Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(3):472-483. doi:10.7150/ijms.67286 This issue Cite
Research Paper
miR-204-5p inhibits cell proliferation and induces cell apoptosis in esophageal squamous cell carcinoma by regulating Nestin
1. Department of Thoracic Surgery, the First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong 510080, China
2. Department of Interventional Medicine, the Fifth Affiliated Hospital Sun Yat-sen University, Zhuhai, Guangdong 519000, China
3. Department of Thyroid and Breast Surgery, the Fifth Affiliated Hospital of Sun Yat sen University, Zhuhai, Guangdong 519000, China
4. Department of Pediatrics, the Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong 519000, China
5. Department of Pharmacy, the Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong 519000suppl, China
6. Department of Thoracic Surgery, the Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong 519000, China
*These authors contributed equally to this work.
Received 2021-9-19; Accepted 2022-2-9; Published 2022-2-28
Abstract
Esophageal cancer (EC) is a highly malignant gastrointestinal tumor, and esophageal squamous cell carcinoma (ESCC) is one of the most common histological types of EC. MicroRNAs (miRNAs) are small noncoding RNAs closely related to tumorigenesis and tumor progression. In addition, Nestin is an intermediate filament protein (class VI) and contributes to the progression of numerous tumors. However, the correlation between miRNAs and Nestin in ESCC remains unclear. This study aimed to investigate the relationship between miR-204-5p and Nestin in ESCC. First, Nestin-related miRNAs in ESCC were explored using RNA sequencing. In ESCC tissues and cell lines, the expression of miR-204-5p was decreased detected by quantitative real-time polymerase chain reaction (qPCR), whereas Nestin protein level was upregulated identified by Western blotting (WB). Besides, Nestin was the direct target of miR-204-5p in ESCC determined via the luciferase reported assay. Moreover, miR-204-5p regulated Nestin to inhibit ESCC cell proliferation detected by the colony formation assay and promote ESCC cell apoptosis identified using the flow cytometry and TUNEL assay. Furthermore, miR-204-5p suppressed tumorigenesis in vivo evaluated by the murine xenograft tumor model. In conclusion, these results indicated that miR-204-5p inhibited cell proliferation and induced cell apoptosis in ESCC through targeting Nestin, which might provide novel therapeutic targets for ESCC therapy.
Keywords: miR-204-5p, Nestin, esophageal squamous cell carcinoma, proliferation, apoptosis
Introduction
Esophageal cancer (EC) is a highly malignant gastrointestinal tumor. The most common histological types of EC are esophageal adenocarcinoma (EAC) and ESCC. In Asia, especially in China, ESCC is the most common pathological subtype [1]. Even though there have been advances in diagnosis, surgical methods, radiotherapy and chemotherapy, the mortality of ESCC is still high because of the recurrence and drug resistance [2]. Advances in the study of molecular biology and gene technology may help to unravel the complicated pathogenesis of EC, which includes the interaction of genetic variations and other physiological factors. So, studying the molecular mechanisms involved in the development and progression of EC is a research hotspot, and may result in the identification of vital and novel therapeutic targets.
MicroRNAs (miRNAs) are small noncoding RNAs regulating post-transcriptional gene expression [3]. The RNA-induced silencing complex (RISC) is a target recognition element of mature miRNA [4]. The RISC binds to its target mRNA via the 3' untranslated region (UTR), and can lead to the degradation of the mRNA, or inhibition of its translation [5], both of which result in down-regulation of protein translation. Studies have suggested that miRNAs regulate about 30% of genes, and it has been calculated that over 60% of protein-coding genes may be potential targets of miRNAs [6, 7]. The genes regulated by miRNAs are involved in numerous cellular processes, and growing evidence indicates that miRNAs participate in the pathogenesis of many diseases, including cancers, nervous system diseases, autoimmune diseases, metabolic diseases, and cardiovascular diseases [8-13]. Importantly, abnormal miRNA expression has been found to be closely related to tumorigenesis and tumor progression [14].
Nestin is an intermediate filament protein (class VI), and is expressed in normal and pathological cells of a number of different tissues and organs [15]. Recent studies have reported that Nestin is expressed in certain malignant cells, and the high Nestin expression correlates with malignant features in some tumors. Other studies have indicated that Nestin is a novel biomarker of brain, bladder, and pancreatic cancer stem cells.[16-18] To date, rare studies have revealed that Nestin positively contributes to cell proliferation and poor prognosis in esophageal squamous cancer [19]. However, the function of miRNAs on the regulation of Nestin in ESCC was unclear.
Thus, the aim of our study was to identify miRNAs targeting Nestin, determine whether these miRNAs were associated with ESCC, and uncover the mechanism how miRNAs regulate Nestin in ESCC.
Materials and Methods
Tissue samples
Fresh ESCC tissue samples and para-carcinoma tissue samples were collected from 3 patients with ESCC treated in First Affiliated Hospital, Sun Yet-San University in 2018. The Institutional Ethics Committee of the First Affiliated Hospital of Sun Yet-San University approved this study.
Small RNA sequencing
Trizol (Invitrogen, Carlsbad, CA, USA) was applied to extract the total RNA from the ESCC tissues according to the manufacturer's protocol. Next, the RNA molecules ranged from 18nt to 30nt were enriched by polyacrylamide gel electrophoresis (PAGE). Then both the 5` and 3' adapters were ligated to RNAs. Next, the ligation products were reverse transcripted, and the 140-160bp size products were collected to construct the cDNA library. The Agilent 2100 bioanalyzer was used to check the distribution of the size of the fragments for the library while quantitative real-time polymerase chain reaction (qPCR, TaqMan Probe) was used to quantify the library.
