Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(16):3839-3850. doi:10.7150/ijms.66451 This issue Cite
Research Paper
Effects of Low Protein Diet on Modulating Gut Microbiota in Patients with Chronic Kidney Disease: A Systematic Review and Meta-analysis of International Studies
1. Department of Nephrology, Chang Gung Memorial Hospital, Keelung, Taiwan.
2. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
3. Department of Mathematical Sciences, Florida Atlantic University, Florida, US.
4. School of Pharmacy, Institute of Clinical Pharmacy and Pharmaceutical Sciences, College of Medicine, National Cheng Kung University, Tainan, Taiwan.
5. Department of Pharmacy, Keelung Chang Gung Memorial Hospital, Keelung, Taiwan.
6. College of Medicine, Chang Gung University, Taoyuan, Taiwan.
#These authors contribute equally to this manuscript.
Received 2021-8-24; Accepted 2021-10-9; Published 2021-10-25
Abstract
Background: Although associations between low protein diet (LPD) and changes of gut microbiota have been reported; however, systematic discernment of the effects of LPD on diet-microbiome-host interaction in patients with chronic kidney disease (CKD) is lacking.
Methods: We searched PUBMED and EMBASE for articles published on changes of gut microbiota associated with implementation of LPD in CKD patients until July 2021. Independent researchers extracted data and assessed risks of bias. We conducted meta-analyses of combine p-value, mean differences and random effects for gut microbiota and related metabolites. Study heterogeneity was measured by Tau2 and I2 statistic. This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.
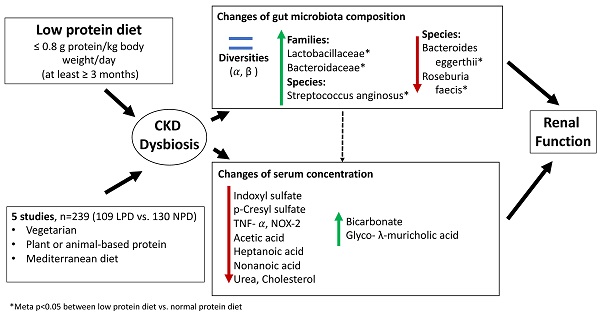
Results: Five articles met inclusion criteria. The meta-analyses of gut microbiota exhibited enrichments of Lactobacillaceae (meta-p= 0.010), Bacteroidaceae (meta-p= 0.048) and Streptococcus anginosus (meta-p< 0.001), but revealed depletion of Bacteroides eggerthii (p=0.017) and Roseburia faecis (meta-p=0.019) in LPD patients compared to patients undergoing normal protein diet. The serum IS levels (mean difference: 0.68 ug/mL, 95% CI: -8.38-9.68, p= 0.89) and pCS levels (mean difference: -3.85 ug/mL, 95% CI: -15.49-7.78, p < 0.52) did not change between groups. We did not find significant differences on renal function associated with change of microbiota between groups (eGFR, mean difference: -7.21 mL/min/1.73 m2, 95% CI: -33.2-18.79, p= 0.59; blood urea nitrogen, mean difference: -6.8 mg/dL, 95% CI: -46.42-32.82, p= 0.74). Other clinical (sodium, potassium, phosphate, albumin, fasting sugar, uric acid, total cholesterol, triglycerides, C-reactive protein and hemoglobin) and anthropometric estimates (body mass index, systolic blood pressure and diastolic blood pressure) did not differ between the two groups.
Conclusions: This systematic review and meta-analysis suggested that the effects of LPD on the microbiota were observed predominantly at the families and species levels but minimal on microbial diversity or richness. In the absence of global compositional microbiota shifts, the species-level changes appear insufficient to alter metabolic or clinical outputs.
Keywords: Chronic kidney disease, Low protein diet, Metabolites, Microbiota, Meta-analysis, Protein, Systematic review
Introduction
The global prevalence of Chronic Kidney Disease (CKD) is 9.1%; however, the burden of disease is increasing affecting 697.5 million people worldwide [1]. The disease has significant impact on metabolic complications, cardiovascular disease, quality of life and mortality. Progression of CKD into end stage renal disease (ESRD) can lead to high medical financial demands. Understanding of pathophysiology of CKD and various management approaches are mandatory in reducing disease burden and its complications. Altered gut-renal interaction and gastrointestinal dysbiosis have been extensively described in CKD patients. The leak gut leads to bacterial translocation, causing micro-inflammation, abnormal immunity and production of noxious metabolites, and further aggravates the uremic toxicity. Significant intestinal bacterial overgrowth and changes of gut microbiota diversity and composition have been observed in CKD patients [2-4]. Dietary counseling, including restriction of salt, potassium and phosphate intakes, represents important components in the care of renal patients. In particular, dietary protein restriction is commonly recommended in moderate to advanced CKD patients to reduce production of uremic wastes. Low protein diet (LPD), defined as daily intake < 0.8 g/Kg body weight, can decrease sodium loading, regulate sympathetic and angiotensin pathway, ameliorate urea and nitrogenous wastes and improve intraglomerular pressure resulting in reduced proteinuria and uremia [5-8]. Clinical studies have indicated that the use of very low protein diet (VLPD, 0.4-0.6 g/Kg body weight/day) supplemented with ketoanalogues amino acids can further retard renal progression and reduce mortality [5, 9-11]. Although associations between dietary protein restriction and preservation of renal function have been reported; however, the results remain ambiguous from diverse studies [12-14]. As such, knowledge on the diet-microbiome-host interaction associated with implementation of LPD remains to be elucidated in CKD patients.
