Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(3):434-445. doi:10.7150/ijms.58067 This issue Cite
Research Paper
CEMIP as a potential biomarker and therapeutic target for breast cancer patients
Department of Oncology, Shengjing Hospital of China Medical University, Shenyang, Liaoning Province, 110004, China.
Received 2021-1-11; Accepted 2022-1-7; Published 2022-2-7
Abstract
Purpose: We aimed to evaluate whether CEMIP plays any role in the survival outcome of breast cancer (BC) patients, as well as to explore the regulatory mechanism of CEMIP in BC.
Methods: We evaluated the expression and prognostic effect of CEMIP in BC patients using the Oncomine, GEPIA, UALCAN, and Kaplan-Meier plotter databases. Additionally, we detected CEMIP mRNA and protein levels in BC and normal tissues via PCR and western blotting analyses. Through immunochemistry analysis, we quantified CEMIP expression in 233 samples from BC patients. We then analyzed the link between the survival outcomes and CEMIP expression based on these clinical samples. Furthermore, we explored the immune-related molecules regulated by CEMIP and its coexpressed genes using the STRING database.
Results: CEMIP expression was higher in BC tissues than in normal tissues. Patients with high CEMIP mRNA levels had a worse survival outcome. Similarly, patients expressing CEMIP had significantly shorter overall survival and disease-free survival than those not expressing the protein (P < 0.01). Some lymphocytes, immune inhibitors, immune stimulators, MHC molecules, chemokines, and chemokine receptors can be regulated by CEMIP, and CEMIP and its coexpressed genes can participate in the hyaluronan biosynthetic process, hyaluronan catabolic process, and other related biological processes in the progression of BC.
Conclusion: Compared to normal tissues, BC tissues had higher number of CEMIP transcripts. CEMIP expression was associated with an adverse prognosis. CEMIP and its coexpressed genes can participate in the progression of BC. Therefore, CEMIP may be a potential biomarker for the treatment of BC patients.
Keywords: Breast cancer, CEMIP, Metastasis, Prognosis, Biomarker
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer worldwide [1]. It is estimated that more than 3.8 million women in the United States have a history of invasive BC, with nearly 270000 women being newly diagnosed every year [2]. Although given the standardization of systemic chemotherapy as the gold-standard approach for most molecular subtypes along with rapid advances in early detection and comprehensive therapy, the number of BC patients continues to grow despite an increasing number of new diagnoses. However, numerous patients suffer from metastasis, relapse, or drug resistance, causing treatment failure [3]. These severe situations urge us to explore more advanced means of early diagnosis and treatment, such as novel therapeutic targets.
Cell migration-inducing and hyaluronan-binding protein (CEMIP), also known as TMEM2L and KIAA1199, is a 150 kDa protein with an N-terminal secretion signal peptide. It was first reported as an inner ear protein, and genetic mutations in CEMIP led to nonsyndromic hearing loss [4]. CEMIP has also been reported to depolymerize hyaluronic acid (HA) [5], and overexpression of CEMIP can lead to resistance to cell immortalization and carcinogenesis in normal human cells [6]. In recent years, its role in carcinogenesis has been observed in various cancers including gastric cancer [7-8], pancreatic cancer [9-10], hepatocellular carcinoma, colorectal cancer, and colon cancer [11-14]. Overexpression of CEMIP has also been observed in BC [15-17] and ovarian cancer [18].
However, further research on CEMIP in BC prognosis has rarely been reported, and the underlying mechanism by which CEMIP affects BC progression is still unknown. Herein, we aimed to explore CEMIP expression in the survival outcomes of BC patients and to determine the relationships between CEMIP expression and other clinicopathological characteristics in BC patients. With the help of public databases, we explored the expression of CEMIP in large sample size cohorts and then examined the expression level of CEMIP in BC tissues collected from surgery patients. These data were then analyzed to explore the clinical implications of CEMIP expression in BC.
Methods
Oncomine database
The Oncomine database (http://www.oncomine.org) was used to analyze the expression of CEMIP mRNA transcripts in different cancer types [19]. Additionally, the expression levels of CEMIP mRNA transcripts in BC and normal tissues among different research subgroups were explored. Finally, we preliminarily analyzed the genes coexpressed with CEMIP in the Ginestier Breast and Schmidt Breast studies.
GOBO database
Expression of CEMIP in the different subtypes of breast tumor and BC cell lines was further explored using the GOBO database (GOBO; http://co.bmc.lu.se/gobo/gsa.pl).
UALCAN database
The UALCAN database (ualcan.path.uab.edu) was used to analyze the levels of CEMIP mRNA transcripts in BC based on the subgroups of sample types, individual cancer stages, major subclasses [with triple-negative BC (TNBC) types], and the effect of CEMIP expression on BC patient survival [20]. In addition, the level of DNA methylation of CEMIP in BC tissues was also explored.
Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com) was used to analyze the prognostic effect of high CEMIP mRNA expression on overall survival (OS) and post-progression survival (PPS) of BC patients [21,22].
RNA extraction and qRT-PCR
Trizol (Thermo Fisher Scientific Company) was used to extract total RNA from BC and normal breast specimens. After reverse transcription, CEMIP mRNA expression was detected by SYBR Premix Taq II (Takara, RR820A, Japan). The primer sequences for CEMIP and GAPDH were: CEMIP primer sequence: forward, 5'-GGAGAGTTCCAAGCAGCA 3'; reverse, 5'-CGTCAATCACCACCACCT-3'; GAPDH primer sequence: forward, 5'-CCTTCCGTGTCCCCACT-3'; reverse, 5'-GCCTGCTTCACCACCTTC-3'.
