Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(4):779-788. doi:10.7150/ijms.71769 This issue Cite
Research Paper
Comprehensive analysis of circRNA expression profiles in rat cerebral cortex after moderate traumatic brain injury
1. Emergency Center, Zhongnan Hospital of Wuhan University, Wuhan 430071, China.
2. Department of Biological Repositories, Zhongnan Hospital of Wuhan University, Wuhan 430071, China.
#These authors contributed equally to this work.
Received 2022-2-7; Accepted 2022-3-31; Published 2022-4-18
Abstract
Traumatic brain injury is a medical event of global concern, and a growing body of research suggests that circular RNAs can play very important roles in traumatic brain injury. To explore the functions of more novel and valuable circular RNA in traumatic brain injury response, a moderate traumatic brain injury in rats was established and comprehensive analysis of circular RNA expression profiles in rat cerebral cortex was done. As a result, 301 up-regulated and 284 down-regulated circular RNAs were obtained in moderate traumatic brain injury rats, the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis were performed based on the circular RNA's host genes, and a circRNA-miRNA interaction network based on differentially expressed circular RNAs was constructed. Also, four circular RNAs were validated by RT-qPCR and Sanger sequencing. This study showed that differentially expressed circular RNAs existed between rat cerebral cortex after moderate traumatic brain injury and control. And this will provide valuable information for circular RNA research in the field of traumatic brain injury.
Keywords: circular RNA, moderate traumatic brain injury, brain injury, gene expression profiles, rat
Introduction
Traumatic brain injury (TBI) is a global public health problem, as an estimated about 50 million people will suffer this injury per year. It is a major cause of death and long-term disabilities in younger patients in developed countries [1,2]. Based on severity, TBI ranges from a mild concussion to severe injury. In moderate to severe TBI, mortality is highest due to the primary injury, secondary insults, for instance, hypoxia or hypotension, and the development of associated complications, including those localized to the cranial vault and systemic problems associated with critical illness [3]. As a complication, impaired cerebral autoregulation after moderate or severe TBI has been associated with a poor prognosis [4]. However, there are currently no effective neuroprotective therapies that have been shown to alleviate TBI-induced injury [5]. Therefore, translation of the many pathogenic mechanisms identified in animal studies to the development of treatment for human TBI remains a considerable challenge, especially in confirming whether the disrupted autoregulation attributes to the changes in specific molecular mediators.
Recently, accumulating evidence indicate that TBI significantly alters the expression of non-coding RNAs (ncRNAs), including long non-coding RNA (lncRNA) and microRNA (miRNA) [6], and neurological damage can be attenuated by normalizing the levels of certain lncRNAs and miRNAs [7-10]. However, the biological functions of ncRNAs in TBI, especially for the circular RNAs (circRNAs), remain largely unknown and enigmatic. CircRNAs are a type of endogenous non-coding RNAs (ncRNAs) with complex stage and tissue-specific expression patterns [11, 12]. Unlike linear RNA, circRNAs are much more stable and form a covalently closed-loop structure without 5'-3' polarity and a poly(A) tail, and might inhibit the function of miRNA as miRNA sponges to regulate the expression of host genes by the competing endogenous RNA (ceRNA) network [12-14]. They are highly enriched in the brain and the changes in circRNA levels are thought to be associated with the development of diverse diseases, including stroke and neurodegenerative diseases [15-19]. In the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats, the circRNA-associated-ceRNA networks are reported as an important regulator of gene expression [20]. Zhang et al. characterized the expression pattern of circRNAs and constructed the circRNA-associated-ceRNA networks in the cerebral cortex of senescence-accelerated prone 8 mice [19]. It also has been shown that the change of circRNA expression pattern may be involved in physiological and pathological processes after traumatic spinal cord injury [21]. All these findings further expanded our knowledge of circRNAs and contributed to understanding their regulation roles in brain diseases.
It has been reported that circRNAs are significantly altered in the hippocampus after TBI. Moreover, circRNAs may not only involve in brain damage but also neural regeneration following TBI through bioinformatics analysis and prediction of circRNA-miRNA interaction [22]. The expression profile of circRNAs in exosomes, which come from the brain extracellular space, could be altered in mice after TBI. In addition, these differentially expressed circRNAs might be related to the growth and repair of neurons, the development of the nervous system, and so on [22]. Many circRNAs are significantly changed in the traumatic cerebral penumbra cortex after TBI. For example, the circRNA chr8_87859283-87904548 potentially promotes neuroinflammation and impedes neurological restoration after TBI [23]. In addition, some researches also show that altered circRNAs are mainly related to inflammation, cell death, repair of injury, and synaptic function, which were involved in the secondary cascade reaction of TBI. CircRNA_01564, circRNA_11926, circRNA_05015, circRNA_16282, and circRNA_05652 are found to play central roles in the crosstalk relationship, while circRNA_16895-miRNA myosin-10 is predicted to modulate fragment crystallizable gamma receptors (FcγR)-mediated phagocytosis pathway [24]. It is also demonstrated that altered circRNA expression patterns might play important roles in post-TBI pathophysiological mechanisms [25].
However, the potential roles of circRNA in the physiological and pathological processes after TBI are still needed to be explored, especially in the role of circRNA-associated-ceRNA networks in the brain cortex after mTBI in rats.