Subsequently, the libraries were sequenced by the BGISEQ-500 platform (BGI-Shenzhen, China). To obtain clean reads, raw reads were filtered using in-house Perl scripts. After discarding dirty reads either containing adapter or over 10% poly-N sequences and low quality reads whose Phred scores were less than 5%, all clean tags were aligned based on GeneBank database and Rfam database (11.0) to remove tRNA, scRNA, snRNA, snoRNA and rRNA. Besides, all clean tags were aligned with human reference genome (Grch37) by TopHat v2.0.9. In addition, those tags mapped to repeat sequences were also discarded. Then clean tags were blasted in miRBase database (Release 21) to identify known miRNAs.
Next, the small RNA expression level was then calculated based on transcripts per million (TPM) using the following formula: TPM=Actual miRNA counts/Total counts of clean tags×106 [20]. Moreover, differential expression analysis was performed using the DEGseq to determine the significance of differences of expression [21]. miRNAs with a fold change ≥2 and P <0.05 in a comparison were determined as significant miRNAs with differently expressions.
Bioinformatics analysis
Based on the sequences of Nestin mRNA and differently expressed miRNAs in ESCC tisssues, the candidate miRNAs targeting Nestin mRNA were predicted by TargetScan website and microRNA.org website [22, 23].
Cell culture
ESCC cell lines including KYSE30, KYSE150, KYSE450 and the human esophageal epithelial cell (HEEC) were purchased from the Cell Bank at the Chinese Academy of Sciences (Shanghai, China). All cells were cultured with DMEM medium (Hyclone, USA) containing 100 units/mL penicillin (Hyclone, USA) and 10% fetal bovine serum (FBS) (Hyclone) at 37 °C in a 5% CO2 humidified incubator.
Cell transfection and stable cell construction
In this study, pCDH-GFP+Puro lentiviral vectors were utilized for the stably transduced cell lines. All vectors used for the stably transduced cell lines were purchased from Hechuang Biotech Ltd. (Guangzhou, China). Nestin cDNA including 3'UTR was used in this study.
The KYSE30 and KYSE450 cell lines were transfected with different lentiviral vectors to establish blank expression vector-transfected stable cells (Blank vec group), miR-204-5p overexpression stable cells (miR-204-5p OE group), Nestin overexpression stable cells (Nestin OE group) and miRNA-204-5p overexpression plus Nestin overexpression stable cells (miR-204-5p OE+Nestin OE group), according to standard protocols. The stable cell lines were selected by measuring the level of green fluorescent protein (GFP) in cells. In addition, the expression of miRNA-204-5p and Nestin in the stably transfected cells was verified by qPCR and Western blotting, respectively.
Total RNA extraction and qPCR
Total RNA from cultured cells or tissues was extracted by TRIzol reagent according to the manufacturer's instructions. For mRNA, 2 mg RNA was used for cDNA synthesis by High-Capacity RNA-to-cDNA™ Kit. For miRNA, 2 mg RNA was reverse transcribed using TaqMan miRNA assays (ABI, Forest City, CA, USA). The qPCR was performed by SYBR-based Roche Light-Cycler® 480II PCR instrument. Moreover, the relative expression levels of target genes were analyzed with 2-ΔΔCt method [24]. In this study, U6 and GAPDH were used as the internal references. The following primer pairs were used for qPCR: miR-204-5p forward: 5'-UUCCCUUUGUCAUCCUAUGCCU-3', reverse: 5'-CTCAACTGGTGTCGTGGA-3'; Nestin: forward, 5'-TGCGGGCTACTGAAAAGTTC-3'; reverse, 5'-GGCTGAGGGACATCTTGAG-3'; U6 forward: 5'-CTCGCTTCGGCAGCACA-3', reverse: 5'-AACGCTTCACGAATTTGCGT-3'; GAPDH forward: 5'-GGGAAACTGTGGCGTGAT-3', reverse: 5'-GAGTGGGTGTCGCTGTTGA-3'.
Western blotting analysis
Total proteins were extracted by RIPA buffer (Cell Signaling Technology, Danvers, MA, USA). Next, proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred onto the PVDF membrane (Millipore). Then membranes were blocked using 5% non-fat milk followed by the incubation with rabbit polyclonal anti-Nestin antibody (1:500, 19483-1-AP, Proteintech, Rosemont, IL, USA) or anti-GAPDH antibody (1:3000. IPVH00010, Millipore, Bedford, MA, USA) overnight at 4 ℃. GAPDH was used as the internal control. The next day membranes were washed by Tris-buffered saline contained 0.1% Tween20 (TBST) and incubated with anti-rabbit secondary antibody (Southern Biotech). The signals of targeted proteins were detected using chemiluminescence detection kit (Beyotime, Shanghai, China). Subsequently, densitometric analyses of bands were performed by Image J (NIH Image, Bethesda, MD, USA), and then the quantification of protein abundance was performed by normalizing the densitometric data of target protein to that of GAPDH.
Luciferase reporter assay
The human wild-type Nestin 3'-UTR sequence or the mutated Nestin 3'-UTR sequence with the predicted target sites was amplified and subcloned into the pmirGLO Dual-Luciferase vector (HeChuang Biotech, Guangzhou, China), while miR-204-5p mimic and NC mimic were purchased from Sango (Shanghai, China). Kyse30 cells were seeded onto 24-well plates (5×105 cells/well) and co-transfected with luciferase reporter vectors (0.125 μg) and miR-204-5p mimic (50 nM) or mimic negative control (50 nM) using Lipofectamine 2000 (Invitrogen).. Briefly, luciferase reporter vectors, miR-204-5p mimic or mimic negative control were added into 250 μL Opti-MEM (A solution), and 4 μL Lipofectamine 2000 was incubated with 250 μL Opti-MEM (B solution) for 5 min at room temperature (RT). Next, A solution was mixed with B solution and added into the well of 24-well plates after 20 min at RT. Then Opti-MEM was replaced with DMEM after 4 h. Finally, luciferase activity was measured by the Luciferase Assay System (Promega, Madison, WI, USA) in a bioluminescence detector (GloMax, Promega) and were normalized by the Renilla luciferase activity according to the manufacturer's protocol.