Western blotting
MCF7, SKBR3, MDA-MB-231, and MCF10A cells were harvested using trypsinization and washed with PBS twice before lysis. Along with the 20 BC and cancer-adjacent specimens, samples were lysed with NP40 lysis and PMSF. Proteins were then separated on a 12% SDS-PAGE gel and transferred to PVDF membrane, followed by blocking with 5% fat-free dry milk for 1 h. Membranes were incubated in a shaker with rabbit monoclonal anti-human CEMIP antibody (Abcam, ab98947) overnight at 4 °C. After washing, the membranes were probed with HRP-conjugated goat anti-rabbit IgG (Zhong Shan Jin Qiao, China) for 1 h at room temperature. These bands were visualized using an enhanced chemiluminescent reagent (Thermo Fisher Scientific, USA).
BC cell lines and cell culture
The human BC cell lines MCF7, MCF-10A, SK-BR3, and MDA-MB-231 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). These BC cells were separately cultured in 10% fetal bovine serum-containing DMEM as well as McCoy's 5A and L15 media, following the manufacturers' instructions, in an atmosphere containing 5% CO2 at 37 °C, according to American Type Culture Collection (ATCC) recommendations.
Patients and BC tissue specimens
For the clinical study, we evaluated breast tissue samples (TMA sample set) from 233 patients who underwent surgery at the China Medical University Affiliated Hospital. Incidences of histologically confirmed ductal BC were recruited from 2006 to 2008. No patient exhibited distant metastases at the time of surgery. Patients with a history of other solid tumors, radiotherapy, chemotherapy, or neoadjuvant chemotherapy were excluded. Clinical information was obtained through a telephone, electronic record system, or in-person visit in outpatient settings. Fresh tumor tissue samples and adjacent normal tissues from 20 patients with breast carcinoma were obtained and stored in liquid nitrogen for western blotting. All patients signed informed consent forms. All samples were validated by expert pathologists. This protocol was approved by China Medical University Institutional Review Board [2020PS171K(X1)].
Immunohistochemistry and evaluation
All 233 BC specimens were fixed in 4% formaldehyde, embedded in paraffin, and then sectioned at 4 μm. Sections were rehydrated with a graded ethanol series after deparaffinization with xylene followed by Tris-buffered saline (TBS). Tissues were then incubated at 4 °C overnight with antibody against CEMIP (Novus, NBP2-50336UV). After that, BC sections were incubated with a secondary antibody at 37 °C for approximately 50 min. Immunohistochemical staining was performed using a DAB kit for 10 min.
Total CEMIP expression was classified semiquantitatively according to the following criteria: 0 if <1 % of tumor cells expressed CEMIP, 1+ if expression occurred in ≥1 and <5% of tumor cells, 2+ if ≥5% and <10% of tumor cells expressed CEMIP, and 3+ if ≥10% did so. The scores of 2+ and 3+ were considered to indicate CEMIP positive expression, whereas scores of 0 and 1+ were considered to indicate CEMIP negative expression.
cBioPortal database
The cBioPortal database (http://www.cbioportal.org) was utilized to analyze the genetic variations of CEMIP [23].
MEXPRESS database
The MEXPRESS database (https://mexpress.be/) was utilized to analyze DNA methylation of CEMIP in primary BC tissues and normal tissues.
TISIDB database
The TISIDB database (http://cis.hku.hk/TISIDB) was utilized to explore the correlations between CEMIP expression and immune-related molecules [24].
Gene coexpressed analysis
Genes coexpressed with CEMIP were constructed from the Coexpedia website (http://www.coexpedia.org/) and STRING database version 10.0. Thereafter, we used the STRING database to perform Gene Ontology (GO) enrichment analyses of all the selected genes.
Statistical analysis
Age, T grade, N grade, menopausal status, ER, PR, HER2, Ki67 index, and other clinicopathological characteristics among BC patients were analyzed using a Chi-square test. Survival curves were generated using the Kaplan-Meier test with the SPSS23.0 software. Disease-free survival (DFS) was defined as the interval from operation to local recurrence or distant metastasis. OS was defined as the interval from surgery to patient death. All statistical tests were two-sided. Significance was set at P <0.05, Analyses were performed using the SPSS software (version 23.0; SPSS Inc., IL, USA). The Kaplan-Meier plotter was used to evaluate how CEMIP mRNA expression influenced the survival outcome of BC patients.