The diet provides substrate for intestinal fermentation and plays a role in modulating gut microbiota composition, altering production of diverse endogenous metabolites and determining disease progression [15, 16]. Although an association between LPD and changes of gut microbiota has been indicated, it is unclear if changes of microbiota induced by dietary protein restriction can have beneficial effects on the outcomes of CKD patients. Diverse studies have indicated changes of gut microbiota associated with dietary interventions [17-21]. Black et al. did not find changes of composition and diversity of gut microbiota [17]; Lai et al have reported variations of relative abundances of gut microbiota at the family-levels [18]; Jiang et al described changes at the genus-level [19]; Di Lorio [20] and Wu et al. [21] revealed alterations in all three taxonomic levels, including bacterial families, genera and species, in patients undergoing LPD compared to those receiving normal diet. In addition to the modifications of microbiota in the gut of patients receiving LPD, the associated alterations of related surrogate indices of outcome, such as clinical, metabolomic and anthropometric parameters, have been described inconsistently in these studies. The interpretation of the results across these studies was limited by imbalanced baseline characteristics, small sample size, and differences in the analytic methodology and taxonomic classification reference. To our knowledge, systematically analysis and meta-analyses of the effects of LPD on gut microbiota in patients with CKD are still lacking. To fill this gap, we conducted a systematic review and meta-analysis which evaluate the roles of LPD in modulating gut dysbiosis in patients with CKD not yet on dialysis.
Materials and Methods
Design and search strategy
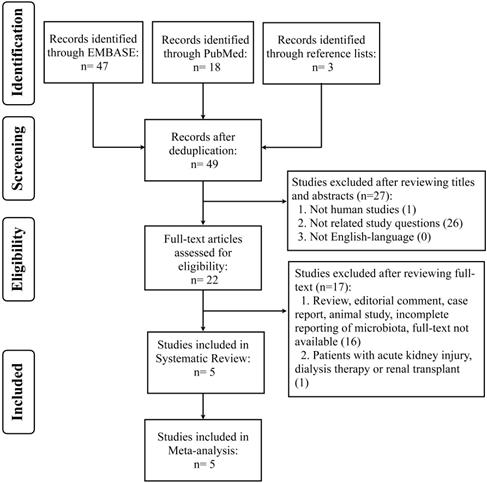
We conducted systematic review and meta-analysis of retrospective, cohort, case-controlled and randomized controlled studies in published literature from PUBMED and EMBASE, until July 17, 2021. The search strategy was based on the Population, Intervention, Comparator, Outcomes, Studies (PICOS) framework and involved the uses of synonyms and medical subject headings (MesH) or Emtree terms, including “renal insufficiency or CKD”, “protein restriction or LPD”, and “microbiota or microflora” (Table S1). Search terms were combined with Boolean operators (OR/AND). Studies were required to provide data on effects of LPD on microbiota (global bacterial composition or abundances of specific bacterial groups across different phylogenetic levels) in CKD patients. The reference lists of included articles were also hand-searched for relevant studies. Articles were managed using EndNote X9 (Analytics, Philadelphia, USA) to remove the duplicates. This study adhered to the reporting guidelines of Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) [22] (Figure 1), and it has been recorded in the International Prospective Register of Systematic Reviews (PROSPERO) database (CRD42021238979).
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow chart on selection and inclusion of studies.
Literature selection
Two independent researchers (CYC and IWW) screened titles and abstracts to identify potentially eligible studies for full-text review. Original articles in English language were reviewed. Those studies involving patients with CKD not yet on dialysis, aged above 18 years old, undergoing LPD as an intervention and describing gut microbiota as outcomes of interest were included for analysis. Normal renal function controls were also included for comparative analysis (non-LPD group), depended on the original design of included studies. On the other hand, those studies enrolling patients aged below 18 years old, studies involving only animal models or reporting incomplete data of gut microbiota were excluded for analysis. Case report, reviews, consensus report, full-text not available and editorial article were also excluded. Studies involving patients with acute kidney injury, dialysis therapy or renal transplant were excluded.
Data extraction and study quality
Data extraction was completed in duplicate by 2 independent reviewers (CKH, KJY). Information pertinent to research protocol, including site location, study design, year, definition, arms and duration of intervention were described. Participants' information regarding ethnicity, sample size, CKD stage, age and gender were extracted. Reporting of primary (microbiota characteristics: analytical methodology, database reference, bacterial diversity, taxonomic abundance) and secondary outcomes (biochemistry profiling or other measurements) were also conducted. For all included studies, mean ± standard deviation (SD) or median and interquartile range were used for data extracted. We contacted the study authors regarding possible incomplete data on the means and of selected outcome reporting. The studies have been excluded if a response was not received after three reminders and/or after attempting to contact another author of the study with no response.
Methodological quality of included studies was assessed independently by 2 authors (LCC, SCSu) based on the Risk of Bias in Non-Randomized Studies of Intervention (ROBINS-I) assessment tool [23]. This tool includes seven specific bias domains, including: (1) assessment of confounding factors; (2) selection of participants; (3) classification of intervention; (4) deviation from interventions; (5) missing outcome data; (6) measurement of outcomes; and (7) selection of reported result. Risk of bias was rated as 1-low risk; 2-moderate risk; 3-serious risk; 4-critical risk; and 0-no information. When the reviewers' assessments differed with regard to data extractions or study quality evaluations, the additional reviewer (SCShao) were drawn in, on a case-by-case basis, to discuss and make the final judgments.
Data synthesis and statistical analysis
We calculated the overall treatment effect for primary and secondary outcomes for each study included, using between-group differences as a measure of treatment effect. Data from cross-over interventions will be analyzed as pooled results (for study of Di Lorio et al. [20]). Meta-analyses were undertaken for outcomes that were reported on by at least 2 studies. Effect sizes between-group differences in bacterial relative abundances and metabolite concentrations were calculated as the mean difference, standardized mean difference or 95% confidential interval (CI). The random effects model and mean differences were used for the meta-analysis. The overall effect of the intervention in relation to statistical significance was based on p < 0.05, and the results of the meta-analysis were presented as forest plots. We also applied meta-analysis methods for combining multiple expression profiles comparisons [24]. For this method, we transformed two-sided p values collected from individual studies to one-sided p values for the consideration of heterogeneity among studies and combined p values analysis using Fisher's method as meta-analysis in microbiota analyses. Statistical heterogeneity among the studies was investigated by calculating Tau2 and the extent of heterogeneity attributable to heterogeneity was measured by the I2 statistic. Meta-analysis was performed using RevMan version 5.3.5 (Cochrane Collaboration, Oxford, UK). Statistical significance was defined as p < 0.05.
