we hope to describe the role of the lncRNA-autophagy axis in the progression and drug resistance of human breast cancer through a systematic review of relevant literature.
Overview of Autophagy
Autophagy is the major intracellular digestion system and dynamic system which provides energy and new building components for cellular renewal and homeostasis. So far there are roughly three classes of autophagy that includes macroautophagy, microautophagy, and chaperone-mediated autophagy [11]. Compared with microautophagy and chaperone-mediated autophagy, the study of macroautophagy has been most extensive and particularly macroautophagy, which is thought to be the main type of autophagy [12, 13]. Autophagy, usually regulated by autophagy-related genes (ATGs), is the process by which cell contents are transported to lysosomes for degradation by a two-membrane vesicle structure called an autophagosome [12]. Autophagy often plays different roles due to different trigger signals [14]. In some cases, autophagy protects cells from death by removing damaged organelles in a self-constructed stress environment and induces cell death when stimulated by other signals [15]. Similarly, autophagy plays a complex role in cancer cells, promoting or inhibiting cancer through different molecular mechanisms. Generally, it is believed that autophagy plays a protective role in the progression of many different kinds of tumors by preventing the toxic accumulation of damaged cellular materials in the early stage of tumor development; in advanced stages, autophagy boosts metabolism to meet cancer's high nutritional requirements and keep tumor cells alive [16].
The role of autophagy in Breast Cancer
There is growing evidence that dysregulation of autophagy can cause a variety of diseases. In 1999, Levine et al.[17]explained that the autophagy gene becline1 regulates the development of breast cancer, first revealing the relationship between autophagy and breast cancer. However, from current studies, the exact role of autophagy in breast cancer is still controversial. It has been suggested that autophagy promotes breast cancer progression by increasing the survival rate of latent breast cancer cells [18], however, some studies have shown that autophagy can also inhibit cancer by protecting the integrity of the genome and inhibiting the metastasis of breast cancer cells [10]. Once tumor cells form, tumor cells can use autophagy to recycle macromolecules to provide energy for the TCA cycle and substrates for biosynthesis to help them survive in adverse microenvironments. Cancer cells seem to maintain high ATP levels to meet their need for high proliferation by activating autophagy. As a result, cancer cells use autophagy to maintain a high metabolic level and thus obtain energy to help the tumor survive.
Loss of autophagy has been found in various subtypes of breast cancer, suggesting that loss of autophagy may promote tumor progression. Paradoxically, elevated levels of autophagy also promote breast cancer progression. William et al. found that autophagy-mediated by nuclear heterotopic action of p53 promoted the progression of breast cancer [19]. Chung et al. found that the STK11/LKB1 signaling pathway regulates ADIPOQ/ adiponectin mediated AMPK activation, map1LC3B-II/LC3B-II release, and ULK1 (Unc-51 like kinase 1) activation, thus inducing autophagy to inhibit tumor progression [20]. In contrast, gene autophagy inhibition led to significant expansion of tumor cells at both primary and metastatic sites, exhibiting invasive and premetastatic basal epithelial differentiation [10]. SRC-3-AKT signaling pathway regulates autophagy of breast cancer cells to promote the proliferation of the tumor cells, and AKT further regulates mTOR signaling to form the AKT-mTOR signaling pathway that affects the poor prognosis in the patients with breast cancer [21]. In addition, Jing et al. found that liensinine, as an alkaloid, can inhibit advanced autophagy/mitochondrial autophagy by blocking autophagosome-lysosomal fusion, thus reducing the activity of breast carcinoma cells and increasing the apoptosis of breast carcinoma cells [22]. In conclusion, autophagy is closely associated with the occurrence and development of breast cancer and the generation of drug resistance. Therefore, it is very important to control its expression in the treatment of malignancy in the breast.
A major regulator of autophagy is the rapamycin (mTOR) pathway target, which is directly regulated by the signaling complex mTORC1. MTORC1 controls autophagy by controlling the phosphorylation of ATG13 and ULK1. AMPK activation occurs when intracellular energy is reduced [6], this protein kinase induces autophagy by two different mechanisms: (1) activating ATG13 and ULK1 by inhibiting the mTOR complex and (2) bypassing the mTOR signaling pathway and directly phosphorylating ULK1, VPS34, and beclin1. Cancer cells appear to activate autophagy to trigger cytotoxic and metabolic stress responses, such as hypoxia and nutrient deprivation, so they can activate autophagy to maintain cancer cells' survival. Studies have found that clinically, small molecule inhibitors can inhibit the PI3K/AKT/mTOR signaling pathway and inhibit the occurrence of autophagy, to treat breast cancer [23]. Interestingly, adriamycin combined with mulandine can prevent breast cancer by inducing the expression of autophagy markers beclin-1 and LC3-II in breast carcinoma cells, reducing the expression level of p62, and inhibiting the PI3K/AKT/mTOR signaling pathway, suggesting that autophagy may be a form of cancer cell death induced by anticancer drugs [24]. As reported, the p62 protein interacts with the autophagy effector protein LC3 and induces breast carcinoma cell death through the autophagy-lysosomal pathway [24]. As autophagy is a survival response of tumor cells after tumor drug therapy, tumor cells enhance their sensitivity to chemotherapy drugs through autophagy [25] and realize tumor immune escape through multiple overlapping mechanisms.
In addition to autophagy-related proteins, autophagy interacts with lncRNA and signal transduction induced by lncRNA as well. Many kinds of studies have proved that lncRNA is involved in the regulation of autophagy.