Colony formation assay
Cell resuscitation and culture of stably transfected KYSE30 cells was performed according to the routine protocol. After trypsinization, the cells were diluted to 1×103 cell/ml and inoculated on the well plate. The cells were then cultured 10 days. Next, cells were fixed by 4% paraformaldehyde solution (PFA, Invitrogen) for 1 h at RT and stained using crystal violet at RT for 30 min. Finally, the number of crystal violet-dyed colonies was calculated. The Enzyme-linked Spot Image Automatic Analyzer (AID, Germany) was used to scan and analyze the result.
Flow cytometric analysis
Apoptosis of the stably transfected cells was evaluated by flow cytometry (BD, Bioscience, U.S.A.) using an Annexin V-FITC Apoptosis Detection Kit (KGA106, Keygen, Jiangsu, China). CellQuest software was used to analyze the results.
TUNEL analysis
Cells were seeded onto a slide and fixed at 4℃ for 25 minutes. The slides were washed with phosphate buffer saline (PBS) twice for 5 minutes, and then treated with 70% ethanol at -20℃ overnight. Next, slides were incubated with equipment buffer for 10 min at RT. Subsequently, the slides were incubated with terminal transferase (TDT) working solution followed with 4', 6-diamidino-2-phenylindole (G3250, Promega) according to standard protocols. The slides were then observed by fluorescence microscopy (MDI6000B, Leica, Germany). The nuclei of apoptotic cells exhibited green fluorescence, and the nuclei of all cells exhibited blue fluorescence.
Nude mice xenograft tumor model
Six-week-old male nude mice were adaptively fed for 3 days. Then 15 mice were randomly divided into 5 groups (3 mice per group). Next, mice were treated with subcutaneous injection of 2×106 wild type (WT) and stable transfected KYSE 30 or KYSE450 cells 3 times as follow: 1) WT cells (NC group); 2) blank vector stable cells (Blank vec group); 3) miR-204-5p stable expression cells (miR-204-5p OE group); 4) Nestin stable expression cells (Nestin OE group); 5) miR-204-5p plus nestin stable expression cells (miR-204-5p OE+Nestin OE group).
After subcutaneous injection, tumor volume in each mouse was measured at day 4, 8, 12, 16, and 20. Then the mice were killed by cervical dislocation at day 21 after injection, and tumor size and tumor weight were measured. All animal experiments were conducted according to established guidelines for the use and care of laboratory animals, and the experiments were approved by the Beijing Deconnor Biotechnology Co.,LTD.
Statistical analysis
All quantitative data were present as mean ± standard deviation (SD). SPSS software version 22 (IBM SPSS Inc. Chicago, IL, USA) was used to conduct the statistical analyses. Briefly, the comparation between two groups was detected using the unpaired Student's t-test. Values of P < 0.05 were considered to be statistically significant.
Results
Identification of Nestin-related miRNAs
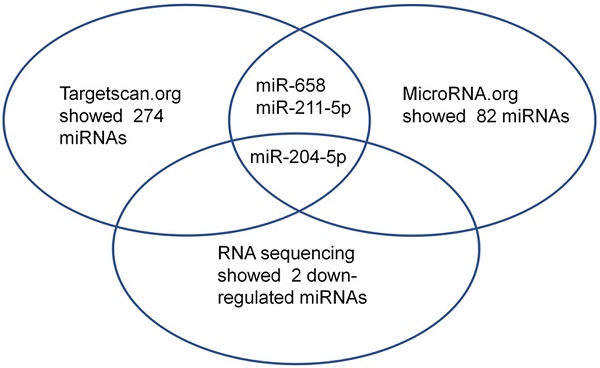
First, small RNA libraries for paracarcinoma tissues (Normal) and ESCC tissues (Tumor) were constructed and sequenced to reveal Nestin-related miRNAs. Potential target miRNAs related to Nestin in ESCC tissues were screened with a BGISEQ500 sequencer (BGI-Shenzhen). Next, miRNAs with a fold change ≥2 and P value <0.05 in a comparison between Normal group and Tumor group were considered as significant differently expressed miRNAs. Among these differently expressed miRNAs, the candidate Nestin-related miRNAs were explored by TargetScan website and the microRNA.org website based on the sequences. The results indicated there were 19 Nestin-related miRNAs that were significantly differentially expressed between ESCC tissues and adjacent para-carcinoma tissues (Table 1). Among them, there were 2 downregulated miRNAs in ESCC tissues, including miR-204-5p and novel-miR-36 (Table 1 and Figure 1). Moreover, results screening from 2 websites (TargetScan website and the microRNA.org website) based on the free energy and score value criteria identified 3 candidate miRNAs: miR-658, miR-211-5p, and miR-204-5p (Figure 1). Taken together, miR-204-5p was the most potential target miRNA of Nestin.
In addition, previous studies have indicated that miR-204 suppresses cell proliferation, apoptosis, invasion and epithelial-mesenchymal transition (EMT) in ESCC [25-27]. Based on these studies, miR-204-5p was selected for this study.
Among the microRNAs screened by Http://www.targetscan.org or Http://www.mircroRNA.org and RNA sequencing of ESCC, one candidate microRNA was overlapped.
Low miR-204-5p expression is inversely correlated with Nestin expression in ESCC tissues and cell lines
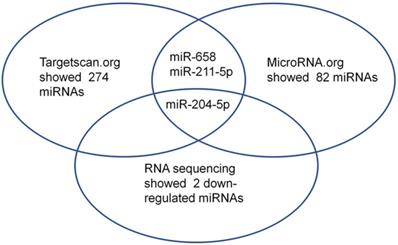
Nestin mRNA expression exhibited a reverse trend with miR-204-5p expression in the comparison between ESCC and para-carcinoma tissues, as well as between ESCC cells and esophageal epithelial cells. In esophageal squamous cancer tissues and cell lines, Nestin mRNA expression was increased, while miR-204-5p expression was decreased; in esophageal epithelial cells and para-carcinoma tissues, Nestin mRNA expression was decreased, while miR-204-5p expression was increased, and the differences were statistically significant (Figure 2). Western blotting indicated that the level of Nestin protein in esophageal squamous cancerous cells and ESCC tissues was higher than in esophageal epithelial cells and para-carcinoma tissues (Figure 3).