Results
We identified 65 records, form PUBMED (n=18) and EMBASE (n=47) databases, and 3 hand-searched references for the initial assessment and only 22 articles were included for full-text review. Finally, five studies which met the inclusion criteria were selected for systematic review and meta-analysis (Figure 1).
Four of them were controlled trials [18-21] and one was prospective observational study [17]. For those studies having more than one nutritional therapy [18, 20], only the LPD group rather than the VLPD or supplemented-LPD was selected as the intervention group for analysis. For the study of Di Lorio et al. [20], the Mediterranean diet, containing protein intake of 0.6-0.8 g/kg body weigh/day, was assigned as the LPD group. Overall, 239 patients (109 patients receiving LPD and 130 patients with normal-protein diet, NPD) were pooled for analysis. Although four of five articles have reported changes of gut microbiota in patients undergoing dietary intervention; however, the descriptions of microbial phylogenetic taxonomy classification were inconsistent, including families, phyla, genera and species-levels information [18-21]. One article reported clustering analysis of gut microbiota rather than microbial taxonomic changes [17]. The variations of indoxyl sulfate (IS) and p-cresyl sulfate (pCS) associated with changes of gut microbiota were described in three of all studies [17, 20, 21]. The characteristics of the studies included are described in Table 1.
The assessments of risks of bias in the included studies are illustrated in Table 2. Three studies included in this review demonstrated serious, moderate and unknown risk of bias in terms of possible confounding effects from baseline and selection of participants [17, 19]. Specifically, Black et al prospectively followed-up 30 pre-dialysis CKD patients who received LPD prescription and assigned the participants into adherent-LPD or non-adherent group according to their compliance to nutritional instructions rather than a pre-determined intervention assignment [17]. Jiang et al. included 36 hospitalized CKD stage 5 patients having different dietary protein regimens [19]; however, causes of inpatient care were not reported and may have affected the study outcome. Lai et al enrolled 16 CKD stage 3G-4G patients and 16 gender and renal function-matched controls to the study; however, the descriptions of baseline characteristics for the participants were lacking [18]. The different domains of assessments of risks of bias of included studied are summarized in Table 2.
All studies applied 16S rRNA sequencing to decipher the composition of gut microbiota in patients undergoing LPD [18-21], except the study from Black et al. [17], where polymerase chain reaction and denaturing gradient gel electrophoresis for microbiota clustering analysis were used. The SILVA database catalogue was used in two studies to determine taxonomic classification of intestinal microbes. The effects of LPD on gut microbiota of patients receiving LPD are outlined in Table 3.
The diversities of gut microbiota community were assessed in four studies and no significant change on the α- and β-diversity among patients undergoing different dietary protein restriction was found, except for the study of Wu et al. [21]. The changes in relative abundances of operational taxonomic units (OTUs) of two microbial families (Lactobacillaceae and Bacteroidaceae), one genus (Escherichia) and six species (Faecalibacterium prausnitzii, Coprococcus eutactus, Streptococcus anginosus, Bacteroides eggerthii, Blautia hydrogenotrophica and Roseburia faecis) were reported in more than two studies (Table S2). The meta-analyses of gut microbiota exhibited enrichments of Lactobacillaceae (meta-p= 0.010), Bacteroidaceae (meta-p= 0.048), Coprococcus eutactus (meta-p= 0.120) and Streptococcus anginosus (meta-p< 0.001) as well as revealed depletion of Bacteroides eggerthii (p=0.017), Blautia hydrogenotrophica (meta-p=0.225) and Roseburia faecis (meta-p=0.019) in patients receiving LPD compared to NPD patients. Although the mean effects of pooled studies have indicated an increase of mean relative abundances of Escherichia (meta-p= 0.444) and Faecalibacterium prausnitzii (meta-p= 0.340), the directions of changes of gut microbiota were inconsistent across the studies (Table 4).