LncRNAs
Long noncoding RNAs (lncRNAs) are transcripts of more than 200 nucleotides in length [26], this kind of RNA has limited protein-coding potential and is widely found in prokaryotes and eukaryotes [27]. RNA-seq studies have shown that there are thousands of unidentified lncRNAs in any given cell type, and lncRNAs may be related to developmental and tissue specificity [28]. LncRNA folds into complex three-dimensional structures that interact with DNA, RNA, and proteins [29]. Many molecular mechanisms of LncRNA activity have been revealed: cell cycle regulation, stem cell pluripotency [30], and regulating gene expression at different levels, including epigenetic regulation and transcriptional regulation [31]. Current studies have shown that lncRNAs are closely associated with some biological processes such as development, differentiation, apoptosis, autophagy, inflammation, and cancer [32]. LncRNA expression in human carcinoma cells is quite different from that in healthy cells. The first confirmed cancer-related lncRNA is HOTAIR, which is overexpressed in the tissue of breast cancer and is closely related to the occurrence and metastasis of this tumor [33]. Involved in cancer and metabolic diseases, many physiological and pathological processes are highly dependent on lncRNAs. For example, some abnormal expressions of lncRNAs in cancers signal the disease progression spectrum and lncRNAs can also be used as an independent predictor of patient prognosis [18, 26, 28].
LncRNAs can be classified into cis-regulatory-lncRNAs (Cis-regulatory-lncRNAs) and trans-regulatory-lncRNAs (Trans-regulatory-lncRNAs) according to their guiding functions. Cis-regulatory-lncRNAs modify specific regions of the genome by recruiting histone complexes leading to local gene expression. It mainly includes H19, AIR, KCNQ1OT1, and XIST, among which H19 is the most widely studied in cancer and has been proved to play a role in the occurrence and inhibition of tumors [34]. However, trans-regulatory lncRNAs represented by HOTAIR are upregulated in breast cancer and hepatocellular carcinoma. HOTAIR lncRNA has been found that is highly upregulated in both primary breast tumors and metastatic breast tumors and can promote cancer metastasis, and its transcription level is 2000 times higher than that of normal breast tissues [28]. In cancer, when GAS5 (another trans-regulatory lncRNA) is overexpressed in breast cancer cell lines, it can induce cell apoptosis and inhibit cell proliferation. However, in human breast tumor samples, the transcription level of GAS5 are significantly reduced, which is statistically significant in stage I and II cancers [35], prompting the decrease of GAS5 expression as an early event of tumorigenesis [36].
Through the ubiquitination-autophagy pathway, lncRNA LINRIS can inhibit the proliferation of colorectal cancer cells, thus inhibiting the progression of colon cancer [37]. CRNDE can increase the sensitivity of gastric cancer patients to 5-FU by inhibiting the expression of autophagy-related proteins LC-Ⅱ and LC-Ⅲ under the condition of overexpression [38]. Overexpression of PVT1 in pancreatic cancer cell lines can enhance sensitivity to Gemcitabine by regulating autophagy, thus inhibiting the growth of pancreatic cancer [39]. Ji et al. [34] demonstrated that in breast cancer cells, through the H19/SAHH/DNMT3B axis, H19 may induce autophagy activation leading to the resistance of tamoxifen in breast cancer. LncRNAs also serve as key regulators of autophagy in breast cancer. Through a review of relevant literature, the regulation of autophagy by breast cancer-related lncRNAs has been identified, [34] which may conduce to the development of new therapeutic strategies for breast cancer and more effectively target breast cancer.
Regulation of autophagy by LncRNAs in Breast Cancer (Table 1)
Long noncoding RNAs (lncRNAs) play a significant role in the progression of tumors and are abnormally expressed in a variety of cancers [40]. Current studies have found that some lncRNAs are associated with the metastasis of breast cancer and the resistance to chemotherapy drugs, and some lncRNAs are overexpressed in breast cancer to promote the progression of breast cancer [40]. LncRNAs regulating autophagy through some specific mechanisms can be divided into three categories: lncRNAs bind to miRNAs as competitive endogenous RNAs (ceRNAs) to regulate miRNAs expression, thus affecting autophagy; lncRNAs may affect the expression of Cis-trans ATG genes as well. LncRNA may promote tumor progression by inhibiting autophagy-mediated apoptosis through AKT/mTOR pathway [16, 41].
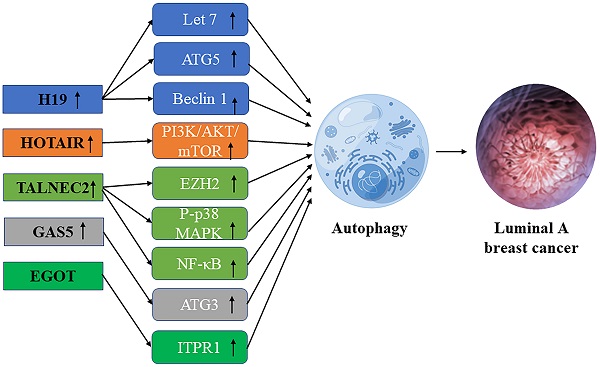
Schematic diagram of the association between lncRNA-mediated autophagy in luminal A breast cancer.