MiR-204-5p directly targets Nestin in ESCC
The Luciferase reporter assay was performed to investigate if miR-204-5p directly targeted Nestin in ESCC using KYSE30 cells (Figure 4). Results showed that the luciferase activity of KYSE30 cells transfected with pmirGLO-Nestin WT and miR-204-5p mimic (mimic+WT group) was dramatically decreased to that of cells transfected with blank pmirGLO and miR-204-5p mimic (mimic+basic group) (Figure 4). Besides, transfection of mimic NC had no effect on the luciferase activity of KYSE30 cells (NC+basic group and NC+WT group) (Figure 4).
Results of related Nestin's target microRNAs from genetic detection in ESCC tissues.
| miRNA | Symbol | Read count (NORMAL) | Read count (TUMOR) | log2 Ratio (TUMOR/NORMAL) | Up down regulation |
|---|---|---|---|---|---|
| hsa-miR-204-5p | NES | 54471 | 3712 | -3.689 | DOWN |
| has-novel-miR-36 | NES | 82 | 0 | -7.172 | DOWN |
| hsa-miR-4443 | NES | 43 | 76 | 1.008 | UP |
| hsa-miR-17-3p | NES | 531 | 953 | 1.030 | UP |
| hsa-miR-432-5p | NES | 421 | 848 | 1.197 | UP |
| hsa-miR-1306-5p | NES | 72 | 154 | 1.283 | UP |
| hsa-miR-1294 | NES | 52 | 121 | 1.404 | UP |
| hsa-miR-1268b | NES | 41 | 99 | 1.458 | UP |
| hsa-miR-4446-3p | NES | 29 | 84 | 1.720 | UP |
| hsa-miR-4516 | NES | 111 | 328 | 1.750 | UP |
| hsa-miR-1293 | NES | 5 | 21 | 2.256 | UP |
| hsa-miR-6720-5p | NES | 11 | 61 | 2.657 | UP |
| hsa-miR-940 | NES | 60 | 357 | 2.759 | UP |
| hsa-miR-1268a | NES | 386 | 2309 | 2.767 | UP |
| hsa-miR-345-5p | NES | 8385 | 62544 | 3.085 | UP |
| hsa-miR-544b | NES | 0 | 10 | 4.508 | UP |
| hsa-miR-509-5p | NES | 1 | 26 | 4.886 | UP |
| hsa-miR-1273h-5p | NES | 0 | 14 | 4.993 | UP |
| hsa-miR-3194-5p | NES | 0 | 19 | 5.434 | UP |
The read count and log2 Ratio of normal and tumor was calculated for the difference between paracancerous and cancerous tissues of ESCC through RNA sequencing. Abbreviation: MiRNA: microRNA; NES: Nestin.
Expression of miR-204-5p and Nestin mRNA in ESCC tissues and para-carcinoma tissues, as well as ESCC cell lines and esophageal epithelial cell lines. A and B: The expression levels of miR-204-5p and Nestin mRNA in ESCC tissues and para-carcinoma tissues were determined by qPCR. C and D: The expression levels of miR-204-5p and Nestin mRNA in ESCC cells and esophageal epithelial cells were determined by qPCR. *P<0.05, **P<0.01 or ***P<0.001 compared to control group (HEEC group or para-carcinoma group). P values were not corrected for multiple testing.
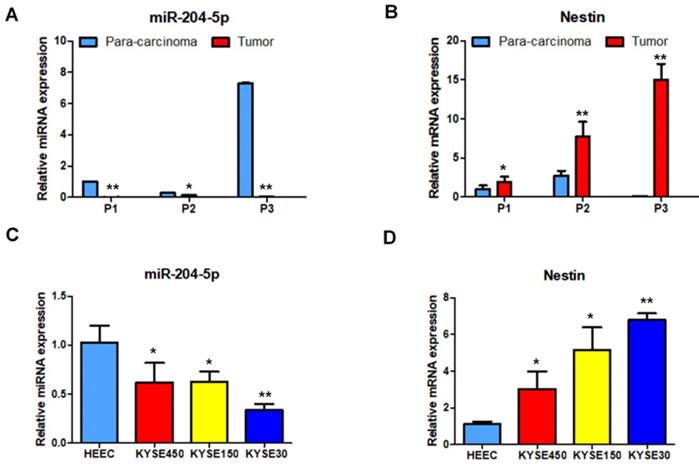
Protein expression of Nestin in ESCC tissues and para-carcinoma tissues, as well as ESCC cells lines and esophageal epithelial cell lines. A: The protein level of Nestin in ESCC tissues and para-carcinoma tissues was determined by Western blotting. B: The protein level of Nestin in ESCC cells and esophageal epithelial cells was determined by western blot. *P<0.05, **P<0.01 or ***P<0.001 compared to control group (HEEC group or para-carcinoma group). P values were not corrected for multiple testing.
Luciferase activity of different groups of cotransfected plasmid cells with different plasmid cotransfected combination (pmirGLO, pmirGLO-Nestin WT, pmirGLO-Nestin mut1, pmirGLO-Nestin mut2, pmirGLO-Nestin mut1+2). A: Schematic representation of the nestin mRNA depicting miR-204-5p binding sites in its 3'-UTR. B: The basic group was cotransfected with pmirGLO and miR-204-5p mimic (mimic+basic group) or mimic NC ((NC+basic group). Nestin-WT (wild type) group was cotransfected with pmirGLO-nestin WT and microRNA-204-5p mimic (mimic+WT group) or NC (NC+WT group). Nestin-Mut1 group was cotransfected with pmirGLO-nestin mut1 (mutant1) with miR-204-5p mimic (mimic+mutant1 group) or NC (NC+mutant1 group). Nestin-mut2 group was cotransfected with pmirGLO-nestin mut2 and miR-204-5p mimic (mimic+mutant2 group) or NC (NC+mutant2 group). Nestin-Mut1+2 was with cotransfected with pmirGLO-nestin mut1+2 and miR-204-5p mimic (mimic+nestin-Mut1+2 group) and NC (NC+nestin-Mut1+2 group). *P<0.05, **P<0.01 or ***P<0.001 compared to NC group. P values were not corrected for multiple testing.