In this study, to explore the target of effective therapeutic strategies for TBI, a high-throughput whole transcriptome sequencing was performed to identify differentially expressed profiles of circRNA in the cerebral cortex of moderate TBI (mTBI) model rats. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed, and a circRNA-miRNA network was also constructed. Finally, four differentially expressed circRNAs were validated by RT-qPCR and Sanger sequencing. Our findings enriched our understanding of mTBI-associated circRNAs and provided significant evidence to further study the function of circRNAs in TBI response.
Materials and methods
Animals
Adult male Sprague-Dawley rats (weight 250-300g) were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). The animal experiment was approved by the Animal Experiment Center and ethics committee of Zhongnan Hospital of Wuhan University and followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Rats were housed for at least 7 days before establishing an animal model in a temperature (22-25 °C) and humidity-controlled (50% relative humidity) animal facility with a 12 h light/dark cycle. Animals had free access to food and water except that food was withheld overnight before surgery.
Moderate traumatic brain injury model in rats
Eight rats were randomly divided into two groups of four rats each. As described previously in our previous study, animal models with moderate TBI (mTBI) were prepared with a weight-drop device [26]. Briefly, the rats received 5% pentobarbital at a dose of 50 mg/kg by intraperitoneal injection. After anesthesia, the hair was shaved, and the skin was disinfected with iodophor. Rats were fixed on the brain stereotaxic device to cut the top of the skull skin and determine the bregma. Then a 5 mm diameter bone window was drilled to expose the dura mater. A 40 g hammer fell at 20 cm vertically along the outer tube leading to mTBI. For the sham-operated group, the animals underwent the same surgical procedure without weight-drop impact. 24 h later, the rats were analyzed for function performance and then were euthanized with sodium pentobarbital (100 mg/kg body weight) via intraperitoneal injection. Rats were housed in individual cages after surgery and placed on heat pads (37 °C) for 24 h to maintain normal body temperature during the recovery periods.
Tissue collection, RNA isolation and sequencing
At 24h after mTBI, animals were anesthetized and transcardially perfused with 100 ml of 4 °C isotonic saline. The ipsilateral cortex around the injury site was dissected rapidly in the mTBI group and the cerebral cortex at the same site was obtained in the sham group. And then they were placed immediately into liquid nitrogen and stored at -80 °C.
Total RNA was isolated from eight ipsilateral cerebral cortex samples, including four mTBI samples and four control samples, using a miRNA Isolation Kit (Ambion, AM1560) following the manufacturer's protocol. The samples with RIN ≥ 7, monitored by the Agilent 2100 Bioanalyzer (Agilent Technologies, USA), were chosen for the following analysis. TruSeq Stranded Total RNA with Ribo-Zero Gold (Illumina, USA) was used for library construction, and the libraries were sequenced on the HiSeqTM 2500 platform.
Data preprocessing and genomic alignment
Raw reads were quality filtered by Trimmomatic software (0.36), removing adapter and filtering out low-quality bases and N-bases or low-quality reads, and the high-quality clean reads were obtained [27]. The FastaQC (v0.11.5) was used for quality control (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
To generate the SAM file, we used BWA software (0.7.5a) to align the sequencing reads of each sample with the reference genome (Rnor_6.0, ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/895/GCF_000001895.5_Rnor_6.0/GCF_000001895.5_Rnor_6.0_genomic.fna.gz) [28].
CircRNA prediction, differentially expressed circRNA analysis and clustering analysis
The circRNA identifier (CIRI) software (v2.0.3) was employed to scan for PCC signals (paired chiastic clipping signals), and circRNA sequences were predicted based on junction reads and GT-AG splicing signals [29].
For screening differential expression profiles of circRNAs, we used the estimateSizeFactors function of the DESeq R package to normalize the counts, and the nbinomTest function was used to calculate p-value and fold change values for the difference comparison [30]. Differentially expressed transcripts with p-value ≤ 0.05 and fold change ≥ 2 or ≤ 0.5 were selected, and these differentially expressed circRNAs between control and TBI groups were identified, respectively. Hierarchical clustering was performed to show the distinguishable circRNAs expression pattern among samples. Moreover, the heatmap was constructed by using the pheatmap R package.
GO annotations and KEGG pathway analyses, and circRNA-miRNA interaction research
For the differentially expressed circRNAs above, GO and KEGG enrichment analyses were done by Hypergeometric Distribution Test. The GO categories are derived from Gene Ontology (http://www.geneontology.org), which comprises three structured networks of defined terms that describe gene product attributes. Pathway analysis for differentially expressed circRNAs was performed, based on the latest KEGG (Kyoto Encyclopedia of Genes and Genomes; https://www.genome.jp/kegg) database, which allowed us to determine the biological pathways with the significantly enriched mRNAs. CircRNAs can serve as miRNA target molecules. And the miRanda software (v3.3a) was used to predict circRNA/miRNA interactions [31].
Identification of miRNAs bound with the circRNAs and construction of circRNA-miRNA interaction network
Because circRNAs contain multiple miRNA binding sites, the circRNAs bound with miRNAs should be identified as the same as the way by which can predict miRNA targeted genes. In this study, miRanda (v3.3a) was used to identify the circRNAs binding with miRNAs. And the parameters were -sc 150 -en -30 -strict. A hypergeometric distribution test was used to screen the miRNAs which the differentially expressed circRNAs enriched. The circRNA-miRNA interaction network was constructed with Cytoscape 3.7.2.