Results
Analysis of differential expression of CEMIP in BC
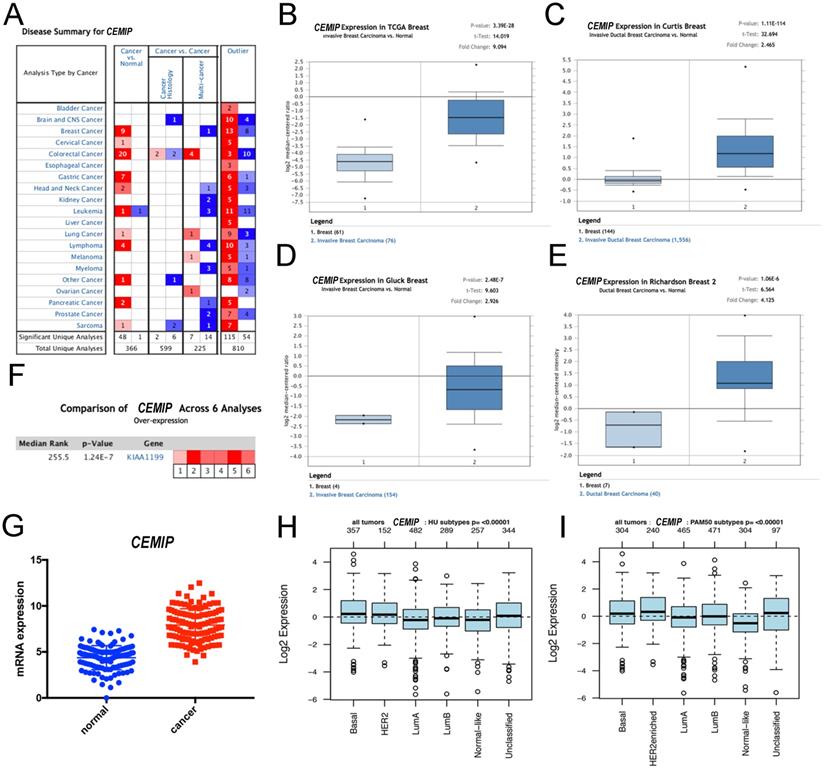
Using Oncomine, we first conducted an overall analysis of the expression of CEMIP in multiple cancers and diseases (Figure 1A). We subsequently investigated the expression of CEMIP in BC, focusing on the differential expression between cancer and normal tissues. Oncomine analysis revealed that CEMIP expression was significantly higher in cancer tissues than in normal samples. Based on the Curtis Breast Statistics dataset, CEMIP transcripts were elevated 2.465-fold in BC samples when compared with normal breast tissues (P = 1.11 × 10-14) [25] (Figure 1C) and were 2.926-fold elevated in ductal carcinoma in situ samples as compared with normal tissues (P = 2.48 × 10-7) in Gluck's study [26] (Figure 1D). In a dataset derived from Richardson's study, CEMIP levels were elevated 4.125-fold in ductal breast carcinoma compared to normal breast tissues (P = 1.06 × 10-6) [2] (Figure 1E). To obtain a more comprehensive conclusion, we conducted a meta-analysis of multiple study datasets including six datasets, TCGA Breast, Cutris Breast, Gluck Breast, Richardson Breast 2, Radvanyi Breast, and Ma Breast 4. This meta-analysis showed a significantly higher expression of CEMIP in BC (Figure 1F). Additionally, we explored CEMIP expression based on TCGA research network, which also displayed higher (9.094-fold) levels of CEMIP in cancer compared to normal breast tissues (Figure 1B,1G). GOBO analyses also validated that CEMIP has a significantly high expression in all subtypes of BC (Figure 1H-1I).
Furthermore, we explored the expression of CEMIP in BC using the UALCAN database. CEMIP mRNA levels were higher in cancer tissues than in normal tissues (Figure 2A). We also found that CEMIP mRNA expression level was significantly higher in breast cancer tissues of different individual stage than in normal tissues (Figure 2B). Out of the four BC subclasses, TNBC patients exhibited the highest CEMIP mRNA expression (Figure 2C).
Exploration the expression of CEMIP in BC tissues and normal breast tissues. A: The level of CEMIP mRNA in different cancer types gained from Oncomine database. The graph presented the number of datasets with statistically significant target genes with increased (red) or reduced expression (blue). B-E: Comparison of CEMIP mRNA expression in BC tissues and normal tissues in different study subgroups. F: Meta-analysis of multiple study datasets for comparison of CEMIP expression between BC tissues and normal breast tissues. G: The expression of CEMIP mRNA in BC tissues and normal breast tissues gained from TCGA database. H-I: The level of CEMIP mRNA expression in BC patients with different molecular subgroups in GOBO database.
Exploration of the effect of high expression of CEMIP mRNA on the survival outcomes of BC
We used the UALCAN database to explore the effect of high CEMIP mRNA expression on the prognosis of BC patients. CEMIP mRNA expression was significantly related to a shorter survival time (P = 0.005, Figure 3A). Furthermore, we explored the Kaplan-Meier plotter database; CEMIP mRNA expression was found to be significantly related to a shorter overall survival (P = 0.03, Figure 3B) and post-progression survival (P = 0.00085, Figure 3C).
Expression levels of CEMIP is associated with progressive BC malignancy
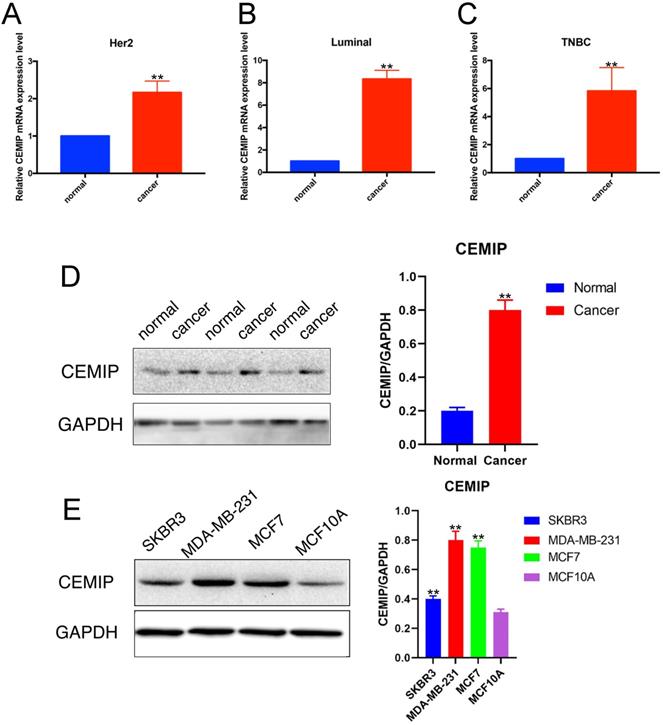
We detected the relative RNA expression of CEMIP in nine fresh paired BC samples and nontumor tissues via qRT-PCR. As shown in Figure 4A-4C, in three subtypes of BC, CEMIP expression was significantly higher in BC tissues than in nontumor tissues of BC different subtypes (P < 0.05). This result predicted that the expression of CEMIP mRNA was higher in BC tissues than in normal tissues regardless of different BC subtypes. And among different BC subtypes, there was little difference. In accordance with our PCR and western blotting analysis results for both cancer cell lines and clinical samples, CEMIP expression was found to be higher in cancer tissues than cancer-adjacent tissues (Figure 4D) and was also higher in the BC cell lines MDA-MB-231, SKBR3, and MCF7 than in the normal BC cell line MCF10A (Figure 4E). The basic clinicopathological characteristics of each cancer patients of Figure 4D were shown in Supplementary Table 3. About these four breast cell lines, MDA-MB-231 belonged to triple negative breast cancer cell lines. SKBR3 was HER2 positive breast cancer cell lines. MCF7 belonged to luminal breast cancer cell lines. MCF10A was normal BC cell line.