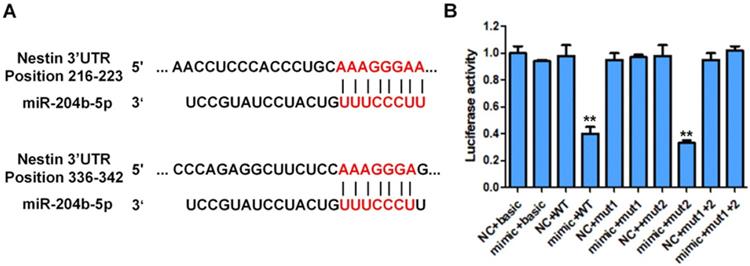
Besides, the luciferase activity of KYSE30 cells transfected with pmirGLO containing the mutated binding site1 or site2 of miR-204-5p on 3' UTR of Nestin mRNA was not affected by the transfection of miR-204-5p mimic (mimic+mutant1 group, mimic+mutant2 group, mimic+ Mut1+Mut2 group) (Figure 4). These results suggested that Nestin was the target of miR-204-5p and miR-204-5p negatively regulated Nestin expression in ESCC.
MiR-204-5p and Nestin expression in stably transfected cells and their relations
Results of qPCR indicated that miR-204-5p and Nestin were stably expressed in KYSE30 and KYSE450 cells (Figure 5).
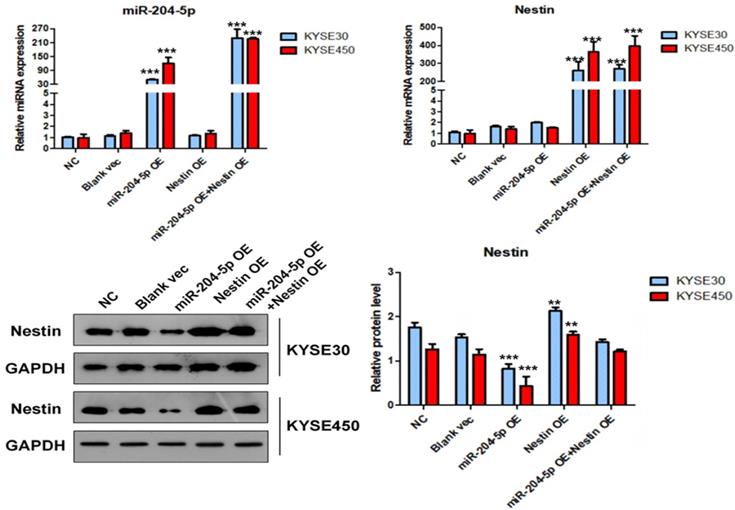
In the miR-204-5p overexpression group, the expression of miRNA-204-5p was increased, while the expression of Nestin mRNA was not affected. In the miR-204-5p overexpression +Nestin overexpression group, miR-204-5p expression was increased, but Nestin mRNA expression was not significantly reduced, indicating that miR-204-5p could not inhibit Nestin mRNA expression. Usually, miRNA inters with target mRNA to suppress the translation of mRNA but not degrade mRNA [28]. Thus, Western blotting was utilized to identify the effect of miR-204-5p on Nestin protein.
Western blotting results indicated that the level of Nestin protein was significantly reduced in the miR-204-5p OE group but was relatively higher in the miR-204-5p overexpression +Nestin overexpression group. This result suggests that miR-204-5p inhibits Nestin protein expression in KYSE30 and KYSE450 cells, and the Nestin overexpression reverses miR-204-5p inhibition.
MiR-204-5p inhibits ESCC cell proliferation by targeting Nestin in vitro
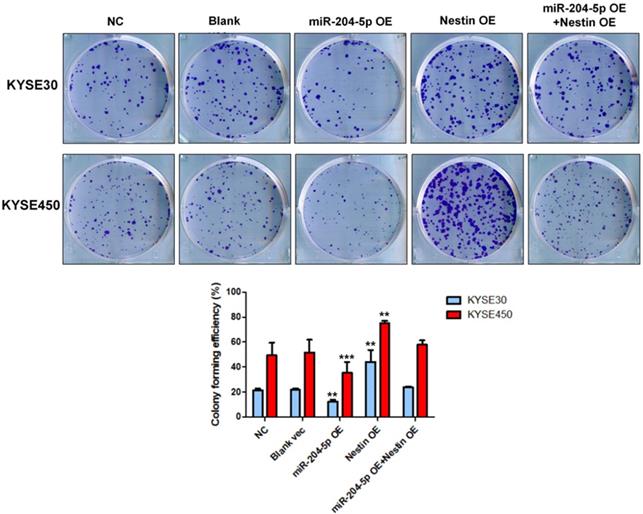
Results of colony formation assay indicated that the number of cell clones was significantly increased in Nestin overexpression group, while that was dramatically reduced in the miR-204-5p overexpression group compared with negative control group, vector control cells, and the miR-204-5p overexpression +Nestin overexpression group (Figure 6). These results suggested that miR-204-5p inhibited the clone formation ability of KYSE30 and KYSE450 cells, while Nestin overexpression reversed miR-204-5p inhibition.