Summary of characteristics of included studies
| First author | Year, country | Study design | Ethnicity | Total, n | CKD, n | CKD stage | Study arms, n | Intervention, n | Comparator, n | Definition of LPD | Age, mean | Male, n (%) | Primary outcome | Secondary outcome | Other measurements |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Black, et al. | 2018, Brazil | Prospective, observational | Latino-American | 30 | 30 | stage 3-4 | 2 (LPD adhesion vs. non-adhesion) | Adhesion group, 14 | Non-Adhesion group, 16 | ≤0.6 g protein/kg/day for 6 months | 55.8 | 16 (53.3%) | N/A (only reported clustering analysis) | Albumin, potassium, sodium, phosphate, urea, creatinine, uric acid, glucose, cholesterol, HDL-C, LDL-C, triglycerides, IS, PCS, IAA | Body fat, Lean mass, BMI, waist circumference |
| Di Lorio, et al. | 2019, Italy | Prosective, randomized, crossover, controlled | Caucasian | 60 | 60 | stage 3B-4 | 3 (free diet, Mediterranean diet, VLPD) | Mediterranean diet group, 60 | Free diet, 60 | 0.6-0.8 g protein/kg/day for 6 months | 68.4 | 49 (81.7%) | Phylum, family, genus, species-levels | Albumin, creatinine, glucose, HbA1C, uric acid, sodium, potassium, calcium, phosphate, bicarbonates, cholesterol, triglycerides, ferritin, PTH, hemoglobin, CRP, urinary indices (sodium, potassium, phosphate, urea, total protein, creatinine), IS, PCS | BMI, systolic and diastolic pressure |
| Lai, et al. | 2019, Italy | prospective and controlled | Caucasian | 32 | 16 | stage 3-4 | 3 (healthy vs. LPD vs. LPD + inulin) | LPD, 7 | Health control, 16 | ≤0.6 g protein/kg/day for 6 months | N/A | N/A | Family-levels | Uric acid, CRP, IL-1β, IL-6, TNF-α, NADPH, NOX2 | SF-36 Health Survey |
| Jiang, et al. | 2020, China | Cross-sectional, controlled | Asian | 40 | 36 | Stage 5 | 3 (NPD vs. VLPD vs. LPD) | LPD, 12 | NPD, 11 | 0.6-0.8 g protein/kg/day (duration: NA) | 54 | 20 (50%) | Phylum, genus-levels | Albumin, creatinine, cholesterol, CRP | BMI |
| Wu, et al. | 2020, Taiwan | Cross-sectional, controlled | Asian | 77 | 43 | Stage 2-5 | 3 (Healthy vs. LPD vs. NPD) | LPD,16 | NPD,27 | ≤0.8 g protein/kg/day for 3 months | 63.4 | 38 (49.4%) | Phylum, family, genus, species-levels | Albumin, urea, creatinine, hemoglobin, sodium, potassium, phosphate, glucose, uric acids, cholesterol, hs-CRP, urine protein-creatinine ratio, 11 saturated short-chain fatty acids, 41 bile acids, IS, PCS | BMI, systolic and diastolic pressure |
Abbreviations: CKD, chronic kidney disease; LPD, low protein diet; VLPD; very-low protein diet; NPD, normal-protein diet; OTU, operational taxonomic units; HDL-C, high-density level cholesterol; LDL-C, low-density level cholesterol; IS, indoxyl sulfate; PCS, p-cresyl sulfate; IAA, indole acetic acid; HbA1C, glycosylated hemoglobin; PTH, parathyroid hormone; CRP, C-reactive protein; IL, interleukin; TNF, tumor necrosis factor alpha; NADPH plasma nicotinamide adenine dinucleotide phosphate; NOX2, NADPH oxidase; BMI, body mass index. N/A: not available.
Risks of bias in included studies according to the Risk of Bias in Non-Randomized Studies of Interventions (ROBINS-I) tool
| Author, year | Pre-intervention | At-intervention | Post-intervention | Overall risk of bias | ||||
|---|---|---|---|---|---|---|---|---|
| Domain 1 | Domain 2 | Domain 3 | Domain 4 | Domain 5 | Domain 6 | Domain 7 | ||
| Bias due to confounding | Bias in selection of participants into study | Bias in classification of interventions | Bias due to deviation from interventions | Bias due to missing data | Bias in measurement of outcomes | Bias in selection of reported results | Unknown/low/moderate/serious/critical | |
| Black et al., 2018 | 3 | 3 | 2 | 1 | 1 | 1 | 2 | Serious |
| Di Lorio et al., 2019 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Low |
| Lai et al., 2019 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | Unknown |
| Jiang et al., 2020 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | Moderate |
| Wu et al., 2020 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Low |
Risk of bias assessment: 0-No information; 1-Low; 2-Moderate; 3-Serious; 4-Critical.
Summary of changes of gut microbiota and clinical parameters associated with low protein diet
| First author | Microbiota methodology | Library catalogues | Changes of gut microbiota | Changes of biochemistry parameters* | Changes of other measurements* | |||
|---|---|---|---|---|---|---|---|---|
| Diversity | Family-levels (RA of OTU) | Genera-levels (RA of OTU) | Species-levels (RA of OTU) | |||||
| Black, 2018 | PCR, DGGE | N/A | ⍺: no change; β: no change | N/A | N/A | N/A | Decreased: Cholesterol, LDL-C, PCS | No significant changes |
| Di Lorio, 2019 | 16S rRNA sequencing | Ribosomal Database Project, ver.10.28 | ⍺: no change; β: no change | Increased: Lachnospiraceae, Ruminococcaceae, Prevotellaceae, Bifidobacteriaceae Decreased: Lactobacillaceae, Streptococcaceae, Verrucomicrobiaceae, Enterobacteriaceae | Increased: Blautia, Bifidobacterium, Clostridium, Faecalibacterium, Coprococcus, Roseburia. Decreased: Ruminococcus, Collisella, Bacteroides, Lactobacillus, Akkermansia, Streptococcus, Escherichia | Increased: B. coccoides, B. hydrogenotrophica, B. obeum, B. wexlerae, B. adolescentis, B. pseudolongum, C. cadaveris, F. prausnitzii, C. eutactus, R. faecis Decreased: R. callidus, C. aerofaciens, B. uniformis, B. vulgatus, L. gasseri, L. salivarius, A. muciniphila, S. bovis, S. mutans, S. sobrinus, S. vestibularis, E. albertii | Increased: Sodium, bicarbonate, urine sodium. Decreased: Urea, phosphate, D-lactate, IS, PCS, urine phosphate | Decreased: Systolic and diastolic pressure |
| Lai, 2019 | 16S rRNA sequencing | SILVA database, ver. 132 | ⍺: N/A; β: no change | Increased: Akkermansiaceae, Bacteroidaceae. Decreased: Christensenellaceae, Clostridiaceae, Lactobacillaceae and Pasteurellaceae | N/A | N/A | Increased: Bicarbonate. Decreased: TNF-α, NOX2 | Increased: Physical role function and general heath perception of SF-36 |
| Jiang, 2020 | 16S rRNA sequencing | Genomes OnLine Database | ⍺: N/A; β: N/A | N/A | Increased: Escherichia, Shigella Decreased: Blautia | N/A | N/A | No significant changes |
| Wu, 2020 | 16S rRNA sequencing | SILVA database, ver. 132 | ⍺: no change; β: increased | Increased: Ruminococcaceae. Decreased: Lachnospiraceae, Bacteroidaceae | Increased: Calditerricola, Coprococcus, Romboutsia, Parabacteroides, Alloprevotella, Subdoligranulum, Ruminococcaceae UCG-010, Faecalibacterium, Subdoligranulum, Cloacibacillus Decreased: Desulfovibrio, Pseudobutyrivibrio, Lachnospira, Eubacterium hallii group, Roseburia, Fusicatenibacter, Anaerostipes, Lachnoclostridium, Prevotellaceae NK3B31 | Increased: Clostridium paraputrificum, Clostridium sordellii, Olsenella uli, Mogibacterium diversum, Blautia hydrogenotrophica, Lactobacillus mucosae, Porphyromonas gingivalis, Streptococcus anginosus, Lactobacillus sp. AB032. Decreased: Bacteroides coprophilus, Bacteroides plebeius, Bacteroides eggerthii | Increased: glyco λ-muricholic acid Decreased: eGFR, albumin, acetic, heptanoic and nonanoic acid | No significant changes |
Abbreviations: LPD, low protein diet; OTU, operational taxonomic units; RA, relative abundances; PCR, polymerase chain reaction; DGGE, denaturing gradient gel electrophoresis LDL-C, low-density level cholesterol; IS, indoxyl sulfate; PCS, p-cresyl sulfate; TNF, tumor necrosis factor alpha; NOX2, nicotinamide adenine dinucleotide phosphate oxidase; eGFR, estimated glomerular filtration rate. N/A: not available.