Real-time quantitative polymerase chain reaction (RT-qPCR) validation
RT-qPCR was used to confirm the differential expression identified in our RNA-seq. Before the reverse transcription reaction, the total RNA sample (≤ 1 µg) was digested with RNase-free DNase I for 30 min at 37 °C, which was stopped by adding 1µL EDTA (50 mM) at 65 °C for 10 min to inactivate DNase I. Then, the cDNA was synthesized from the digested total RNA using the PrimeScript RT reagent Kit (Takara Bio Company, Japan). The real-time qPCR reaction was performed using the ChamQ Universal SYBR qPCR Master Mix Q711 (Vazyme, China) in a QuantStudio 1 real-time PCR system (Thermofisher, USA) with the following conditions: 95 °C, 30 sec for one cycle; then 95 °C 10 sec and 60 °C 30 sec for 40 cycles. The specific quantitative primers were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/). GAPDH was designed as an internal control. The 2-ΔΔCt method was used to determine the relative quantification of gene expression levels. Each experiment had at least three replicates. The primers used in this study were listed in Table S3.
Statistical analysis
All data were analyzed using GraphPad Prism 8 and presented as mean ± standard error of the mean (SEM). Student's t-tests were used for comparisons between two groups. False discovery rates (FDR) were calculated to correct p-values in RNA-seq analysis. Differences with p < 0.05 were considered to be statistically significant.
Results
Identification characterization of circRNAs in the rat cerebral cortex after mTBI
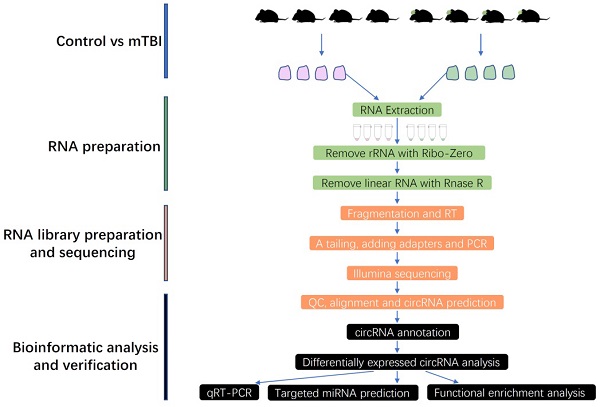
To obtain more and precise circRNAs, the rRNA and linear RNAs were removed during RNA library preparation in this study, and the full workflow was shown in Figure 1. Finally, the circRNAs were systematically identified and annotated in the rat cerebral cortex after mTBI, and a total of 51995 circRNAs were identified from 2 groups including control and mTBI (Supplemental Table S1).
All the 51995 circRNAs can be classified into 2 main categories: Sense circRNA and antisense circRNA. By type and location, these two categories also can be divided into 4 subgroups respectively. For sense circRNAs, there were 96.12% genic_exonic, 1.87% genic_intronic, 1.06% intergenic_downstream, 0.95% intergenic_upstream, while for antisense circRNAs, the proportion were 30.27%, 11.54%, 15.16%, 43.03%, respectively (Supplemental Table S1, Supplemental Figure S1). The length of the circRNAs varied greatly. The longest circRNA was 99986 bp, the shortest one was only 45 bp, and about 63.39% cicrRNAs were shorter than 1000 bp (Supplemental Table S1). Moreover, about 70.00% cicrRNAs had 1 to 5 exons (Supplemental Figure S2), and most of circRNAs enriched at chromosomes 1 to 10 (Supplemental Figure S3). The amount of circRNA expression was estimated by RPM (reads per million) based on the number of back-spliced reads, and the distribution of circRNA expression was shown by a density distribution map (Supplemental Figure S4). This result showed that circRNAs generally had low expression levels.
Differential expression of circRNAs in the rat cerebral cortex induced by mTBI
To clarify which circRNAs were essential for mTBI in the rat cerebral cortex. An analysis with differentially expressed circRNAs (DECRs) was carried out in this study. Finally, a total of 585 DECRs were identified and showed in the Volcano Plot based on a screening threshold p < 0.05, |log2FC| > 1 (Figure 2A, Supplemental Table S1), in which 301 up-regulated and 284 down-regulated circRNAs in mTBI rats compared to control (Figure 2B). There were 275 host genes in upregulated circRNAs, and 251 host genes in down-regulated circRNAs (Supplemental Table S1). The top 10 up-regulated and down-regulated circRNAs were listed in Table 1. Cluster analysis of DECRs based on the circRNAs expression was shown with a heatmap (Figure 2C). These DECRs may play important roles in the rat cerebral cortex induced by mTBI.
Workflow chart in this study. mTBI: Traumatic Brain Injury, RT: Reverse Transcription, QC: Quality Control, qRT-PCR: Quantitative Real Time Polymerase Chain Reaction.
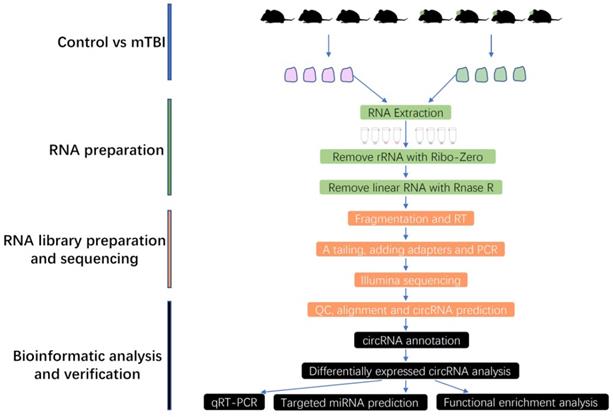
Differentially expressed circRNAs in rat cerebral cortex after mTBI. Volcano plot for DECRs in rat cerebral cortex after mTBI based on RNA_seq analysis. (B) Histogram plot for DECRs in mTBI compare to control. (C) Heatmap plot for DECRs. p < 0.05, |log2FC| > 1.