MiR-204-5p induces cell apoptosis of ESCC by targeting Nestin in vitro
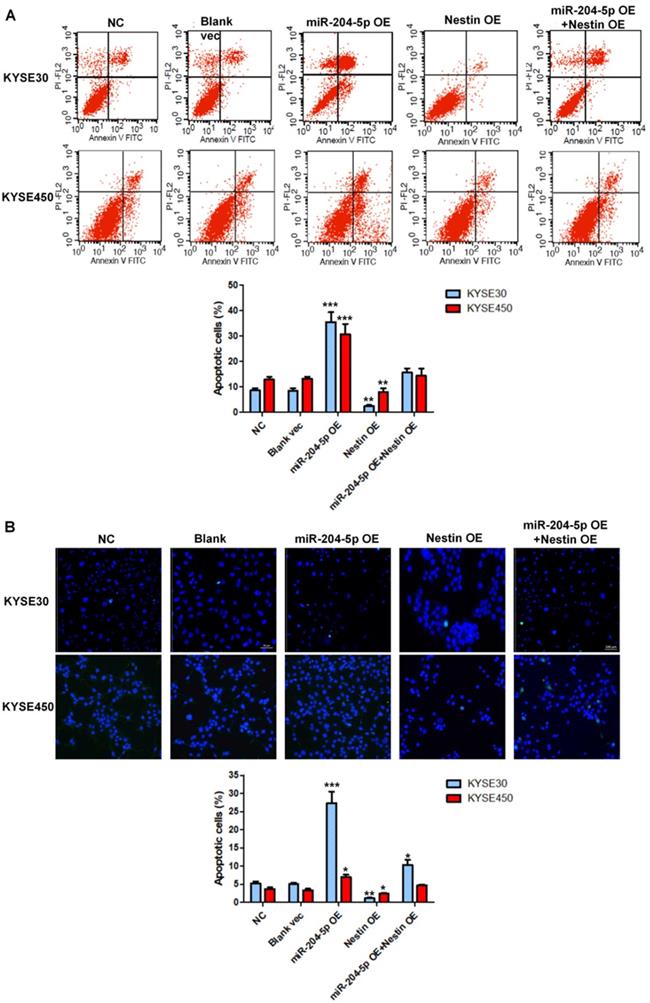
Cell apoptosis in stably transfected KYSE30 cells and KYSE450 cells was analyzed by flow cytometry. Results showed that miR-204-5p overexpression significantly induced apoptosis of KYSE30 cells and KYSE450 cells whereas Nestin overexpression suppressed cell death of KYSE30 cells and KYSE450 cells (Figure 7A). Consistent with the results of flow cytometry, the number of TUNEL-positive cells was highest in the miR-204-5p overexpression group while that was lowest in Nestin overexpression group (Figure 7B). Moreover, Nestin overexpression could abolish the effect of miR-204-5p overexpression on cell apoptosis (Figure 7A and B). These results suggested that miR-204-5p could induce apoptosis in KYSE30 and KYSE450 cells through regulating Nestin.
The expression level of miR-204-5p, Nestin mRNA and protein in KYSE30 and KYSE450 stably transfected cells, including NC group, Blank vec group, miR-204-5p OE group, Nestin OE group and miR-204-5p OE+Nestin OE group. A and B: The expression levels of miR-204-5p and Nestin mRNA in stably transfected cells were explored using qPCR. C: The protein levels of Nestin in stably transfected cells were determined by Western blotting, *P<0.05, **P<0.01 or ***P<0.001 compared to NC group. P values were not corrected for multiple testing.
Colony formation assay was performed in KYSE30 and KYSE450 stably transfected cells, including NC group, Blank vec group, miR-204-5p OE group, Nestin OE group and miR-204-5p OE +Nestin OE group. *P<0.05, **P<0.01 or ***P<0.001 compared to NC group. P values were not corrected for multiple testing.
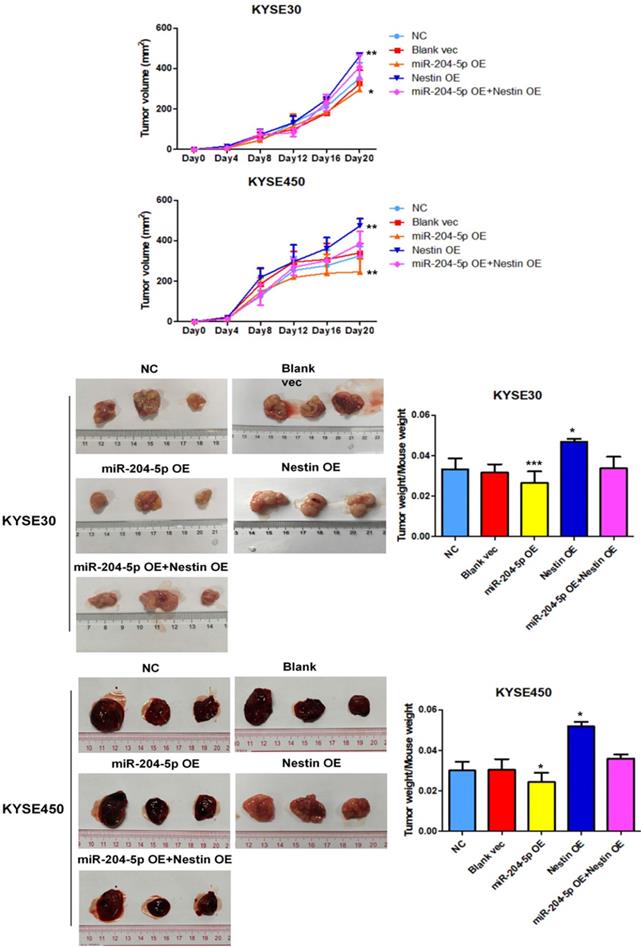
MiR-204-5p inhibits xenograft tumor growth in vivo
A murine xenograft tumor model was used to assess the effect of miR-204-5p on tumor growth. Tumor volume was measured at set time points after injection with stably transfected KYSE30 and KYSE450 cells. Of the 5 groups, tumor volume was the smallest in the miR-204-5p overexpression group while that was largest in the Nestin overexpression group (Figure 8A). The mice were killed on day 21 after injection, and then tumor size and weight were measured. Of the 5 groups, the weight of the group injected with miR-204-5p overexpression stable cells was the lightest while that was heaviest in the group injected with Nestin overexpression stable cells (Figure 8B). Furthermore, Nestin overexpression reversed the effect of miR-204-5p overexpression on suppressing the tumor growth (Figure 8A and B). These results suggest that xenograft tumor growth is inhibited by miR-204-5p in vivo, and the inhibition is attenuated by Nestin overexpression.