The Role of Autophagy Mediated by LncRNAs in Initiation, Development, and Drug Resistance of Breast Cancer
| Cancer Specificity | LncRNAs | Expression | Anti-/Pro autophagy | Role | References |
|---|---|---|---|---|---|
| Luminal A breast cancer | TALNEC2 | Upregulated | Pro | OncoLncRNA | [49] |
| GAS5 | Downregulated | Pro | TsLncRNA | [35, 36, 71-74] | |
| HOTAIR | Upregulated | Pro | OncoLncRNA+ Promoting DR | [44, 48] | |
| H19 | Upregulated | Pro | OncoLncRNA+ Promoting DR | [34] | |
| EGOT | Upregulated | Pro | Promoting DS | [53] | |
| Luminal B breast cancer | ROR | Downregulated | Pro | OncoLncRNA+ Promoting DR | [58] |
| HER-2 positive breast cancer | AGAAP2-AS1 | Upregulated | Pro | Promoting DR | [63, 66] |
| EPIC1 | Upregulated | -- | OncoLncRNA | [84] | |
| UCA1 | Upregulated | Pro | OncoLncRNA+ Promoting DR | [62] | |
| ZNF649-AS1 | Upregulated | Pro | OncoLncRNA+ Promoting DR | [64] | |
| Basal-like breast cancer | snaR | Upregulated | -- | OncoLncRNA | [85] |
| TALNEC2 | Upregulated | Pro | OncoLncRNA | [49] | |
| OTUD6B | Upregulated | Pro | Promoting DR | [83] | |
| NAMPT | Upregulated | Anti | OncoLncRNA | [80] | |
| XIST | Downregulated | --- | OncoLncRNA | [86] | |
| LUCAT1 | Upregulated | --- | OncoLncRNA | [29] | |
| GAS5 | Downregulated | Pro | TsLncRNA | [35, 36, 71-74] | |
| HOTAIR | Upregulated | Pro | OncoLncRNA | [44, 48] | |
| WDFY3-AS2 | Downregulated | --- | TsLncRNA | [87] | |
| DRHC | Downregulated | --- | TsLncRNA | [88] | |
| DANCR | Upregulated | Anti | OncoLncRNA | [78] | |
| EGOT | Upregulated | Pro | Promoting DS | [53] |
DR: drug resistance; DS: drug sensitivity; BC: breast cancer; OncolncRNA: oncology lncRNA; TslncRNA: tumor suppression lncRNA.
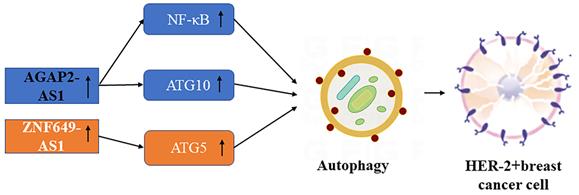
Schematic picture of the association in lncRNAs mediated autophagy of HER-2+ breast cancer.
Interaction of lncRNA and autophagy in luminal A breast cancer
LncRNA H19
LncRNA H19 is a 2.3kb imprinted lncRNA [28]. LncRNA H19 was found to be less abundant in ER-negative breast cancer than in ER-positive breast cancer. Overexpression of lncRNA H19 promotes autophagy in hormone receptor-positive breast cancer cells [42]. H19 inhibits EMT (epithelial-mesenchymal transformation) of BC cells and promotes autophagy through the H19/Let-7/Lin28 signaling pathway [43]. Through the H19/SAHH/DNMT3B axis, H19 may induce autophagy activation, which may help to produce tamoxifen resistance in breast cancer. Overexpression of beclin1, a key mediator of autophagy, leads to estrogen-induced desensitization of signal transduction, contributing to tamoxifen resistance in ER-positive breast cancer. Inhibition of some autophagy genes including ATG5 (autophagy-related 5), ATG7 (autophagy-related 7), and beclin1 leads to hypersensitization of tamoxifen-resistant breast cancer cells [34].
LncRNA HOTAIR
HOTAIR, mainly HOX antisense RNA plays an important role in chromatin dynamics and may cause transcriptional gene silence by interacting with histone modifiers [44]. Bhan et al. found that HOTAIR overexpression promotes the development and metastasis of ER-positive breast cancer through E2 regulation [45]. Since HOTAIR regulates the progression of breast cancer through the autophagy of ubiquitination and degradation of proteins in the proteasome [44], it plays a crucial role in the development of breast cancer and drug resistance in the patients with breast cancer, so it is promising to become a useful biomarker and potential therapeutic target. Studies have found that HOTAIR can regulate oral squamous cell autophagy by regulating the mTOR pathway and inhibiting cell apoptosis and cisplatin sensitivity [46]. Li et al. found that by inhibiting the PI3K/AKT/mTOR pathway, lncRNA-HOTAIR down-regulation could effectively weaken the resistance of breast carcinoma cells to Doxorubicin [47]. Xue et al. found in their study that HOTAIR promoted the resistance of ER-positive breast cancer to tamoxifen by up-regulating its expression in ER+ breast cancer patients [48], it is suggested that HOTAIR may be a useful biomarker and a potential therapeutic target.
LncRNA TALNEC2
Qiao et al. found that TALNEC2, a long non-coding RNA expressed on chromosome 2, targets p57KIP2 by binding EZH2 and is involved in the P-p38 MAPK and NF-кB pathways. It plays a carcinogenic role in luminal A and triple-negative breast cancers, and some studies have shown that TALNEC2 knockout can inhibit cell viability and community integration in luminal A and triple-negative breast cancers [49] Polycomb histone methyltransferase enhancer 2 (Polycomb histone methyltransferase enhancer of zeste homolog 2, EZH2) is considered as a key marker of invasive breast cancer [50]. In addition, EZH2 regulates the NF-κB pathway by interacting with RelA and RelB complexes to regulate the development of breast cancer [51]. As a major regulator in the cell cycle progression, autophagy, and apoptosis, EZH2 plays an important role in drug-resistant tumor types, suggesting that TALNEC2 may influence the progression of breast cancer and the generation of drug resistance by promoting autophagy through EZH2 [52], But the specific regulation mechanism is not clear.