The characterization of top 10 up-regulated and top 10 down-regulated circRNAs
| circRNA_id | log2FC | pval | up-down | circRNA_chr | circRNA_strand | circRNA_start | circRNA_end | genomic_length | circRNA_length | Type | gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| circRNA_04099 | Inf | 1.90E-16 | Up | Chr1 | + | 278661823 | 278749829 | 88007 | 758 | sense-overlapping | Atrnl1 |
| circRNA_35079 | Inf | 1.22E-08 | Up | Chr7 | - | 98606921 | 98682856 | 75936 | 63953 | sense-overlapping | Tmem65 |
| circRNA_18531 | Inf | 3.38E-06 | Up | Chr19 | + | 46552916 | 46559348 | 6433 | 346 | sense-overlapping | Vat1l |
| circRNA_03118 | 5.22 | 9.08E-06 | Up | Chr1 | - | 221185801 | 221186464 | 664 | 664 | intergenic | --- |
| circRNA_15598 | 2.47 | 1.78E-05 | Up | Chr17 | + | 36602134 | 36627167 | 25034 | 267 | sense-overlapping | Cdkal1 |
| circRNA_10508 | 2.96 | 5.83E-05 | Up | Chr13 | + | 101274882 | 101280792 | 5911 | 363 | sense-overlapping | Susd4 |
| circRNA_17935 | 2.65 | 2.84E-04 | Up | Chr19 | - | 12691066 | 12713398 | 22333 | 509 | sense-overlapping | Large1 |
| circRNA_27089 | 11.94 | 4.62E-04 | Up | Chr4 | + | 97812913 | 97830459 | 17547 | 17547 | intergenic | --- |
| circRNA_15434 | 2.88 | 6.74E-04 | Up | Chr17 | + | 20098292 | 20136179 | 37888 | 455 | sense-overlapping | Dtnbp1 |
| circRNA_40399 | 11.22 | 8.63E-04 | Up | Chr9 | - | 119615345 | 119617261 | 1917 | 1917 | exonic | Emilin2 |
| circRNA_15695 | 0.27 | 2.04E-06 | Down | Chr17 | - | 48657102 | 48679275 | 22174 | 793 | sense-overlapping | Vps41 |
| circRNA_33112 | -lnf | 2.56E-06 | Down | Chr6 | + | 112684486 | 112689839 | 5354 | 823 | sense-overlapping | Nrxn3 |
| circRNA_22458 | 0.25 | 3.64E-04 | Down | Chr20 | - | 31723858 | 31736111 | 12254 | 11458 | sense-overlapping | RGD1305587 |
| circRNA_24421 | 0.08 | 4.11E-04 | Down | Chr3 | + | 104889557 | 104936645 | 47089 | 356 | sense-overlapping | Fmn1 |
| circRNA_13425 | 0.21 | 7.07E-04 | Down | Chr15 | + | 61667133 | 61683055 | 15923 | 809 | sense-overlapping | Mtrf1 |
| circRNA_27617 | 0.18 | 7.58E-04 | Down | Chr4 | + | 140372957 | 140379514 | 6558 | 576 | sense-overlapping | Itpr1 |
| circRNA_26650 | 0.39 | 1.82E-03 | Down | Chr4 | - | 64265934 | 64270334 | 4401 | 453 | sense-overlapping | Ptn |
| circRNA_19958 | 0.42 | 2.13E-03 | Down | Chr2 | - | 98509848 | 98586772 | 76925 | 3386 | sense-overlapping | Zfhx4 |
| circRNA_13422 | 0.37 | 2.48E-03 | Down | Chr15 | + | 61665894 | 61683055 | 17162 | 1235 | sense-overlapping | Mtrf1 |
| circRNA_21502 | 0.22 | 2.60E-03 | Down | Chr2 | + | 217971903 | 217983563 | 11661 | 523 | sense-overlapping | Olfm3 |
GO and KEGG pathway enrichment analysis on the differentially expressed circRNAs
To learn about the related functions of DECRs, the GO and KEGG enrichment analyses were done based on the circRNA's host genes.
For all DECRs, there were 1281 GO terms were enriched (p < 0.05), including 890 GO BP (biological process), 132 GO CC (cellular component), 259 GO MF (molecular function). We further screened the terms including 3 genes at least and found that the DECRs were enriched in regulation of T cell differentiation, immunological synapse formation, positive regulation of axon regeneration, positive regulation of endocytosis and cell proliferation, and regulation of ERK1/ERK2, JNK, and MAPK cascade. (Figure 3A, Supplemental Table S1).