Discussion
ESCC is a common and prevalent malignant tumor of digestive system worldwide, especially in China. The disease is associated with a poor prognosis and high mortality, and thus researches are focused on new and novel treatments. Targeted therapies have been shown to be promising for many cancers, such as lung cancer and breast cancer and so on. It is implied that studies of the pathogenic mechanisms of esophageal cancer may lead to the development of novel targeted treatments.
Nestin is originally found to be expressed in progenitor cells of the nervous system, and subsequently its expression has been found to be elevated in several different malignant tumors. Our prior study showed that Nestin is expressed in ESCC, and 32 of 93 cases of ESCC patients (34.4%) are positive for Nestin expression [19]. This finding is similar with that of Shinichiro et al who reported that Nestin is expressed in 35.5% of lung cancers. At the same time, Nestin is also detected in Eca-109 and TE-1 cells.[29]
Our prior study has confirmed the association between Nestin and ESCC, and the results of that study lead us to 3 conclusions: 1) Nestin can be expressed in ESCC tissues and cell lines; 2) Nestin-positive ESCC is associated with a poor prognosis; 3) A positive correlation between Nestin phenotype and tumor cell proliferation indexes (Ki67 and PCNA) indicated that Nestin may be involved in the malignant proliferation of ESCC. However, only one study has revealed that Nestin positively contributes to cell proliferation and poor prognosis in ESCC [19]. Thus, the current study was performed to explore the mechanism how Nestin regulates the proliferation of ESCC cells.
Cell apoptosis in the KYSE30 and KYSE450 stably transfected cells were analyzed by flow cytometry (A) and TUNEL assay (B). *P<0.05, **P<0.01 or ***P<0.001 compared to NC group. P values were not corrected for multiple testing.
The effect of miR-204-5p on tumor growth was assessed by a nude mice xenograft tumor model. A: Growth curve of tumor volumes were calculated. Data were shown as mean ± SD. B: Photographs of tumors size and weight obtained from the different groups of nude mice. *P<0.05, **P<0.01 or ***P<0.001 compared to NC group. P values were not corrected for multiple testing.
In recent years, several studies have revealed the associations between miRNAs and esophageal carcinoma. For example, it has been reported that miR-186 regulates cell proliferation and apoptosis in human ESCC by targeting SKP2 [30]. MiR-134 suppresses the development of ESCC through blocking MAPK pathway via targeting PLXNA1.[31] Moreover, circular RNA ciRS-7 enhances the growth and metastasis of ESCC through inhibiting the effect of miR-7 on HOXB13 [32]. To identify Nestin-related miRNAs in ESCC, differently expressed miRNAs in ESCC tissues compared with paracarcinoma tissues were identified by small RNA sequencing. Paracarcinoma tissues are normal tissues and serve as controls for tumor tissues to reveal differently expressed genes in tumor tissues. Subsequently, Nestin-related miRNAs identified by small RNA sequencing were compared with results screening from 2 websites (TargetScan website and the microRNA.org website) to further demonstrate the potential Nestin-related miRNAs. Results found that the number of Nestin-related miRNAs predicted by the two online resources was different (TargetScan website showed 274 miRNAs while microRNA.org website showed 82 miRNAs), which might be due to the different rules utilized by different online resources to predict the interaction between target mRNA and miRNA. Thus, these data suggested that the prediction of miRNA-mRNA interaction should be based on overlapped information obtained from different resources.
To date, the role of miR-204-5p in the development of esophageal cancer is still not clear. More and more researches reveal that miR-204-5p plays an important role in many diseases, especially in malignant tumors. Wang et al. investigated the expression and functional roles of miR-204-5p in OSCC [33]. They found that miR-204-5p could enhance OSCC cell proliferation and metastasis by targeting CXCR4. In addition, some studies have shown that miR-204-5p is regulated by long noncoding RNA (lncRNA). Yu et al. reported that LncRNA TUG1 positively regulated the expression via sponging miR-204-5p to promote osteogenic differentiation in calcific aortic valve disease (CAVD) [34]. Recently, the study of miR-204-5p in ESCC has also been reported. Tang et al. investigated the potential role of miR-204-5p in ESCC. They found that miR-204-5p functioned by directly targeting IL-11 [26]. However, the molecular mechanism and signal transduction mechanism of miR-204-5p in ESCC are still unknown. In the current study, we demonstrated for the first time that miR-204-5p regulates ESCC via targeting Nestin.
Our results suggest that miR-204-5p targets the Nestin 3'UTR, and the target recognition element of RISC bound to the 3'UTR of targeted mRNA. Because miRNA-204-5p is not tightly bound to Nestin, it only inhibits Nestin protein translation, but does not degrade Nestin mRNA, resulting in down-regulation of protein expression. As an upstream regulator of Nestin, miR-204-5p may inhibit cancer cell proliferation and induce cancer cell apoptosis in esophageal squamous cancer cells.
Conclusions
The results of this study showed that miR-204-5p may target Nestin mRNA, and miR-204-5p could inhibit the proliferation and induced apoptosis of ESCC cells by regulating Nestin. These results suggested that miR-204-5p maybe a key regulator of Nestin in ESCC and targeting miR-204-5p might be a novel treatment for ESCC.
Abbreviations
EC: esophageal cancer; EAC: esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma; RISC: RNA-induced silencing complex; UTR: untranslated region; HEEC: human esophageal epithelial cell lines; qPCR: quantitative real-time polymerase chain reaction; RT-PCR: reverse transcription polymerase chain reaction; GFP: Green fluorescent protein; SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis; PVDF: polyvinylidene fluoride; GAPDH: glyceraldehydes-3-phosphate dehydrogenase; TUNEL: terminal deoxynucleotidyl transferase mediated nick end labeling; PBS: phosphate buffer saline; NC: negative control; PCNA: proliferating cell nuclear antigen; CDK5: cyclin dependent kinase-5.