LncRNA EGOT
LncRNA EGOT may enhance the sensitivity of triple-negative breast cancer cells and Luminal A breast cancer cells to paclitaxel by upregulating ITPR1 cis and trans expression to enhance autophagosome accumulation. On the one hand, EGOT induces pre-ITPR1 accumulation by forming pre-ITPR1/EGOT dsRNA, and increases ITPR1 protein expression in a cis-manner, thus upregulating the ITPR1 level. On the other hand, two binding motifs of EGOT 2 segments (324-645 nucleotides) are enhanced the alternative splicing of trans pre-ITPR1 of EGOT in the exon [53]. ITPR1 contains 59 exons and multiple GGGA/C/G motifs distributed in these exons. HnRNPH1 mediates pre-ITPR1 splicing in human cancers by binding to these GGGA/C/G motifs [54, 55]. ITPR1 protein expression is decreased in an estrogen-receptor-dependent manner, and estrogen-induced GROWTH of MCF7 cells is sensitive to drug inhibitors of ITPR1. In conclusion, lncRNA enhances the sensitivity of breast cancer cells to paclitaxel therapy through enhancing autophagy.
Interaction of lncRNA and autophagy in luminal B breast cancer
LncRNA ROR
As shown in many studies, LncRNA ROR can contribute to the maintenance of induced pluripotent stem cells and embryonic stem cells. In addition, lncRNA ROR (ROR, reprogramming regulator of reprogramming) is up-regulated in nasopharyngeal carcinoma, hepatocellular carcinoma, and breast cancer [56-58]. Li et al. found that the proliferation, invasion, and migration of Luminal B breast cancer cells were inhibited by down-regulation of long non-coding RNA ROR, and the effect of tamoxifen on Luminal B breast cancer cells was reversed [59]. LncRNA ROR may promote autophagy by targeting autophagy-associated proteins LC3 and beclin1, thereby promoting the progression of breast cancer and tamoxifen resistance [59]. Hou et al. found that lncRNA ROR regulates the EMT process of breast cancer by interacting with miR-205 [57]. Gabriel et al. found that ROR could also regulate the development and metastasis of triple-negative mammary glands by interacting with miR145 [60]. ROR is an important target in the progression of treatment in breast cancer.
Interaction of lncRNA and autophagy in HER-2+ breast cancer
Human epidermal growth factor receptor 2(HER-2) positive breast cancer is the most common type of breast cancer, accounting for about 15%-20% of breast cancer patients. Trastuzumab is the first-line drug in the treatment of HER-2 breast cancer [61]. In 1998, Trastuzumab, approved by FDA in the United States, is designed to target and inhibit HER2 signaling in cancer cells for treating HER2-positive breast cancer [62]. But about 60 percent of patients develop resistance after drug treatment, significantly reducing the clinical usefulness and efficacy of trastuzumab, which has become one of the most pressing challenges in treating breast cancer [63]. So as shown in some research, lncRNA plays a vital role in some drug sensitivity of breast cancer [64].
LncRNA AGAAP2-AS1
ATG10 (autophagy-related 10) expression induces autophagy, leading to lncRNA AGAP2-AS1 mediated trastuzumab resistance in breast cancer, the expression of ectopic ATG10 is significantly related to lymph node metastasis and poorer prognosis in this cancer [65]. Mechanically, AGAP2-AS1 is associated with ELAVL1 protein, and they are easy to form as AGAP2-AS1-ELAVL1 complex directly binding to the promoter region of ATG10, inducing enrichment of H3K27ac and H3K4me3, and ultimately activate transcription of ATG10 [63]. Trastuzumab-resistant breast cancer cells show higher autophagy activity, with increased LC3-Ⅱ expression and decreased P62 protein expression compared with BC parental cells, which confirms the close relationship between autophagy and tumor drug resistance. Through activating the NF-κB signaling pathway and up-regulating MyD88 expression, AGAP2-AS1 can promote the growth and trastuzumab resistance of breast tumors [66]. Down-regulation of AGAP2-AS1 reverses trastuzumab resistance in HER-2 positive breast cancer [67].
LncRNA ZNF649-AS1
Han et al. found that lncRNA ZNF649-AS1 associated with PTBP1 protein induced by H3K27ac modification can promote the transcriptional activity of the ATG5 gene [64]. PTB was found to be a splicing regulator that can effectively cross-link with RNA by shortwave UV light [68]. AS one of the key regulators of autophagy-mediated cell death, autophagy-related gene-5 (ATG-5) is widely regarded as a protective molecular mechanism in various cancer treatments [69]. Down-regulation of ZNF649-AS1 can inhibit autophagy and inhibit ATG5 expression to reverse trastuzumab resistance in breast cancer with positive HER-2.
Interaction of lncRNA and autophagy in basal-like breast cancer
Triple-negative breast cancer, also called basal-like breast cancer, accounts for about 15%-20% of breast cancer. Due to its lack of progesterone receptor (PR), estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER-2), triple-negative breast cancer patients have a worse prognosis and tumors are more aggressive, and higher mortality [70]. Triple-negative breast cancer has the characteristics of strong invasion and early recurrence. Studies have proved that compared with other subtypes of breast cancer, autophagy is more intense in triple-negative breast cancer. For example, autophagy-related microtubule-associated proteins Beclin1, LC3A, and LC3B are highly expressed in this cancer cell, promoting the progression and metastasis of breast cancer [23].