The correlations between CEMIP expression and clinicopathological characteristics
| Variables | CEMIP positive (%) | CEMIP negative (%) | P-value |
|---|---|---|---|
| No. of patients | 90 (38.6) | 143 (61.4) | |
| Age | 0.615 | ||
| ≤65 | 80 (88.9) | 130 (90.9) | |
| >65 | 10 (11.1) | 13 (9.1) | |
| T grade | 0.110 | ||
| 1 | 24 (26.7) | 51 (35.7) | |
| 2 | 55 (61.1) | 84 (58.7) | |
| 3 | 11 (12.2) | 8 (5.6) | |
| N grade | 0.367 | ||
| 0 | 48 (53.3) | 81 (56.6) | |
| 1 | 19 (21.1) | 38 (26.6) | |
| 2 | 7 (7.8) | 9 (6.3) | |
| 3 | 16 (17.8) | 15 (10.5) | |
| Menopausal status | 0.380 | ||
| Premenopausal | 40 (44.4) | 72 (50.3) | |
| Postmenopausal | 50 (55.6) | 71 (49.7) | |
| ER status | 0.518 | ||
| Positive | 61 (67.8) | 91 (63.6) | |
| Negative | 29 (32.2) | 52 (36.4) | |
| PR status | 0.816 | ||
| Positive | 53 (58.9) | 82 (57.3) | |
| Negative | 37 (41.1) | 61 (42.7) | |
| HER2 status | 0.316 | ||
| Positive | 30 (33.3) | 57 (39.9) | |
| Negative | 60 (66.7) | 86 (60.1) | |
| Ki67 index | 0.676 | ||
| ≤20% | 39 (43.3) | 58 (64.4) | |
| >20% | 51 (56.7) | 85 (94.4) | |
| Distant metastasis | <0.001 | ||
| Yes | 52 (57.8) | 42 (29.4) | |
| No | 38 (42.2) | 101 (70.6) | |
| Death | 0.014 | ||
| Yes | 29 (32.2) | 26 (18.2) | |
| No | 61 (67.8) | 117 (81.8) |
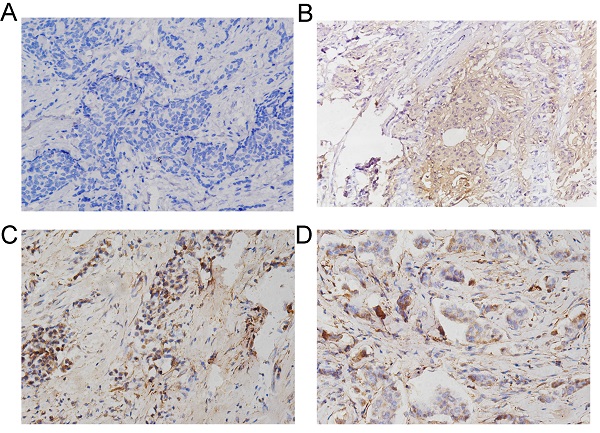
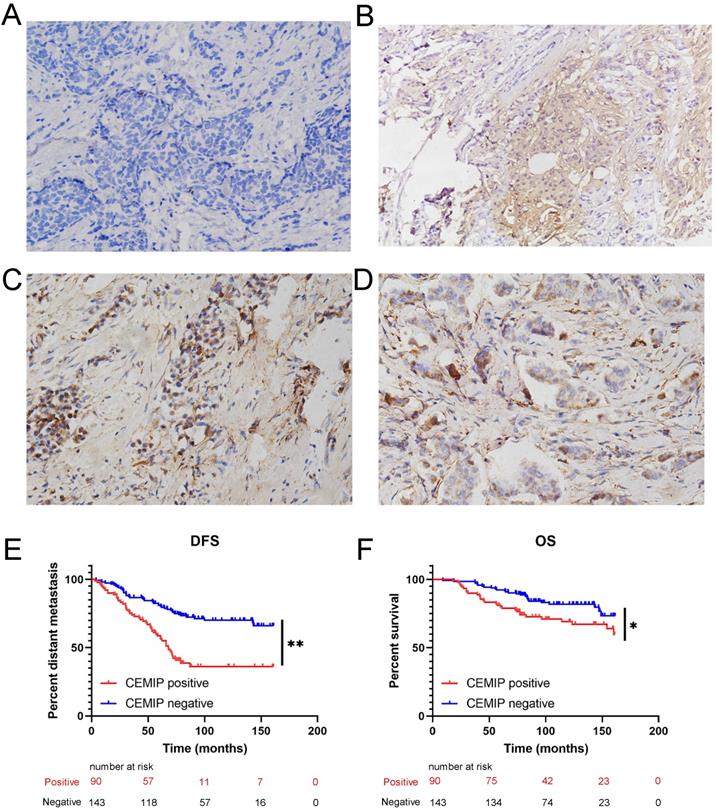
Furthermore, a total of 233 female patients who underwent BC surgery were analyzed. The correlations between CEMIP expression and clinical characteristics are shown in Table 1. CEMIP expression was significantly associated with a higher ratio of distant metastasis (P < 0.001) and death (P = 0.014). A total of 90 out of the 233 patients (38.6%) were found to be CEMIP-positive via immunohistochemistry; typical pictures of CEMIP-negative and CEMIP-positive results in luminal, HER2+ and TNBC are shown in Figure 5A-5D. We subsequently compared the clinical and histological characteristics between positive (scores 1-3) and negative CEMIP expression (score 0) groups. However, no significant differences were identified in age, menopausal status, or other characteristics as shown in Table 1.