The characterization of circRNAs verified by RT-qPCR
| circRNA_id | log2FC | pval | up_down | circRNA_chr | circRNA_strand | circRNA_start | circRNA_end | genomic_length | circRNA_length | Type | gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| circRNA_17935 | 1.40 | 0.00028 | Up | Chr19 | - | 12691066 | 12713398 | 22333 | 509 | sense-overlapping | Large1 |
| circRNA_15434 | 1.52 | 0.00067 | Up | Chr17 | + | 20098292 | 20136179 | 37888 | 455 | sense-overlapping | Dtnbp1 |
| circRNA_19958 | -1.24 | 0.00217 | Down | Chr2 | - | 98509848 | 98586772 | 76925 | 3386 | sense-overlapping | Zfhx4 |
| circRNA_26562 | -2.24 | 0.00347 | Down | Chr4 | + | 58627457 | 58669806 | 42350 | 42350 | intergenic | - |
For KEGG pathway analysis, there were 47 KEGG pathways were enriched (p < 0.5) in all DECRs. And found that Natural killer cell mediated cytotoxicity, Th1, Th2, and Th17 cell differentiation, C-type lectin receptor signaling pathway, NOD-like receptor signaling pathway, MAPK signaling pathway, and some neurodegenerative disease pathway, for example, Huntington's disease and Alzheimer's disease, were enriched in the DECRs (Figure 3B, Supplemental Table S1).
GO and KEGG enrichment analyses on DECRs in the rat cerebral cortex after mTBI. GO analysis on differentially expressed circRNAs (DECRs) in rat cerebral cortex after mTBI. (B) GO analysis on differentially expressed circRNAs (DECRs) in rat cerebral cortex after mTBI.
These results suggested that cell proliferation and differentiation, immune response, and kinase-induced signal transduction may play essential roles in the mTBI response in the rat cerebral cortex.
Prediction of circRNA-miRNA interaction network
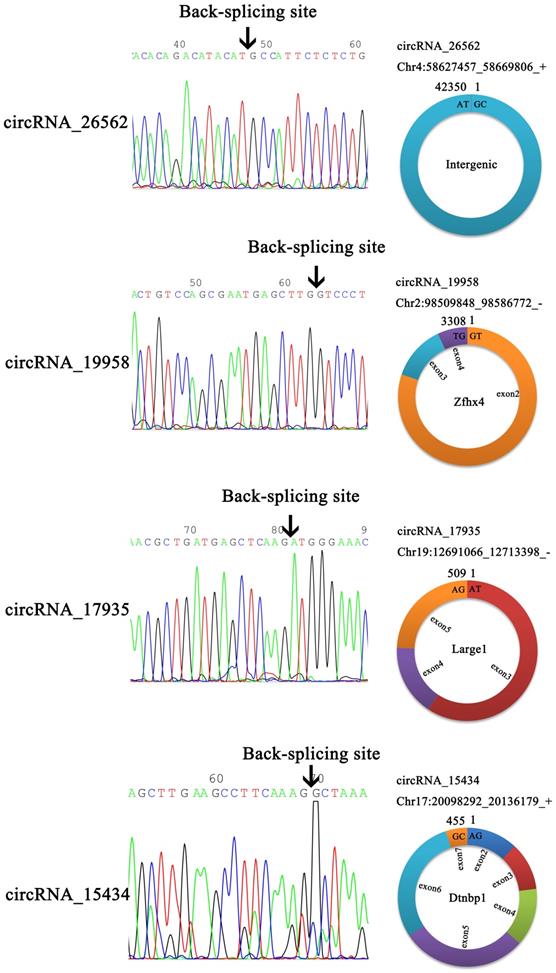
CircRNAs, as sponges for miRNAs, have been reported to indirectly modulate the expression level of other related RNAs by miRNA response elements [32]. Therefore, it is very important to identify the interaction of circRNAs and miRNAs. In this study, 576 targeted miRNAs were identified for the DECRs (Supplemental Table S2). To understand which miRNAs were more effective for the DECRs, an enrichment analysis was done for all the miRNAs by a hypergeometric distribution test. As a result, the DECRs were significantly enriched in 49 miRNAs (p < 0.05) (Supplemental Table S2). 36 circRNAs and 17 miRNAs were selected to construct a circRNA-miRNA network (Figure 4, Supplemental Table S2). The result showed rno-miR-667-5p and rno-miR-466b-3p were regulated by a greater number of circRNAs (Figure 4). This result suggested rno-miR-667-5p and rno-miR-466b-3p may be participated in the mTBI response by interacting with these circRNAs.
Validation of differentially expressed circRNAs by RT-qPCR
To validate the transcription data, 10 genes were randomly chosen to test the expression level by the RT-qPCR using divergent primers. In our result, we found that four circRNAs were differentially expressed in the mTBI sample compared to the control. CircRNA_19958 and circRNA_26562 were down-regulated, while circRNA_15434 and circRNA_17935 were up-regulated significantly (Figure 5, Table 2). However, there are still 6 circRNAs were not significantly changed in expression level (Supplemental Figure S3).
CircRNA-miRNA interaction network. The blue ovals show the targeted miRNAs, the green triangles show down-regulated circRNAs and the light red triangles show up-regulated circRNAs.