*Only those parameters showing significant differences between LPD vs. NPD (p<0.05) in the original articles were illustrated in this table.
Meta-analysis of changes of microbiota in patients receiving low protein diet
| Operational Taxonomic Units | p of study 1 | p of study 2 | Same direction | Mean effect | Fisher statistics | Meta p |
|---|---|---|---|---|---|---|
| Families | ||||||
| Lactobacillaceae | 0.0235 | 0.0615 | Yes | LPD > NPD | 13.07894592 | 0.010896423 |
| Bacteroidaceae | 0.013 | 0.639 | Yes | LPD > NPD | 9.581313492 | 0.048102986 |
| Genera | ||||||
| Escherichia | 0.202 | 0.769 | No | LPD > NPD | 3.724303782 | 0.44460095 |
| Species | ||||||
| Faecalibacterium prausnitzii | 0.8625 | 0.121 | No | LPD > NPD | 4.519769727 | 0.340209734 |
| Coprococcus eutactus | 0.2175 | 0.1185 | Yes | LPD > NPD | 7.316797494 | 0.120064405 |
| Streptococcus anginosus | 0.0175 | 0.00232 | Yes | LPD > NPD | 20.22348498 | 0.000451137 |
| Bacteroides eggerthii | 0.2065 | 0.012 | Yes | LPD < NPD | 12.00060699 | 0.017346752 |
| Blautia hydrogenotrophica | 0.0815 | 0.72 | Yes | LPD < NPD | 5.671312651 | 0.225076313 |
| Roseburia faecis | 0.0665 | 0.044 | Yes | LPD < NPD | 11.66823795 | 0.019996632 |
Abbreviations: LPD, low protein diet; NPD, normal-protein diet. “p” denoted p values.
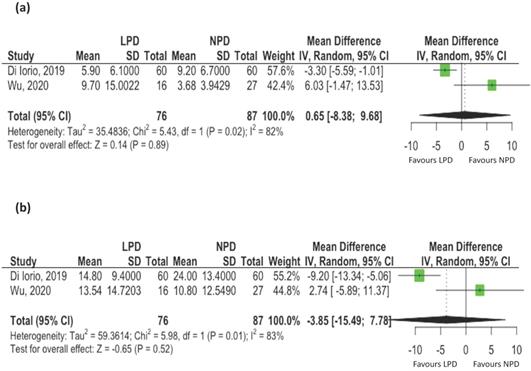
Three studies reported changes of uremic toxins, including IS and pCS [17, 20, 21]. Two studies reporting mean values were included for meta-analyses. The serum levels of total IS [mean difference: 0.68 ug/mL, 95% CI: -8.38-9.68, p= 0.89, Figure 2A) and total pCS (mean difference: -3.85 ug/mL, 95% CI: -15.49-7.78, p = 0.52, Figure 2B) did not change with LPD compared to NPD patients. However, there was evidence of statistical heterogeneity among the studies (I2 = 82%, p=0.02 for IS; I2 = 83%, p=0.01 for pCS).
The effects of low protein diet on uremic toxins associated with changes of gut microbiota. (A) Indoxyl sulfate; (B) p-cresyl Sulfate (Black et al. conducted observational study and the mean values of uremic toxins were not available. Only these two controlled studies were pooled for metaanalysis).