Analysis of CEMIP mRNA expression in BC by UALCAN database. Expression of CEMIP in BC based on different sample types (A), individual cancer stages (B), and BC subclasses (C).
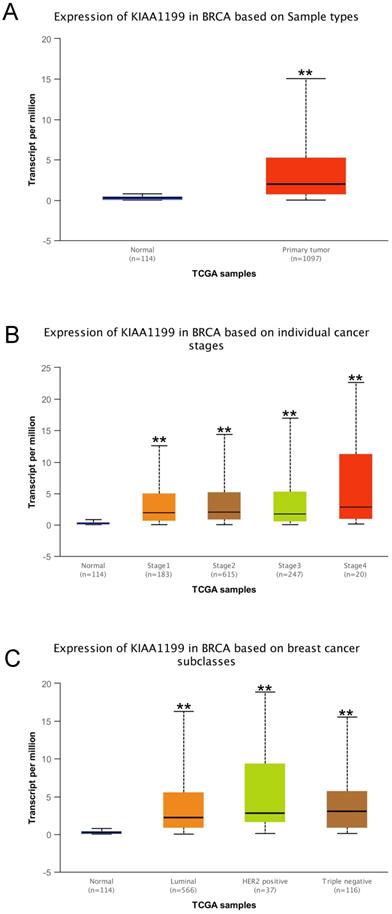
Effect of CEMIP expression on the prognosis of BC patients. A: Effect of CEMIP expression in the overall survival of BC patients in UALCAN database. B: Effect of CEMIP expression in the overall survival of BC patients in Kaplan-Meier plotter database. C: Effect of CEMIP expression in the post-progression survival of BC patients in Kaplan-Meier plotter database.
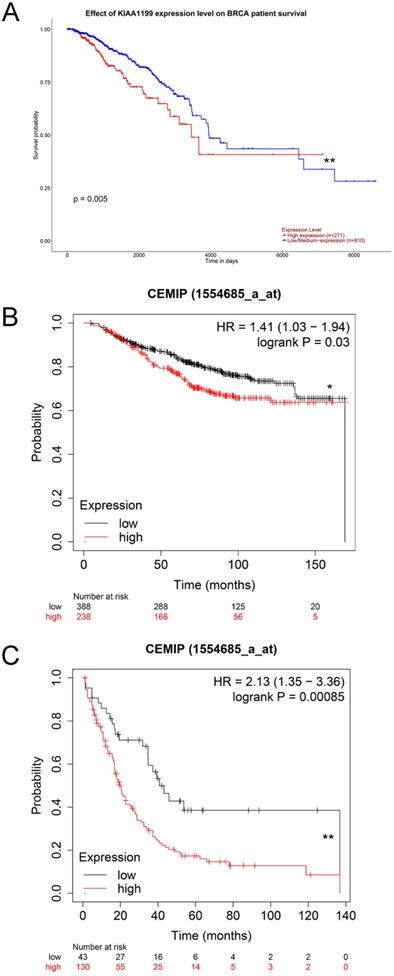
Furthermore, we evaluated the effect of CEMIP expression on the prognosis of 233 BC patients. We found that high CEMIP expression was related to a significantly shorter DFS (P < 0.001, Figure 5E) and OS (P = 0.016, Figure 5F).
Genetic variations of CEMIP gene in BC
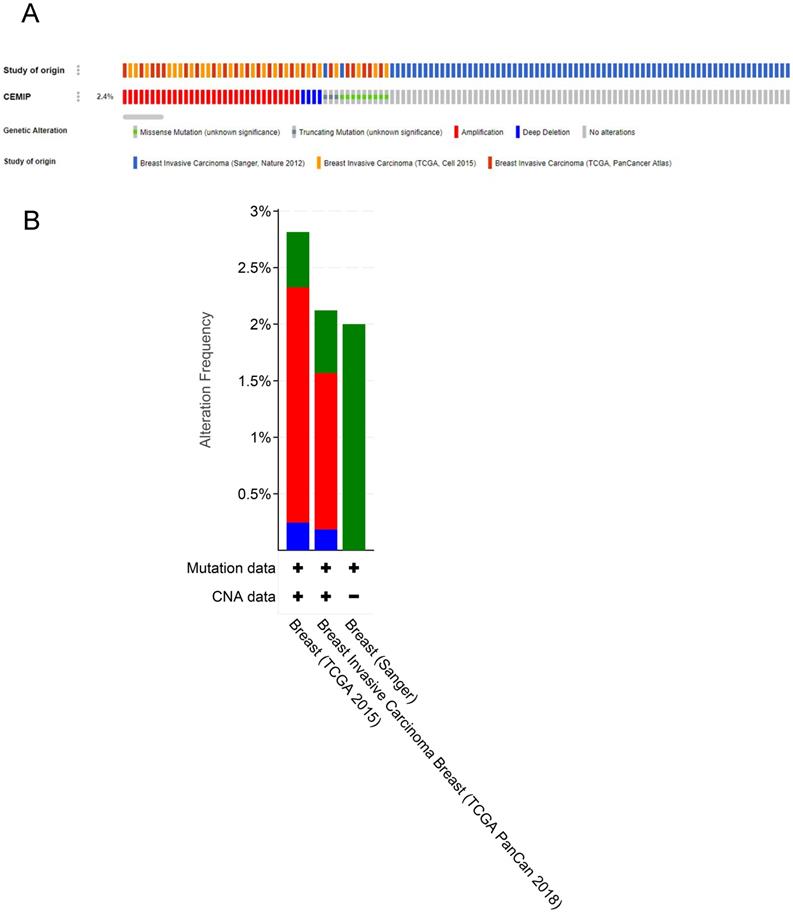
Genetic variations of CEMIP in three studies (Sanger, Nature 2012; TCGA, Cell 2015; and TCGA, PanCancer Atlas) were analyzed using the cBioPortal database (Figure 6A). In these cases from TCGA, Cell 2015, the ratio of CEMIP mutation, amplification, and deep deletion was 2.8%, whereas in these cases from TCGA, PanCancer Atlas, the ratio of CEMIP mutation, amplification, and deep deletion was 2.1%. In these cases from Sanger, Nature 2012, the ratio of CEMIP mutations was 2% (Figure 6B).