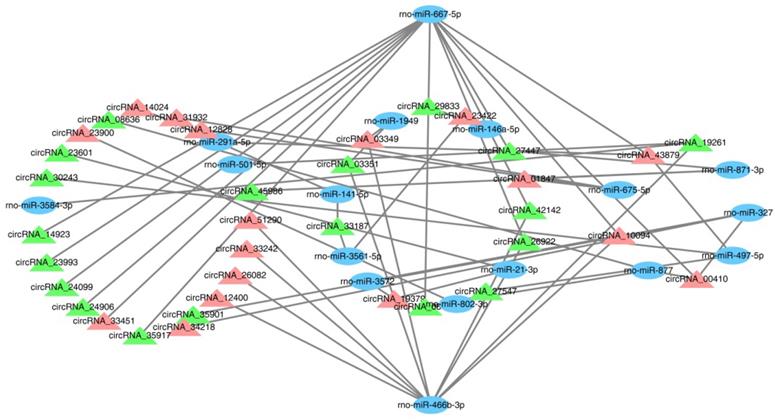
Expression level analysis on circRNAs by RT-qPCR. The expression of circRNA_19958, circRNA_26562, circRNA_17935 and circRNA_15434 were tested in at least 3 control s and 3 mTBI rat cerebral cortex samples by RT-qPCR. *: P < 0.05, **: P < 0.01.
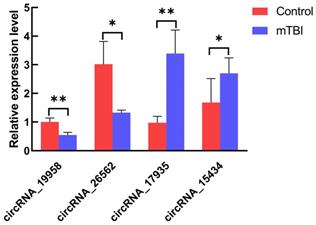
In order to verify the back-splicing site and make clear the structure of the four circRNAs, the RT-qPCR products were used for Sanger sequencing. The results showed circ_19958, circ_17935, and circ_15434 were originated from a different number of exons, although circ_26562 was originated from an intergenic sequence (Figure 6).
Discussion
As the body's most vulnerable organ, the brain is the center of human consciousness, controlling how we act, feel, speak, and so on [33-35]. TBI is an important cause of morbidity and mortality worldwide. Many drug therapies have been developed to protect the brain after injury, however, none of those are successful in TBI outcomes [36-38]. Therefore, TBI will bring heavy burdens to the patient's family. At present, a large number of studies have shown that circRNAs are very important in brain diseases such as Alzheimer's disease, Parkinson's disease, and glioma [39,40]. It also has been reported that circRNA expression profiles of the TBI brain are different from that of the normal brain in both intracellular and extracellular space, suggesting that circRNAs may be involved in the pathogenesis of TBI and act as a regulator [22-24,41,42]. As a result, exploring the novel circRNAs and making clear their function in recovering brain injuries have great significance. In this study, we predicted many novel and meaningful circRNAs in the rat mTBI model by high-throughput sequencing and verified 4 circRNAs by RT-qPCR and Sanger sequencing. This will provide new knowledge of circRNAs' functions on brain injuries in rats.
CircRNAs can regulate gene expression in different ways. Firstly, circRNAs can affect parental gene expression. This is mainly because the formation of circRNAs can influence the typical splicing of their precursor transcripts, leading to changes in gene expression levels [43]. Some nuclear-located circRNAs may even regulate gene expression at the transitional and splicing levels, including CircSEP3 from exon 6 of SEPALLATA3, which can regulate the splicing of its linear counterpart [44,45]. Secondly, circRNAs can be translated due to internal ribosome entry sites or N6-methyladenosine modification enriched in circRNAs [45,46]. Besides, circRNAs also can act as a sponge for miRNAs to regulate gene expression indirectly. It's reported that as a sponge for miR-138, circRNA sex-determining region Y can prevent miR-138 from interacting with its target genes [13]. To know the functions of predicted circRNAs in this study, we identified the target miRNAs and constructed a network between circRNA and miRNA. The network showed that rno-miR-466b-3p and rno-miR-667-5p may serve as a potential target of multiple circRNAs and be involved in their expression regulation. However, their relationship and downstream genes still needed to be confirmed and explored by further studies.
Head-to-tail splicing in the RT-qPCR product of circRNA.
CircRNAs also play important roles in brain injury. A wide variety of studies have shown that circRNAs are enriched in brain tissues, which include the cortex, cerebellum, striatum, hippocampus, and olfactory bulbs, and are closely connected with neuronal development such as the development of neural stem cells [47-50]. Besides the functions on neuronal development, they are also complexly linked to brain disorders, for example, Alzheimer's disease and temporal lobe epilepsy [51-53]. Studies have shown that stroke-induced brain damage is mediated by multiple synergistic pathophysiologic mechanisms, including autophagy, mitochondrial dysfunction, apoptosis, inflammation, and so on [54,55]. KEGG pathway enrichment analysis revealed that the cell cycle, mitogen-activated protein kinase (MAPK) signaling, focal adhesion, and regulation of the actin cytoskeleton are core pathways associated with circRNAs [15]. In brain ischemia-reperfusion injury, apoptosis-related, immune-related, and metabolism-related pathways may have critical roles [56]. However, in traumatic brain injury, it's also found that circRNAs can participate in the immune response, inflammation response, and neuronal apoptosis [24, 57-60]. In our study, the GO analysis results showed differentially expressed circRNAs in the mTBI group were enriched immunological synapse formation, positive regulation of axon regeneration, positive regulation of ERK1 and ERK2 cascade, MAPK cascade, regulation of JNK cascade. On the other hand, the KEGG results showed that immune-related pathways such as NOD-like receptor signaling pathway, Natural killer cell mediated cytotoxicity, C-type lectin receptor signaling pathway, and Th1/2/17 cell differentiation, and some neurorelated diseases, Huntington's disease and Alzheimer's disease were also enriched in mTBI. As a result, circRNAs may participate in mTBI response regulation via regulating immune, inflammation, and apoptosis and it's very necessary for further exploring the molecular mechanisms.