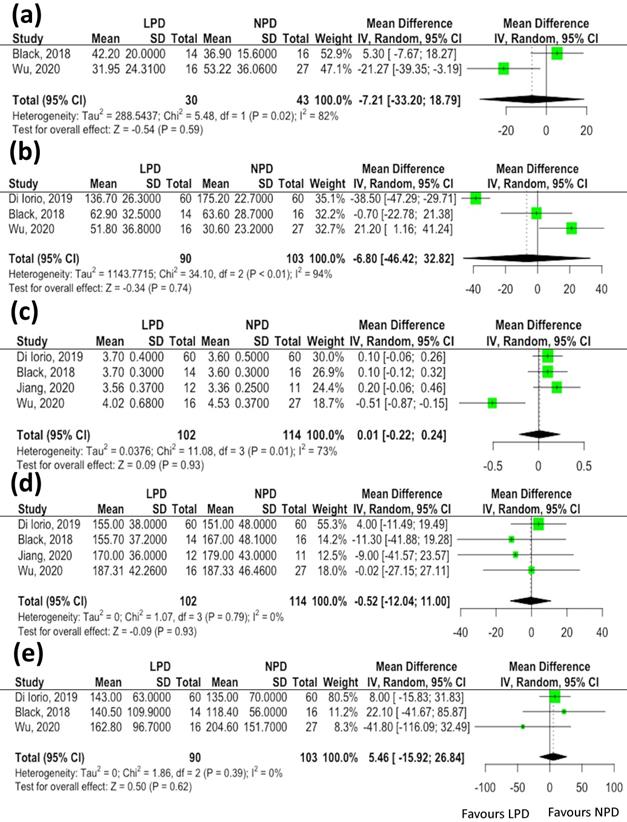
Twelve serum clinical parameters (blood urea nitrogen, serum creatinine, sodium, potassium, phosphate, albumin, fasting sugar, uric acid, total cholesterol, triglycerides, C-reactive protein and hemoglobin levels) and three anthropometric estimates (body mass index, systolic blood pressure and diastolic blood pressure) associated with changes of gut microbiota were reported in more than two studies. We did not find significant differences in data indicative of renal function in association with changes of microbiota in patients receiving LPD compared to the NPD group [estimated glomerular filtration rate (eGFR), mean difference: -7.21 mL/min/1.73 m2, 95% CI: -33.2-18.79, p= 0.59, Figure 3A; blood urea nitrogen, mean difference: -6.8 mg/dL, 95% CI: -46.42-32.82, p= 0.74, Figure 3B]. Similarly, the serum albumin (mean difference: 0.01 mg/dL, 95% CI: -0.22-0.24, p= 0.93, Figure 3C), total cholesterol (mean difference: -0.52 mg/dL, 95% CI: -12.04-11.00, p= 0.93, Figure 3D) and triglycerides (mean difference: -15.92 mg/dL, 95% CI: -116.09-32.49, p= 0.62, Figure 3E) levels did not vary between the two groups. The meta-analyses of other clinical parameters (sodium, p=0.43; potassium, p=0.62; phosphate, p= 0.18; fasting sugar, p=0.39; uric acid, p=0.61; C-reactive protein, p=0.83 and hemoglobin, p=0.95) and three anthropometric estimates (body mass index, p=0.32; systolic blood pressure, p=0.31 and diastolic blood pressure, p=0.77) revealed not significant differences in these parameters associated with changes of gut microbiota between patients undergoing LPD vs. NPD.
Overall, significant changes of gut microbiota, predominantly at the families and species levels but minimal on microbial diversity or richness, were associated with use of LPD in CKD patients. In the absence of modification in the architecture of global microbiota community but presence of heterogeneity and bias of some studies, the changes of abundances of selected gut microbes appeared insufficient to shift metabolic or clinical output.
Discussion
Comprehensive discernment of effects of dietary therapy on the intestinal dysbiosis remains incompletely elucidated in CKD patients. Protein restriction is the most frequent dietary intervention given to renal patients, in addition to the salt and water restriction. The results of this systematic review and meta-analysis have revealed significant changes of gut microbiota, mainly in the enrichments for Lactobacillaceae, Bacteroidaceae and Streptococcus anginosus and depletion of Bacteroides eggerthii and Roseburia faecis, in patients receiving LPD compared to NPD group. In contrast, the microbiota changes were not associated with significant variations in gut-producing uremic toxins, renal function, and other clinical indices.
Although several studies have described dysbiosis of gut microbiota associated with LPD, the results are varied and were inconclusive, in terms of taxonomic classification, participants setting regarding to the disease severity and small sample size which limited statistical power. To fill this gap, the present work, using rigorous criteria on the assessments of eligibility of included studies, has recapitulated all literatures relevant to the changes of gut microbiota associated with LPD and has combined information across multiple studies to increase sensitivity. Common meta-analysis methods mainly combine three different types of statistics: combine p-values, combine effect sizes and combine ranks [25]. Because of irregularity in reporting bacterial taxonomic abundances across studies, we applied a meta-analysis method combining p values using Fisher's statistics. This method has demonstrated satisfactory performance on the detection capability, biological association, model stability and robustness of study [24] warranting sufficient statistical power to the study.
The effects of low protein diet on clinical parameters associated with changes of gut microbiota. (A) estimated glomerular filtration rate; (B) blood urea nitrogen; (C) serum albumin; (D) total cholesterol; (E) triglycerides.
Intestinal dysbiosis and compositional differences of gut microbiota have been described in renal patients [4, 26, 27]. We did not find changes in microbiota diversity with dietary protein restriction. However, the LPD was associated with restoration of some but not all of the abundances of microbiota of CKD patients compared to health subjects. The abundances of butyrate-producing bacterial families, including Lactobacillaceae, Bacteroidaceae and Prevotellaceae, were lower in patients with uremia, whereas the population of indole- or p-cresol-producing bacterial families, such as Enterobacteriaceae, Clostridiaceae, and Verrocomicrobiaceae, were expanded compared to health individuals [4, 28, 29]. While information on these tryptophanase possessing bacteria was incompletely unraveled from the included studies, this meta-analysis study found an increase in the abundances of Lactobacillaceae and Bacteroidaceae in patients receiving LPD. Lactobacillus species can metabolize tryptophan into indole-3-aldehyde, which increase the production of interleukin-22 through acting on the aryl hydrocarbon receptor in intestinal immune cells, ultimately controlling intestinal epithelial homeostasis [30, 31]. The indole can further be metabolized by the liver into IS and became accumulated in CKD patients because of reduced renal excretion leading to renal progression [32]. Administration of strains of Lactobacillus salivarius was associated with reduction of serum levels of both IS and pCS [33]. The enrichment of Lactobacillaceae found in patients receiving LPD may in part explain the decreased serum levels of IS and pCS found in various included articles of this study [17, 20]. Faecalibacterium prausnitzii is another proposed gut microbe associated with serum levels of IS or pCS; however, the abundance of this bacteria did not differ between LPD vs. NPD groups in our meta-analysis study. Two of three included studies [17, 20, 21] have shown reduction of uremic toxins after implementation of LPD in Caucasian population [17, 20]. However, our findings did not suggest lowering of uremic toxins in association with dietary protein restriction. Furthermore, study heterogeneity, especially on stages of CKD, residual renal function, and discrepancy of dietary composition among participants across different countries, may have impacts on the actual association of serum levels of uremic toxins with variation of microbiota in CKD patients receiving LPD.