Effect DNA methylation on CEMIP mRNA expression
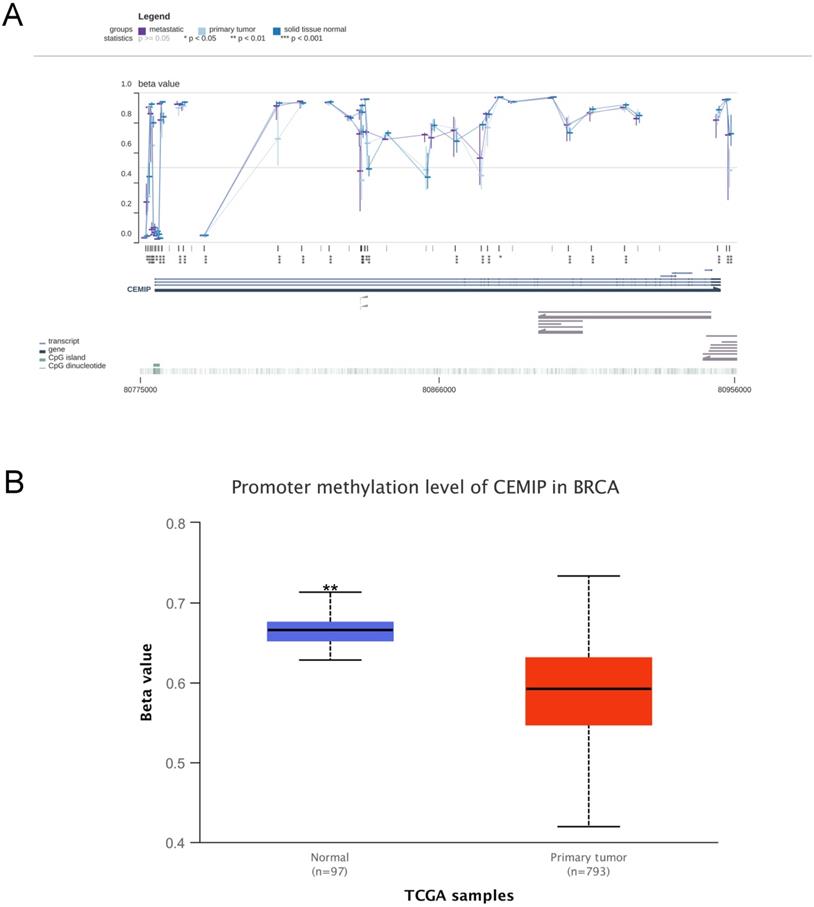
In the MEXPRESS and UALCAN databases, the level of DNA methylation in CEMIP was higher in normal tissues than in cancer tissues (Figure 7A-7B). This result was consistent with the theory that DNA methylation is negatively related to mRNA expression [27]. This result led to the prediction that CEMIP mRNA expression is regulated by DNA promoter methylation in BC.
Regulation of immune-related molecules by CEMIP in BC
The immune-related molecules, including lymphocytes, immune inhibitors, immune stimulators, MHC molecules, chemokines, and chemokine receptors, which were regulated by CEMIP in BC, were further explored by using the TISIDB database.
Supplementary Figure 1A shows the relationship between CEMIP expression and tumor-infiltrating lymphocytes. The tumor-infiltrating lymphocytes most closely related with CEMIP expression in BC were CCL7, CCL14, CCL20, and CXCL14 (Supplementary Figure 1B). Supplementary Figure 1C shows the relationship between CEMIP expression and immune inhibitors. The immune inhibitors most closely related with CEMIP expression in BC were CD160, HAVCR2, PDCD1LG2, and TGFBR1 (Supplementary Figure 1D). Supplementary Figure 1E shows the relationship between CEMIP expression and immune stimulators. The immune stimulators most closely related with CEMIP expression in BC were CD276, TNFSF4, NT5E, and ULBP1 (Supplementary Figure 1F). Supplementary figure 1G shows the relationships between CEMIP expression and MHC molecules. The MHC molecules most closely related with CEMIP expression in BC were HLA-DMB, HLA-DOB, HLA-DOA1, and TAPBP (Supplementary Figure 1H).
Detection of CEMIP expression in BC samples and cell lines. A-C: Detection of CEMIP mRNA expression in BC samples of different molecular subgroup. D: Detection of CEMIP expression in BC tissues and normal breast tissues specimens. E: Detection of CEMIP expression in different BC cell lines.