Conclusion
In conclusion, we report a circRNA expression profile in the rat cerebral cortex after moderate traumatic brain injury. In our study, 585 differentially expressed circular RNAs (DECRs) were identified, and the functional enrichment analysis was performed which may reveal the potential and important roles of circular RNAs in mTBI regulation. Besides that, A network between the DECRs and their target miRNAs was constructed and found that rno-miR-667-5p and rno-miR-466-3p may have an important role in the regulation of circular RNAs expression in mTBI response. Our study also confirmed four circRNAs expression levels between sham and mTBI, and further make clear their sequence composition. We believe that this study will bring valuable information to researchers in this field.
Supplementary Material
Supplementary figures and table 3.
Supplementary table 1: Annotation of circRNA and DECRs.
Supplementary table 2: Interaction between circRNA and miRNA.
Acknowledgements
This study was supported by the Research Fund from Medical Sci-Tech Innovation Platform of Zhongnan Hospital, Wuhan University (PTXM2020001).
Author Contributions
Conceptualization and design of experiments, H.M. and Y.Z.; experiments and data analysis, G.L., S.L., and R.L.; Writing-original draft preparation, G.L. and S.L.; writing-review and editing, H.M. and Y.Z.; visualization, G.L., S.L., and J.Y. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Maas AI, Menon DK. Integrated approaches to paediatric neurocritical care in traumatic brain injury. Lancet Neurol. 2013;12(1):26-8
2. Meissner L, Gallozzi M, Balbi M, Schwarzmaier S, Tiedt S, Terpolilli NA. et al. Temporal Profile of MicroRNA Expression in Contused Cortex after Traumatic Brain Injury in Mice. J Neurotrauma. 2016;33:713-20
3. Carney N, Totten AM, O'Reilly C. et al. Guidelines for the Management of Severe Traumatic Brain Injury, Fourth Edition. Neurosurgery. 2017;80(1):6-15
4. Zeiler FA, Donnelly J, Smielewski P, Menon DK, Hutchinson PJ, Czosnyka M. Critical Thresholds of Intracranial Pressure-Derived Continuous Cerebrovascular Reactivity Indices for Outcome Prediction in Noncraniectomized Patients with Traumatic Brain Injury. J Neurotrauma. 2018;35(10):1107-15
5. Zhu H, Xing Z, Zhao Y, Hao Z, Li M. The Role of Circular RNAs in Brain Injury. Neuroscience. 2020;428:50-9
6. Chandran R, Mehta SL, Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int. 2017;111:12-22
7. Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191-201
8. Liu L, Sun T, Liu Z, Chen X, Zhao L, Qu G. et al. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS One. 2014;9:e103948
9. Wang CF, Zhao CC, Weng WJ, Lei J, Lin Y, Mao Q. et al. Alteration in Long Non-Coding RNA Expression after Traumatic Brain Injury in Rats. J Neurotrauma. 2017;34:2100-8
10. Zhong J, Jiang L, Huang Z, Zhang H, Cheng C, Liu H. et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun. 2017;65:183-94
11. Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163-8
12. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-85
13. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
14. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603-10
15. Mehta SL, Pandi G, Vemuganti R. Circular RNA Expression Profiles Alter Significantly in Mouse Brain After Transient Focal Ischemia. Stroke. 2017;48:2541-8
16. Kumar L, Shamsuzzama, Haque R, Baghel T, Nazir A. Circular RNAs: the Emerging Class of Non-coding RNAs and Their Potential Role in Human Neurodegenerative Diseases. Mol Neurobiol. 2017;54:7224-34
17. Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271-81
18. Lukiw W, Zhao Y, Rogaev E, Bhattacharjee S. A Circular RNA (circRNA) ciRS-7 in Alzheimer's disease (AD) targets miRNA-7 trafficking and promotes deficits in the expression of the ubiquitin conjugase (UBE2A) and the epidermal growth factor receptor (EGFR). FASEB JOURNAL. 2016 30
19. Zhang S, Zhu D, Li H, Li H, Feng C, Zhang W. Characterization of circRNA-Associated-ceRNA Networks in a Senescence-Accelerated Mouse Prone 8 Brain. Mol Ther. 2017;25:2053-61
20. Wang Z, Xu P, Chen B. et al. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging (Albany NY). 2018;10(4):775-88
21. Qin C, Liu CB, Yang DG. et al. Circular RNA Expression Alteration and Bioinformatics Analysis in Rats After Traumatic Spinal Cord Injury. Front Mol Neurosci. 2019;11:497
22. Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC, Dong JF. et al. Circular Ribonucleic Acid Expression Alteration in Exosomes from the Brain Extracellular Space after Traumatic Brain Injury in Mice. J Neurotrauma. 2018;35:2056-66
23. Chen Z, Wang H, Zhong J. et al. Significant changes in circular RNA in the mouse cerebral cortex around an injury site after traumatic brain injury. Exp Neurol. 2019;313:37-48
24. Jiang YJ, Cao SQ, Gao LB. et al. Circular Ribonucleic Acid Expression Profile in Mouse Cortex after Traumatic Brain Injury. J Neurotrauma. 2019;36(7):1018-28
25. Xie BS, Wang YQ, Lin Y. et al. Circular RNA Expression Profiles Alter Significantly after Traumatic Brain Injury in Rats. J Neurotrauma. 2018;35(14):1659-66