We found enrichment for intestinal Bacteroidaceae families but depletion of Bacteroides eggerthii species in LPD patients. The later bacteria were found to increase in non-CKD subjects [4]. These anaerobic bacteria produce enzymes responsible for breakdown of complex plant polysaccharides (such as cellulose and hemicellulose) and host-derived polysaccharides (such as mucopolysaccharides) generating phenolic acids [34]. Main phenolic metabolites (phenylacetic acid, 4‐hydroxylphenylacetic acid and indole‐3‐acetic acid), can modulate mucosal glycosylation and promote angiogenesis and immune maturation. Yet, levels of these metabolites were only described in one included study [17]. The relationships among changes of Bacteroidaceae abundance, related metabolites and renal outcome remain to be determined. Streptococcus anginosus, which increased in condition of unhealthy microbiome [35], was associated with tumorigenesis of intestinal tract, such as colon or esophageal cancer, and lupus activity [36-38]. The abundance of this bacterium was lower in healthy subjects compared to CKD patients [4]. The variation of this microbe observed in our study may confirm existence of intestinal dysbiosis of CKD patients. Finally, Roseburia faecis, a butyrate-producing anaerobic bacteria, can derive short-chain fatty acids (SCFAs) from fermentation of dietary fiber. Common SCFAs, including acetate, propionate and butyrate, may exert anti-inflammation, anti-atherosclerosis and anti-oxidative functions leading to amelioration of kidney damage [39, 40]. The abundances of Roseburia were higher in healthy subjects than CKD patients [4]. Only one included study reported associations of SCFAs with changes of gut microbiota in patients having LPD [21]. The changes of abundances of Bacteroidaceae families, Bacteroides eggerthii and Roseburia faecis species found in this study may in part explain the variation of SFCA observed in patients receiving LPD [17, 21]. The absence of tangible changes of abundance of these bacteria may in part explain the scanty impact on renal function associated with LPD. Although the inferences of effectiveness of nutritional intervention on the microbiome-metabolite axis were present in this meta-analysis; however, the subsequent impacts on the renal outcome of CKD patients should be validated in further large-scale studies.
Dietary components constitute important modulators of microbiota composition and function, which in turn affects the absorption, metabolism and storage of ingested nutrients, promotes intestinal barrier integrity, regulates mucosa inflammation and produces endogenous metabolites, resulting in intense effects on host physiology [41, 42]. Although the adulthood microbiota is resilient, significant interindividual variability and plasticity of the gut microbiota have been observed. Changes in abundance or richness of certain gut microbial groups were identified in many chronic metabolic disease compared to healthy subjects [42]. Microbiota remodeling creates an exceptional opportunity, by manipulating various external factors such as the diet or by transferring candidate gut microbiota, to reshape the architecture and biological functions of gut microbes for improved human health [15]. Implementation of dietary intervention consisting of 12-week energy-restricted high-protein and weight-maintenance diet was able to elevate gut microbial gene richness and to alter enterotypes in obese and overweight individuals [43]. The mechanisms by which changes in the dietary protein components and communities of gut microbiota regulate metabolic control are still evolving. The amino acids of diet provide gut microbes essential carbon and nitrogen for their metabolism function [15]. Global serum metabolomics analysis has detected alterations of 130 metabolites in patients undergoing LPD compared with those receiving moderate-protein diet and has reported significant differences in the serum levels of 32 metabolites between participants assigned to the VLPD compared to those receiving LPD [44]. Lobel et al. demonstrated that high sulfur amino acid-containing diet can not only induce posttranslational modification of microbial tryptophanase activity, leading to reduced cecal levels of IS, but also can mitigate kidney injury without altering microbial community composition in adenine induced-CKD mice [45]. Gut microbial gene enrichments of D-alanine metabolism, synthesis/degradation of ketone bodies and glutathione metabolism were also observed in the CKD patients receiving protein restriction [21]. Other possible mechanisms have been also documented, including altering circulating levels of uremic toxins [17, 20], tumor necrosis factor alpha (TNF-⍺), plasma nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX2) [18], SCFAs and bile acids [21] in association with changes of gut microbiota in patients receiving LPD. Further efforts to establish causal relationships of diet-microbiome interaction and to design prospects of personalized nutrition with specific dietary component manipulation should be necessary for CKD patients.