Supplementary Figure 2A shows the relationship between CEMIP expression and chemokines. The chemokines most closely related with CEMIP expression in BC were CCL1, CCL19, CXCL18, and CXCL12 (Supplementary Figure 2B). Supplementary Figure 2C shows the relationships between CEMIP expression and chemokine receptors. The chemokine receptors most closely related with CEMIP expression in BC were CCR1, CCR4, CCR7, and CCR8 (Supplementary Figure 2D).
Exploration of CEMIP molecular functions and regulation mechanism
In the Oncomine coexpression analysis derived from Ginestier's research, CEMIP expression was significantly correlated with SULF1 (r = 0.811) and PLAU (r = 0.830) [28] (Supplementary Figure 3A); Schmidt's research showed the same result for PLAU (r = 0.681) and SULF1 (r = 0.622) (Supplementary Figure 3B) [29]. Lu's study shows that SULF1 has a poor survival outcome in ER-positive BC in the Chinese population [30], and PLAU is a key pathway protein in aggressive BC [31]. Therefore, as a coexpression protein of SULF1 and PLAU, we speculate that CEMIP also plays an important role in TNBC. Using the bioinformatic databases, we selected several neighboring genes that were related to CEMIP from Coexpedia to explore the potential molecular mechanisms of the role of CEMIP in cancer and other diseases (Supplementary Figure 3C). The results show that PLAU is the most relevant protein with regard to CEMIP, which is consistent with our finding that CEMIP is a novel biomarker in BC.
Validation the prognostic effect of CEMIP expression in clinical BC specimens. A: Representative negative CEMIP immunohistochemical results in BC specimens (×200 magnification). B: Representative positive CEMIP immunohistochemical results in luminal BC specimens (×200 magnification). C: Representative positive CEMIP immunohistochemical results in HER2+ BC specimens (×200 magnification). D: Representative positive CEMIP immunohistochemical results in triple negative BC specimens (×200 magnification). E: Validation the prognostic effect of CEMIP expression on the disease-free survival of BC patients. F: Validation the prognostic effect of CEMIP expression on the overall survival of BC patients.
Further, we explored the CEMIP molecular function and regulation pathways in BC. First, we explored these genes that interacted with CEMIP by the STRING database (Supplementary Figure 3D). These selected genes were then subjected to GO enrichment analysis (Supplementary Figure 3E). The GO analysis suggested that these proteins were mainly involved in the hyaluronan biosynthetic process, hyaluronan catabolic process, and other related processes. These findings may assist us to determine the exact regulatory mechanisms of CEMIP in BC.
Discussion
CEMIP which has a PbH1 domain consisting of parallel β-helix repeats, is predicted to take part in polysaccharide hydrolysis process. This protein structure suggested that CEMIP is a kind of secreted factor that may play an important role in extracellular ligand binding and processing. Deregulated expression of CEMIP has been found in pancreatic tumors, prostate tumors, renal tumors, and breast tumors [32-35]. However, the effect of CEMIP expression on the survival of BC patients, and related regulation mechanism remains unclear. In the study, CEMIP expression negatively influenced the survival outcomes of BC patients was found. The regulatory mechanism of CEMIP in BC was also explored.
Analyses of genetic variations of CEMIP by cBioPortal database. A: OncoPrint visual summary of genetic variations of CEMIP in BC. B: Analyses of genetic variations of CEMIP in different BC studies.
Most of the aforementioned results strongly suggest that CEMIP serves as a negative factor for cancer prognosis. Specifically, the Oncomine analysis and TCGA data showed a higher level of CEMIP mRNA transcripts in BC tissues than in noncancerous tissues. Using clinical BC specimens, we found CEMIP expression was associated with a larger tumor size, distant metastasis, and even death. Additionally, CEMIP expression can also significantly shorten DFS and OS. Furthermore, we validated CEMIP mRNA levels in both BC cell lines and clinical samples. The results revealed that the CEMIP mRNA level was increased in BC tissues compared with normal breast tissues. Western blotting and database analysis showed that CEMIP was significantly highly expressed in carcinomas compared to nontumor tissues. CEMIP was overexpressed in MDA-MB-231, SKBR3, and MCF7 cells than MCF10A cells, indicating that CEMIP may be associated with metastasis and invasion of BC.
Analyses of CEMIP promoter methylation in BC. A: Analysis the level of CEMIP promoter methylation in primary BC tissues and normal breast tissues in MEXPRESS database. B: Analysis the level of CEMIP promoter methylation in primary BC tissues and normal breast tissues in UALCAN database.
In addition, we found that DNA promoter methylation was involved in the transcription of CEMIP, although it was not a direct result. And CEMIP can regulate immune-related molecules to participate in BC progression. Additionally, Oncomine coexpression analysis showed expression of CEMIP as well as other BC oncogenes. Finally, using GO analysis, we found that CEMIP was significantly enriched in hyaluronan biosynthetic process and hyaluronan catabolic process and related processes. All these findings predicted that CEMIP is an adverse prognostic factor of BC and may participate in the regulation of BC progression.
This research also has some limitations. Firstly, this was a small sample study. Studies that include more BC patients are needed to explore the precise influence of CEMIP expression in BC. Secondly, we only explored the regulatory pathway of CEMIP based on bioinformatic tools and did not validate by in vitro cell experiments and in vivo animal experiments. Thus, future studies should focus on molecule experiments to elucidate the mechanisms by which CEMIP expression can affect the progression of BC.
Overall, this study found that CEMIP expression is higher in cancer tissues than in noncancerous tissues and that high CEMIP expression is associated with distant metastasis and death. And CEMIP can regulate immune-related molecules in BC. In conclusion, CEMIP was an adverse independent predictor of BC prognosis and may serve as a potential biomarker for BC patients.