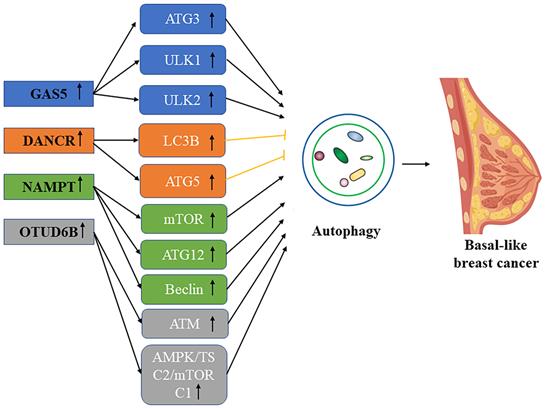
LncRNA GAS5
Growth arrest‑specific 5 (GAS5) is known as a tumor suppressor, negatively regulating cell survival and malignancy in different cancer cell types. GAS5 lncRNA can not only inhibit the proliferation of various types of tumors, but also promote their apoptosis, and the mechanism of these cells may jointly constitute the basis of their tumor inhibition [71]. LncRNA GAS5 regulates autophagy in breast cancer through the GAS5-miR-23a-ATG3 axis, and GAS5 expression is down-regulated in breast cancer. GAS5 can act as a “molecular sponge” blocking and binding miR-23a, it can positively regulate miR23A-targeting autophagy-related gene ATG3 as well, thus up-regulating GAS5 in breast cancer to inhibit tumor cell progression through autophagy [72]. Mark et al. found in their study that lncRNA GAS5 could induce apoptosis of all breast cancer types, especially in triple-negative breast cancer [73]. Li et al. proved in their study that GAS5 can hold up the formation, proliferation, and migration of breast cancer by mediating the autophagy-related promoter ULK1 and ULK2 (Unc-51 like kinase 2) [74].
LncRNA DANCR
LncRNA DANCR (Differentiation antagonist non-protein coding RNA) is carcinogenic in a variety of tumors and compared with the expression of normal issues this kind of LncRNA is higher in the tumor of breast [75]. The studies have shown that miR-758-3p works as a tumor suppressor in multiple cancers, such as ovarian cancer and non-small cell lung cancer [76, 77]. Research by Zhang et al. found that lncRNA mediating the DANCR-mir-758-3P-PAX6 molecular network can regulate apoptosis and autophagy in breast cancer. Studies have found that DANCR is negatively correlated with miR-758-3p, and down-regulation of DANCR can activate tumor suppressor miR-758-3p to play a tumor-suppressive role and promote apoptosis and autophagy of breast cancer cells: Promote the transcription and protein expression of Caspase-3, Caspase-9, Bcl-2, LC3B and ATG5 to inhibit the malignant proliferation of breast cancer cells [78].
LncRNA NAMPT
NAMPT-AS has potential RNA enhancer properties and can be co-transcribed with NAMPT from bidirectional promoters [79], NAMPT degradation was reduced by regulating miR-548B-3p. AS a carcinogenic lncRNA in triple-negative breast cancer, NAMPT-AS/NAMPT promotes the progression of tumors and metastasis by regulating autophagy by the mTOR pathway. Zhang et al. found that an increased proportion of NAMPT-AS in triple-negative breast cancers was associated with poor prognosis compared with receptor hormone-positive breast cancers. RNA-seq and Western blot analysis demonstrate that the down-regulation of the NAMPT-AS gene importantly led to the inactivation of the mTOR pathway, the up-regulation of ATG5, ATG12, and beclin, and the vital transformation of LC3-Ⅰ to LC3-II. Therefore, NAMPT-AS can inhibit the mTOR pathway and cause autophagy in TNBC cells [80]. In conclusion, the NAMPT-AS/POU2F2/NAMPT and NAMPT-6 AS/miR-548B-3p/NAMPT axis activate the mTOR pathway and inhibit autophagy and apoptosis-related genes, thereby promoting the survival and invasion ability of triple-negative breast cancer cells.
LncRNA OTUD6B
Paclitaxel (PTX) -based combination chemotherapy remains the key to triple-negative breast cancer (TNBC) because lacking the HER-2 targeted therapy and endocrine therapy. However, chemotherapeutic resistance of up to 30% to 50% limits the effectiveness of combination drug therapy strategies, resulting in a poor prognosis [81]. Direct loss, expression imbalance, or abnormal function of DDR proteins (TP53, BRCA1, ATM, etc.) all increase the risk of breast cancer, the occurrence of malignant subtypes, and the chemotherapy resistance of tumors. ATM activates autophagy through the AMPK/TSC2/mTORC1 pathway and it could also directly phosphorylate and stabilizes nuclear TP53 (tumor protein p53) to promote autophagy [82]. Li et al. found that miR-26a-5p is a protective element to protect breast cancer from attack, in contrast, OTUD6B-AS1 and MTDH are destructive elements of breast cancer, and miR-26a-5 is down-regulated through OTUD6B-AS1/miR-26a5p/MTDH pathway. MTDH and OTUD6B-AS1 overexpression promote autophagy and DNA damage, and autophagy defects can increase the short-term sensitivity of tumor cells to chemotherapeutic drugs [83]. In conclusion, OTUD6BAS1/miR-26A-5p/MTDH promotes PTX resistance in triple-negative breast cancer by upregulation of autophagy and DDR inhibition mediated genomic instability.