26. Zhang Z, Yu J, Wang P, Lin L, Liu R, Zeng R, Ma H, Zhao Y. iTRAQ-based proteomic profiling reveals protein alterations after traumatic brain injury and supports thyroxine as a potential treatment. Mol Brain. 2021;14(1):25
27. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114-20
28. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
29. Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16(1):4
30. Anders S. Analysing RNA-Seq data with the DESeq package. Mol Biol. 2010;43:1-17
31. Schuldt AJ, Adams JHJ, Davidson CM. et al. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes & Development. 1998;12(12):1847-57
32. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
33. Zeki S, Shipp S. The functional logic of cortical connections. Nature. 1998;335:311-317
34. Tononi G, Edelman GM, Sporns O. Complexity and coherency: integrating information in the brain. Trends Cogn Sci. 1998;2:474-84
35. Friston K. Functional integration and inference in the brain. Prog Neurobiol. 2002;68:113-43
36. Rubiano AM, Carney N, Chesnut R, Puyana JC. Global neurotrauma research challenges and opportunities. Nature. 2015;527:S193-7
37. Brazinova A, Rehorcikova V, Taylor MS, Buckova V, Majdan M, Psota M, Peeters W, Feigin V, Theadom A, Holkovic L, Synnot A. Epidemiology of traumatic brain injury in europe: living systematic review. J Neurotrauma. 2021;38(10):1411-40
38. Cancelliere C, Coronado VG, Taylor CA, Xu L. Epidemiology of isolated versus nonisolated mild traumatic brain injury treated in emergency departments in the United States, 2006-2012: sociodemographic characteristics. J Head Trauma Rehabil. 2017;32:E37-46
39. Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8-18
40. Akhter R. Circular RNA and Alzheimer's disease. Adv Exp Med Biol. 2018;1087:239-43
41. Xie BS, Wang YQ, Lin Y, Zhao CC, Mao Q, Feng JF, Cao JY, Gao GY, Jiang JY. Circular RNA expression profiles alter significantly after traumatic brain injury in rats. J Neurotrauma. 2018;35:1659-66
42. Jiang L, Li H, Fan Z, Zhao R, Xia Z. Circular RNA expression profiles in neonatal rats following hypoxic-ischemic brain damage. Int J Mol Med. 2019;43(4):1699-708
43. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
44. Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554-65
45. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214-27
46. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626-41
47. Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu G, Yao H. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14:404-18
48. Yang Q, Wu J, Zhao J, Xu T, Zhao Z, Song X, Han P. Circular RNA expression profiles during the differentiation of mouse neural stem cells. BMC Syst Biol. 2018;12:128
49. Yang X, Ji H, Yao Y, Lai X, Jiang Y, Wu D, Cai L, Zhu W, Gu X, Hu R, Li L, Xu L, Jiang M. Downregulation of circ_008018 protects against cerebral ischemia-reperfusion injury by targeting miR-99a. Biochem Biophys Res Commun. 2018;499:758-64
50. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018 110
51. Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's Disease (AD) is linked to deficits in a natural circular miRNA-7 Sponge (circRNA; ciRS-7). Genes (Basel). 2016 7
52. Shi Z, Chen T, Yao Q, Zheng L, Zhang Z, Wang J, Hu Z, Cui H, Han Y, Han X, Zhang K, Hong W. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-kappaBdependent manner. FEBS J. 2017;284:1096-109
53. Li J, Lin H, Sun Z, Kong G, Yan X, Wang Y, Wang X, Wen Y, Liu X, Zheng H, Jia M, Shi Z, Xu R, Yang S, Yuan F. Highthroughput data of circular RNA profiles in human temporal cortex tissue reveals novel insights into temporal lobe epilepsy. Cell Physiol Biochem. 2018;45:677-91
54. Balog J, Mehta SL, Vemuganti R. Mitochondrial fission and fusion in secondary brain damage after CNS insults. J Cereb Blood Flow Metab. 2016;36:2022-33
55. Kim T, Vemuganti R. Mechanisms of Parkinson's diseaserelated proteins in mediating secondary brain damage after cerebral ischemia. J Cereb Blood Flow Metab. 2017;37:1910-26
56. Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, Ma QJ. Circular RNA expression alterations are involved in OGD/Rinduced neuron injury. Biochem Biophys Res Commun. 2016;471:52-6
57. Saika R, Sakuma H, Noto D, Yamaguchi S, Yamamura T, Miyake S. MicroRNA-101a regulates microglial morphology and inflammation. J Neuroinflammation. 2017;14:109
58. Mellios N, Feldman DA, Sheridan SD. et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry. 2018;23:1051-65
59. Deng ZF, Zheng HL, Chen JG, Luo Y, Xu JF, Zhao G, Lu JJ, Li HH, Gao SQ, Zhang DZ, Zhu LQ, Zhang YH, Wang F. miR-214-3p targets beta-catenin to regulate depressive-like behaviors induced by chronic social defeat stress in mice. Cereb Cortex. 2019;29:1509-19
60. Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C, Dorsey SG, Faden AI. Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci. 2014;34:10055-71
Author contact
![]() Corresponding authors: Haoli Ma, E-mail: mahaoliedu.cn; Yan Zhao, E-mail: doctoryanzhaoedu.cn.
Corresponding authors: Haoli Ma, E-mail: mahaoliedu.cn; Yan Zhao, E-mail: doctoryanzhaoedu.cn.

 Global reach, higher impact
Global reach, higher impact