Acknowledgements
This study was supported by grant from the Medical Science and Technology Research Fund Project of Guangdong Province (No. 201619211359549).
Author Contributions
BLZ, HHL, and FFZ conceived the study. BLZ, HHL, FFZ, and WZL participated in the analysis of ESCC specimens and cell lines. BLZ, WZL, and HYZ performed qPCR, Western blotting and constructed stably transfected cell lines. BLZ, HYZ, CXL, and FL conducted the cell proliferation assay and apoptosis test. BLZ, HHL, and FFZ performed statistical analysis of all data. BLZ and HHL designed the experiments, coordinated the study, and drafted the manuscript. All authors have read and approved the final version of the manuscript.
Data Availability Statement
The datasets used during this study are available from the corresponding author on request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y. et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:1915-22
2. Alder H, Taccioli C, Chen H, Jiang Y, Smalley KJ, Fadda P. et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 2012;33:1736-44
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
4. Kawamata T, Tomari Y. Making RISC. Trends in biochemical sciences. 2010;35:368-76
5. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54
6. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20
7. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92-105
8. Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia. 2012;26:404-13
9. Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637-41
10. Esteller M. Non-coding RNAs in human disease. Nature reviews Genetics. 2011;12:861-74
11. Teng GG, Wang WH, Dai Y, Wang SJ, Chu YX, Li J. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PloS one. 2013;8:e56709
12. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M. et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980-4
13. Cacchiarelli D, Incitti T, Martone J, Cesana M, Cazzella V, Santini T. et al. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO reports. 2011;12:136-41
14. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6:259-69
15. Guerette D, Khan PA, Savard PE, Vincent M. Molecular evolution of type VI intermediate filament proteins. BMC evolutionary biology. 2007;7:164
16. Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267-73
17. Bentivegna A, Conconi D, Panzeri E, Sala E, Bovo G, Vigano P. et al. Biological heterogeneity of putative bladder cancer stem-like cell populations from human bladder transitional cell carcinoma samples. Cancer science. 2010;101:416-24
18. Jimeno A, Feldmann G, Suarez-Gauthier A, Rasheed Z, Solomon A, Zou GM. et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Molecular cancer therapeutics. 2009;8:310-4
19. Zhong B, Wang T, Lun X, Zhang J, Zheng S, Yang W. et al. Contribution of nestin positive esophageal squamous cancer cells on malignant proliferation, apoptosis, and poor prognosis. Cancer cell international. 2014;14:57
20. Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nature methods. 2011;9:72-4
21. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics (Oxford, England). 2010;26:136-8
22. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015 4
23. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149-53
24. Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagnostic molecular pathology: the American journal of surgical pathology, part B. 2006;15:56-61
25. Sun Y, Yu X, Bai Q. miR-204 inhibits invasion and epithelial-mesenchymal transition by targeting FOXM1 in esophageal cancer. International journal of clinical and experimental pathology. 2015;8:12775-83
26. Tang J, Li Z, Zhu Q, Wen W, Wang J, Xu J. et al. miR-204-5p regulates cell proliferation, invasion, and apoptosis by targeting IL-11 in esophageal squamous cell carcinoma. Journal of cellular physiology. 2020;235:3043-55
27. Shen Z, Chai T, Luo F, Liu Z, Xu H, Zhang P. et al. Loss of miR-204-5p Promotes Tumor Proliferation, Migration, and Invasion Through Targeting YWHAZ/PI3K/AKT Pathway in Esophageal Squamous Cell Carcinoma. OncoTargets and therapy. 2020;13:4679-90
28. Ni WJ, Leng XM. miRNA-Dependent Activation of mRNA Translation. Microrna. 2016;5:83-6
29. Ryuge S, Sato Y, Wang GQ, Matsumoto T, Jiang SX, Katono K. et al. Prognostic significance of nestin expression in resected non-small cell lung cancer. Chest. 2011;139:862-9
30. He W, Feng J, Zhang Y, Wang Y, Zang W, Zhao G. microRNA-186 inhibits cell proliferation and induces apoptosis in human esophageal squamous cell carcinoma by targeting SKP2. Laboratory investigation; a journal of technical methods and pathology. 2016;96:317-24
31. Wang WW, Zhao ZH, Wang L, Li P, Chen KS, Zhang JY. et al. MicroRNA-134 prevents the progression of esophageal squamous cell carcinoma via the PLXNA1-mediated MAPK signalling pathway. EBioMedicine. 2019;46:66-78
32. Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG. et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell death & disease. 2018;9:838
33. Wang X, Li F, Zhou X. miR-204-5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;82:202-7
34. Yu C, Li L, Xie F, Guo S, Liu F, Dong N. et al. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovascular research. 2018;114:168-79
Author contact
![]() Corresponding author: Beilong Zhong, E-mail: zhongblsysu.edu.cn; Address 1: No.58, ZhongShan Er Lu, Guangzhou, Guangdong 510080, China; Address2: No.52, MeiHua Dong Road, XiangZhou District, ZhuHai, Guangdong 519000, China. Fangfang Zheng, E-mail: zhuhaizhengfangcom; Address: No.52, MeiHua Dong Road, XiangZhou District, ZhuHai, Guangdong 519000, China.
Corresponding author: Beilong Zhong, E-mail: zhongblsysu.edu.cn; Address 1: No.58, ZhongShan Er Lu, Guangzhou, Guangdong 510080, China; Address2: No.52, MeiHua Dong Road, XiangZhou District, ZhuHai, Guangdong 519000, China. Fangfang Zheng, E-mail: zhuhaizhengfangcom; Address: No.52, MeiHua Dong Road, XiangZhou District, ZhuHai, Guangdong 519000, China.

 Global reach, higher impact
Global reach, higher impact