Schematic picture of the association in lncRNA mediated autophagy in basal-like breast cancer.
The therapeutic potentials and the relevant mechanisms associated with the interference between LncRNAs and autophagy
According to current studies, lncRNA not only mediates the occurrence and development of the tumor in the breast through autophagy but also affects the metastasis and prognosis of this tumor, as well as the sensitivity of breast cancer cells to drug therapy. Autophagy activation is a response of tumor cells to chemoradiotherapy [18, 23]. In most cancer patients, abnormal autophagy leads to cancerous changes that promote tumor cells' resistance to radiation and chemotherapy [25, 89]. LncRNA is the main autophagy regulator affecting drug resistance to radiotherapy and chemotherapy in breast tumors [34] [53]. For example, Jing et al. found that liensinine can treat breast cancer by promoting autophagy to induce apoptosis of breast cancer cells [22]. H19 contributes to the tamoxifen resistance in ER-positive breast cancer by overexpression of Beclin1, a key mediator of autophagy, leading to estrogen-induced desensitization of signal transduction [43]. We found that lncRNA-mediated expression of autophagy-associated proteins promotes trastuzumab resistance in HER-2-positive breast cancer [63, 64]. Li et al. found that the OTUD6B-AS1/miR-26a-5p/MTDH signaling pathway promotes autophagy, thereby promoting the occurrence of paclitaxel resistance in triple-negative breast cancer [83]. These findings provide new insights into lncRNA-mediated autophagy as therapeutic methods for breast cancer. Abnormal tumor-associated lncRNAs in the breast system may be consistent with autophagy to identify the specific mechanisms by which tumor cells respond to drug therapy, a key area requiring further exploration.
Limitations and future perspectives
It is the first systematic review of all evidence on the association between lncRNA and autophagy in breast tumors. A large number of specific lncRNAs-mediated-autophagy might work in the malignant transformation of breast tumors, affecting the ability of tumor cells to proliferation, apoptosis, cell cycle, migration, invasion, angiogenesis, and cell senescence. In Luminal A breast cancer, TALNEC2, GAS5, HOTAIR, H19, EGOT mediated autophagy plays a role in tumor development by influencing cancer cell development. For Luminal B breast cancer, ROR can regulate the proliferation potential of cancer cells through autophagy. For HER-2+ breast cancer, ZNF649-AS1 and AGAAP2-AS1 regulate trastuzumab resistance of breast cancer cells through autophagy, while TALNEC2 has been reported to connect with invasion and migration of breast cancer cells. However, there are some limitations in explaining the role of lncRNA in drug resistance through autophagy. First, due to limited clinical studies, the lncRNA coefficient and its clinical diagnostic, therapeutic and prognostic value still need to be further studied. Secondly, the molecular mechanisms of lncRNA-mediated autophagy, breast tumorigenesis, and drug resistance need further illustration and exploration. In addition to targeted genes, related proteins, and corresponding signaling pathways, other factors such as the microenvironment of the tumor and tumor immune checkpoints may also act a role in the pathogenesis and evolution of breast cancer. Finally, we are supposed to note that autophagy plays a dual role in cancer, not only acting as a tumor promoter but also inhibiting tumor cell development and metastasis in some breast cancer subtypes. Once the pathogenesis of breast cancer is addressed, it will be helpful to discover more useful diagnostic and prognostic biomarkers and innovative therapeutic approaches to better treat breast systemic malignancies. LncRNA-mediated autophagy has a great influence on tumor genesis, development, metastasis, and anti-tumor treatment resistance. Given this, autophagy regulation targeting the above lncRNAs may be a reliable biomarker for diagnosis and prognosis or provide a therapeutic strategy with great promise in breast cancer.