Abbreviations
BC: breast cancer; CEMIP: cell migration-inducing and hyaluronan-binding protein; HA: hyaluronic acid; TNBC: triple-negative BC; OS: overall survival; PPS: post-progression survival; ATCC: American Type Culture Collection; TBS: Tris-buffered saline; DFS: disease-free survival.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by China National Natural Science Foundation (81872159).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Samuel CA, Pinheiro LC, Reeder-Hayes KE. et al. To be young, black, and living with breast cancer: a systematic review of health-related quality of life in young black breast cancer survivors. Breast Cancer Res Treat. 2016;160:1-15
3. Anastasiadi Z, Lianos GD, Ignatiadou E. et al. Breast cancer in young women: an overview. Updates Surg. 2017;3:313-7
4. Abe S, Usami S, Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters' cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J Hum Genet. 2003;11:564-70
5. Yoshida M, Sai S, Marumo K. et al. Expression analysis of three isoforms of hyaluronan synthase and hyaluronidase in the synovium of knees in osteoarthritis and rheumatoid arthritis by quantitative real-time reverse transcriptase polymerase chain reaction. Arthritis Res Ther. 2004;6:R514-20
6. Michishita E, Garcés G, Barrett JC. et al. Upregulation of the KIAA1199 gene is associated with cellular mortality. Cancer Lett. 2006;1:71-7
7. Jia S, Qu T, Wang X. et al. KIAA1199 promotes migration and invasion by Wnt/β-catenin pathway and MMPs mediated EMT progression and serves as a poor prognosis marker in gastric cancer. PLoS One. 2017 4
8. Matsuzaki S, Tanaka F, Mimori K. et al. Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann Surg Oncol. 2009;7:2042-51
9. Koga A, Sato N, Kohi S. et al. KIAA1199/CEMIP/HYBID overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Pancreatology. 2017;1:115-22
10. Kohi S, Sato N, Koga A. et al. KIAA1199 is induced by inflammation and enhances malignant phenotype in pancreatic cancer. Oncotarget. 2017;10:17156-63
11. Liu J, Han P, Gong J. et al. Knockdown of KIAA1199 attenuates growth and metastasis of hepatocellular carcinoma. Cell Death Discov. 2018;4:102
12. Zhang D, Zhao L, Shen Q. et al. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int J Cancer. 2017;10:2298-309
13. Evensen NA, Li Y, Kuscu C. et al. Hypoxia promotes colon cancer dissemination through up-regulation of cell migration-inducing protein (CEMIP). Oncotarget. 2015;24:20723-39
14. Fink SP, Myeroff LL, Kariv R. et al. Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget. 2015;31:30500-15
15. Evensen NA, Kuscu C, Nguyen HL. et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst. 2013;18:1402-16
16. Kuscu C, Evensen N, Kim D. et al. Transcriptional and epigenetic regulation of KIAA1199 gene expression in human breast cancer. PLoS One. 2012;9:e44661
17. Jami MS, Hou J, Liu M. et al. Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC Cancer. 2014;14:194
18. Shen F, Zong ZH, Liu Y. et al. CEMIP promotes ovarian cancer development and progression via the PI3K/AKT signaling pathway. Biomed Pharmacother. 2019;114:108787
19. Rhodes DR, Kalyana-Sundaram S, Mahavisno V. et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;2:166-80
20. Chandrashekar DS, Bashel B, Balasubramanya SAH. et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649-58
21. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients, Breast Cancer Res Treatment, 2010; 3:725-31
22. Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets, Scientific Reports, 2018;8:9277
23. Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:41-4
24. Ru B, Wong CN, Tong Y. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;20:4200-2
25. Curtis C, Shah SP, Chin SF. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;7403:346-52
26. Glück S, Ross JS, Royce M. et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat. 2012;3:781-91
27. Klutstein M, Nejman D, Greenfield R, Cedar H. DNA Methylation in Cancer and Aging. Cancer Res. 2016;12:3446-50
28. Ginestier C, Cervera N, Finetti P. et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;15:4533-44
29. Stickeler E, Pils D, Klar M. et al. Basal-like molecular subtype and HER4 up-regulation and response to neoadjuvant chemotherapy in breast cancer. Oncol Rep. 2011;4:1037-45
30. Zhou Q, Jiang Y, Yin W. et al. Single-nucleotide polymorphism in microRNA-binding site of SULF1 target gene as a protective factor against the susceptibility to breast cancer: a case-control study. Onco Targets Ther. 2016;9:2749-57
31. Moquet-Torcy G, Tolza C, Piechaczyk M. et al. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014;17:11011-24
32. Wang XD, Lu J, Lin YS. et al. Functional role of long non-coding RNA CASC19/miR-140-5p/CEMIP axis in colorectal cancer progression in vitro. World J Gastroenterol. 2019;14:1697-714
33. Yoshida H, Nagaoka A, Komiya A. et al. Reduction of hyaluronan and increased expression of HYBID (alias CEMIP and KIAA1199) correlate with clinical symptoms in photoaged skin. Br J Dermatol. 2018;1:136-44
34. Zhang P, Song Y, Sun Y. et al. AMPK/GSK3β/β-catenin cascade-triggered overexpression of CEMIP promotes migration and invasion in anoikis-resistant prostate cancer cells by enhancing metabolic reprogramming. FASEB J. 2018;7:3924-3935
35. Chen Y, Li L, Zhang J. Cell migration inducing hyaluronidase 1 (CEMIP) activates STAT3 pathway to facilitate cell proliferation and migration in breast cancer. J Recept Signal Transduct Res. 2021;2:145-152
Author contact
![]() Corresponding author: Caigang Liu, Tel: 86-18940254967; Fax: +86-24-22834060; Email: liucgorg.
Corresponding author: Caigang Liu, Tel: 86-18940254967; Fax: +86-24-22834060; Email: liucgorg.

 Global reach, higher impact
Global reach, higher impact