Conclusions
In this review, lncRNA-related autophagy may be a vital molecular mechanism of breast tumor genesis and development and play a key role in breast cancer drug resistance. Up-regulation or down-regulation of lncRNAs can induce some complex procession in breast cancer through influencing autophagy. LncRNAs target genes, proteins, and different signaling pathways constituting a complex network, coordinate autophagy, affecting the carcinogenic and inhibitory effects of breast malignancies and affecting the sensitivity of cancer drug therapy.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81902138), the Project of Public Welfare Technology Application of Zhejiang Province (LGF22H200016), and the Zhejiang Medical and Health Science and Technology Project (2019KY243, 2021KY1222).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
2. Mariotto AB, Yabroff KR, Shao Y, Feuer EJ, Brown ML. Projections of the cost of cancer care in the United States: 2010-2020. J Natl Cancer Inst. 2011;103:117-28
3. Chodosh LA. Breast cancer: current state and future promise. Breast Cancer Res. 2011;13:113
4. Yeo SK, Guan JL. Breast Cancer: Multiple Subtypes within a Tumor? Trends Cancer. 2017;3:753-60
5. Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M. et al. Tailoring therapies-improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533-46
6. Niklaus NJ, Tokarchuk I, Zbinden M, Schlafli AM, Maycotte P, Tschan MP. The Multifaceted Functions of Autophagy in Breast Cancer Development and Treatment. Cells. 2021 10
7. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70
8. Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC. et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161:279-87
9. Hutchinson L. Breast cancer: progress from NOAH study comes in twos. Nat Rev Clin Oncol. 2014;11:122
10. Marsh T, Debnath J. Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy. 2020;16:1164-5
11. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728-41
12. Li W, He P, Huang Y, Li YF, Lu J, Li M. et al. Selective autophagy of intracellular organelles: recent research advances. Theranostics. 2021;11:222-56
13. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-42
14. Deng R, Zhang HL, Huang JH, Cai RZ, Wang Y, Chen YH. et al. MAPK1/3 kinase-dependent ULK1 degradation attenuates mitophagy and promotes breast cancer bone metastasis. Autophagy. 2021;17:3011-29
15. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741-52
16. Tiessen I, Abildgaard MH, Lubas M, Gylling HM, Steinhauer C, Pietras EJ. et al. A high-throughput screen identifies the long non-coding RNA DRAIC as a regulator of autophagy. Oncogene. 2019;38:5127-41
17. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672-6
18. Li X, Jin F, Li Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med. 2021;25:4-14
19. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L. et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829-42
20. Chung SJ, Nagaraju GP, Nagalingam A, Muniraj N, Kuppusamy P, Walker A. et al. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy. 2017;13:1386-403
21. Yang M, Jing F. FBXL16 modulates the proliferation and autophagy in breast cancer cells via activating SRC-3-AKT signaling pathway. Reprod Biol. 2021;21:100538
22. Zhou J, Li G, Zheng Y, Shen HM, Hu X, Ming QL. et al. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy. 2015;11:1259-79
23. Cocco S, Leone A, Piezzo M, Caputo R, Di Lauro V, Di Rella F. et al. Targeting Autophagy in Breast Cancer. Int J Mol Sci. 2020 21
24. Wei T, Xiaojun X, Peilong C. Magnoflorine improves sensitivity to doxorubicin (DOX) of breast cancer cells via inducing apoptosis and autophagy through AKT/mTOR and p38 signaling pathways. Biomed Pharmacother. 2020;121:109139
25. Sisinni L, Pietrafesa M, Lepore S, Maddalena F, Condelli V, Esposito F. et al. Endoplasmic Reticulum Stress and Unfolded Protein Response in Breast Cancer: The Balance between Apoptosis and Autophagy and Its Role in Drug Resistance. Int J Mol Sci. 2019 20
26. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407
27. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46
28. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38
29. Xing C, Sun SG, Yue ZQ, Bai F. Role of lncRNA LUCAT1 in cancer. Biomed Pharmacother. 2021;134:111158
30. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-19
31. Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416-25
32. Xiong H, Ni Z, He J, Jiang S, Li X, He J. et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528-40
33. Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106-16
34. Wang J, Xie S, Yang J, Xiong H, Jia Y, Zhou Y. et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12:81
35. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8
36. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195-208
37. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174
38. Zhang F, Wang H, Yu J, Yao X, Yang S, Li W. et al. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. 2021;20:6
39. Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X. et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 2020;19:118
40. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L. et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer. 2020;19:85
41. Zhang R, Zhu Q, Yin D, Yang Z, Guo J, Zhang J. et al. Identification and Validation of an Autophagy-Related lncRNA Signature for Patients With Breast Cancer. Front Oncol. 2020;10:597569
42. Huang Y, Du J, Mi Y, Li T, Gong Y, Ouyang H. et al. Long Non-coding RNAs Contribute to the Inhibition of Proliferation and EMT by Pterostilbene in Human Breast Cancer. Front Oncol. 2018;8:629
43. Xiong H, Shen J, Chen Z, Yang J, Xie B, Jia Y. et al. H19/let7/Lin28 ceRNA network mediates autophagy inhibiting epithelialmesenchymal transition in breast cancer. Int J Oncol. 2020;56:794-806
44. Pawlowska E, Szczepanska J, Blasiak J. The Long Noncoding RNA HOTAIR in Breast Cancer: Does Autophagy Play a Role? Int J Mol Sci. 2017 18
45. Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707-22
46. Wang X, Liu W, Wang P, Li S. RNA interference of long noncoding RNA HOTAIR suppresses autophagy and promotes apoptosis and sensitivity to cisplatin in oral squamous cell carcinoma. J Oral Pathol Med. 2018;47:930-7
47. Li Z, Qian J, Li J, Zhu C. Knockdown of lncRNA-HOTAIR downregulates the drug-resistance of breast cancer cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp Ther Med. 2019;18:435-42
48. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B. et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746-55
49. Qiao E, Chen D, Li Q, Feng W, Yu X, Zhang X. et al. Long noncoding RNA TALNEC2 plays an oncogenic role in breast cancer by binding to EZH2 to target p57(KIP2) and involving in p-p38 MAPK and NF-kappaB pathways. J Cell Biochem. 2019;120:3978-88
50. Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606-11
51. Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK. et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798-810
52. Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13:104
53. Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J. et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer. 2019;18:89
54. Li X, Qian X, Peng LX, Jiang Y, Hawke DH, Zheng Y. et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol. 2016;18:561-71
55. Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009-15
56. Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458-67
57. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y. et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287
58. Li L, Gu M, You B, Shi S, Shan Y, Bao L. et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016;107:1215-22
59. Li Y, Jiang B, Zhu H, Qu X, Zhao L, Tan Y. et al. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumour Biol. 2017;39:1010428317705790
60. Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330-8
61. Amadori D, Milandri C, Comella G, Saracchini S, Salvagni S, Barone C. et al. A phase I/II trial of non-pegylated liposomal doxorubicin, docetaxel and trastuzumab as first-line treatment in HER-2-positive locally advanced or metastatic breast cancer. Eur J Cancer. 2011;47:2091-8
62. Zhu HY, Bai WD, Ye XM, Yang AG, Jia LT. Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem Biophys Res Commun. 2018;496:1308-13
63. Qian X, Qu H, Zhang F, Peng S, Dou D, Yang Y. et al. Exosomal long noncoding RNA AGAP2-AS1 regulates trastuzumab resistance via inducing autophagy in breast cancer. Am J Cancer Res. 2021;11:1962-81
64. Han M, Qian X, Cao H, Wang F, Li X, Han N. et al. lncRNA ZNF649-AS1 Induces Trastuzumab Resistance by Promoting ATG5 Expression and Autophagy. Mol Ther. 2020;28:2488-502
65. Cao QH, Liu F, Yang ZL, Fu XH, Yang ZH, Liu Q. et al. Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am J Transl Res. 2016;8:3831-47
66. Dong H, Wang W, Mo S, Chen R, Zou K, Han J. et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37:202
67. Han M, Gu Y, Lu P, Li J, Cao H, Li X. et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19:26
68. Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47:360-78
69. Ge J, Chen Z, Huang J, Chen J, Yuan W, Deng Z. et al. Upregulation of autophagy-related gene-5 (ATG-5) is associated with chemoresistance in human gastric cancer. PLoS One. 2014;9:e110293
70. Liu J, Zhao G, Liu XL, Zhang G, Zhao SQ, Zhang SL. et al. Progress of non-coding RNAs in triple-negative breast cancer. Life Sci. 2021;272:119238
71. Pickard MR, Williams GT. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes (Basel). 2015;6:484-99
72. Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M. et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 Axis in Regulating Autophagy in Patients with Breast Cancer. Cell Physiol Biochem. 2018;48:194-207
73. Pickard MR, Williams GT. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7:10104-16
74. Li G, Qian L, Tang X, Chen Y, Zhao Z, Zhang C. Long noncoding RNA growth arrestspecific 5 (GAS5) acts as a tumor suppressor by promoting autophagy in breast cancer. Mol Med Rep. 2020;22:2460-8
75. Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: A valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract. 2018;214:801-5
76. Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94-100
77. Hu X, Li Y, Kong D, Hu L, Liu D, Wu J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol. 2019;234:10800-8
78. Zhang XH, Li BF, Ding J, Shi L, Ren HM, Liu K. et al. LncRNA DANCR-miR-758-3p-PAX6 Molecular Network Regulates Apoptosis and Autophagy of Breast Cancer Cells. Cancer Manag Res. 2020;12:4073-84
79. Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR. et al. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018;32:42-57
80. Zhang H, Zhang N, Liu Y, Su P, Liang Y, Li Y. et al. Epigenetic Regulation of NAMPT by NAMPT-AS Drives Metastatic Progression in Triple-Negative Breast Cancer. Cancer Res. 2019;79:3347-59
81. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938-48
82. Li Y, Zhang J, Liu T, Chen Y, Zeng X, Chen X. et al. Molecular machinery of autophagy and its implication in cancer. Am J Med Sci. 2012;343:155-61
83. Li PP, Li RG, Huang YQ, Lu JP, Zhang WJ, Wang ZY. LncRNA OTUD6B-AS1 promotes paclitaxel resistance in triple negative breast cancer by regulation of miR-26a-5p/MTDH pathway-mediated autophagy and genomic instability. Aging (Albany NY). 2021;13:24171-91
84. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y. et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018;33:706-20 e9
85. Lee J, Jung JH, Chae YS, Park HY, Kim WW, Lee SJ. et al. Long Noncoding RNA snaR Regulates Proliferation, Migration and Invasion of Triple-negative Breast Cancer Cells. Anticancer Res. 2016;36:6289-95
86. Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY. et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2018;78:4316-30
87. Rodrigues de Bastos D, Nagai MA. In silico analyses identify lncRNAs: WDFY3-AS2, BDNF-AS and AFAP1-AS1 as potential prognostic factors for patients with triple-negative breast tumors. PLoS One. 2020;15:e0232284
88. Yu F, Wang L, Zhang B. Long non-coding RNA DRHC inhibits the proliferation of cancer cells in triple negative breast cancer by downregulating long non-coding RNA HOTAIR. Oncol Lett. 2019;18:3817-22
89. Clarke R, Cook KL, Hu R, Facey CO, Tavassoly I, Schwartz JL. et al. Endoplasmic reticulum stress, the unfolded protein response, autophagy, and the integrated regulation of breast cancer cell fate. Cancer Res. 2012;72:1321-31
Author contact
![]() Corresponding authors: Yichao Wang, Taizhou Central Hospital (Taizhou Univesity Hospital), No.999 Donghai Road, Jiaojiang District, Taizhou, Zhejiang, 318000, China. E-mail: wangyichaobeicom; Hongsheng Lu, Taizhou Central Hospital (Taizhou Univesity Hospital), No.999 Donghai Road, Jiaojiang District, Taizhou, Zhejiang, 318000, China. E-mail: luhscom.
Corresponding authors: Yichao Wang, Taizhou Central Hospital (Taizhou Univesity Hospital), No.999 Donghai Road, Jiaojiang District, Taizhou, Zhejiang, 318000, China. E-mail: wangyichaobeicom; Hongsheng Lu, Taizhou Central Hospital (Taizhou Univesity Hospital), No.999 Donghai Road, Jiaojiang District, Taizhou, Zhejiang, 318000, China. E-mail: luhscom.

 Global reach, higher impact
Global reach, higher impact