Although five studies were included in the overall systematic review and meta-analysis, merely two and three studies were included in the meta-analysis of gut microbiota and circulating markers, respectively. This could limit the generalizability of findings. Various possible reasons may contribute to the rarity of pertinent references on the research of gut microbiota in CKD patients undergoing LPD. First, good adherence to such a diet should need supports of multidisciplinary team (consisted of nephrologist, dietitian, case management nurse) to maximizing the compliance of patients. Second, research of microbiota requires sophisticated methodological collaboration of not only clinicians but also microbiologists, computing scientists and experts of bioinformatics and statistics to manage the megadata sets. Finally, the high cost of 16S rRNA sequencing may sometimes represent a barrier for large sample size studies. From all the included studies, Di Lorio et al. have conducted largest study enrolling 60 patients, by adopting cross-over design, to compensate the shortage in sample size of the study. We believed that the systematic review and meta-analysis may have an exceptional advantage to overcome the interpretation of individual studies having small sample size. Although the overall number of included studies remained low, the pooling of patients from all these researches may certainly increase the power and significance of studies. Therefore, more studies are needed in this area of research. In addition, differences in the reporting levels of gut bacterial abundances and in the taxonomic catalogue may hamper the comparison of actual effects of LPD across the studies included. Further high-throughput sequencing technology may facilitate in-depth studies of the gut microbiota associated with nutritional therapy. We used the “Mediterranean diet” as surrogate of LPD in the study of Di Lorio et al. [20]; however, this diet is characterized by consumption of high contents of vegetables, fruits, legumes, nuts, beans, cereals, grains, fish and unsaturated fats, as well as low intake of meat and dairy foods. Extreme dietary protein restriction, namely the VLPD (< 0.4-0.6 g/ kg body weigh/day) often supplemented with ketoanalogues, may have additional effects on microbiota modulation [20, 46]. We did not assess effects of VLPD on gut microbiota because of the scarce data presented in only two included studies. One inherent limitation to studying the health effects of particular diet is that some nutrients are rarely consumed in isolation. In consequence, experimentally handling of a specific nutrient can alter incorporation of other food components that may have metabolic effects unto themselves. For example, high-fat diets are commonly associated with LPD to avoid caloric malnutrition, which can also trigger effects on microbiota composition and metabolic consequences. Given the limitations of studying nutrients in isolation, a body of knowledge is accumulating to emphasize health effects of particular dietary pattern rather than experimental reductionism [41, 47]. Lastly, in spite of the use of the random effects model in the analysis of clinical and metabolite markers, the high heterogeneity of included studies may have also affected the results of this meta-analysis. Thus, combination of different conventional meta-analysis methods may strengthen the robustness of models and the power of our study. We did not find variations of tangible renal outcome with LPD manipulation in CKD patients. Differences in patient selection, duration and composition of dietary intervention, and limited sample size may all contribute to the observed discrepancies with previous studies [5-8]. Further prospective longitudinal studies with breakthrough methodologies, such as shotgun metagenomic sequencing, may help to elucidate the function of LPD intervention on mysterious intestinal microbiome-host metabolite synergies in order to preserve renal function of CKD patients.
Conclusions
In conclusion, this systematic review and meta-analysis found that LPD can significantly alter the relative abundances of specific bacterial groups, including enrichments of Lactobacillaceae, Bacteroidaceae and Streptococcus anginosus as well as depletion of Bacteroides eggerthii and Roseburia faecis, in patients receiving LPD compared to NPD group. Our study suggested that the effects of LPD on the microbiota were observed predominantly at the families and species levels and relatively minimal on microbial diversity and richness. In the absence of global compositional microbiota shifts, the species-level changes appear insufficient to alter metabolic or clinical outputs. Furthermore, methodological aspects should be standardized to reduce potential bias and study heterogeneity and to allow interpretation of data in future studies. The findings of the present study, focusing on diet-microbiome-host interactions, provide insights for personalized diet recommendation potentially beneficial for CKD patients.
Abbreviations
CKD: chronic kidney disease; ESRD: end stage renal disease; LPD: low protein diet; VLPD: very low protein diet; PICOS: Population, Intervention, Comparator, Outcomes, Studies; MesH: medical subject headings; PRISMA: Preferred Reporting Items for Systematic Reviews and Meta-analyses; PROSPERO: International Prospective Register of Systematic Reviews; SD: standard deviation; ROBINS-I: Risk of Bias in Non-Randomized Studies of Intervention; CI: confidential intervals; NPD: normal protein diet; IS: indoxyl sulfate; pCS: p-Cresyl sulfate; OTUs: operational taxonomic units; SCFAs: short-chain fatty acids.
Supplementary Material
Supplementary tables.
Acknowledgements
We thank all researchers and clinicians involved in the individual trials.
Funding
This research was funded by Chang Gung Memorial Hospital, grant number CGRPG2F0081 and the Ministry of Science and Technology, Taiwan (MOST 108-2314-B-182A-029 and MOST 109-2314-B-182A-127).
Trial registration
PROSPERO registration number CRD42021238979.
Author Contributions
Conceptualization, I.W.W., S.C.Su; methodology, L.C.C, S.C.Shao; validation, K.J.Y, C.Y.C and Y.T.C; formal analysis, L.CC, S.C.Su; writing - original draft preparation, C.K.H; writing - review and editing, I.W.W., S.C.Su. All authors have read and agreed to the published version of the manuscript.
Availability of data and material
The primary data for this study is available from the authors on direct request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Global regional, national burden of chronic kidney disease 1990-2017. a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709-33
2. Andersen K, Kesper MS, Marschner JA, Konrad L, Ryu M, Kumar Vr S. et al. Intestinal Dysbiosis, Barrier Dysfunction, and Bacterial Translocation Account for CKD-Related Systemic Inflammation. J Am Soc Nephrol. 2017;28:76-83
3. Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res. 2017;179:24-37
4. Wu IW, Lin CY, Chang LC, Lee CC, Chiu CY, Hsu HJ. et al. Gut Microbiota as Diagnostic Tools for Mirroring Disease Progression and Circulating Nephrotoxin Levels in Chronic Kidney Disease: Discovery and Validation Study. Int J Biol Sci. 2020;16:420-34
5. Garneata L, Stancu A, Dragomir D, Stefan G, Mircescu G. Ketoanalogue-Supplemented Vegetarian Very Low-Protein Diet and CKD Progression. J Am Soc Nephrol. 2016;27:2164-76
6. Klahr S, Levey AS, Beck GJ, Caggiula AW, Hunsicker L, Kusek JW. et al. The effects of dietary protein restriction and blood-pressure control on the progression of chronic renal disease. Modification of Diet in Renal Disease Study Group. N Engl J Med. 1994;330:877-84
7. Milovanova L, Fomin V, Moiseev S, Taranova M, Milovanov Y, Lysenko Kozlovskaya L. et al. Effect of essential amino acid кetoanalogues and protein restriction diet on morphogenetic proteins (FGF-23 and Кlotho) in 3b-4 stages chronic кidney disease patients: a randomized pilot study. Clin Exp Nephrol. 2018;22:1351-9
8. Kalantar-Zadeh K, Fouque D. Nutritional Management of Chronic Kidney Disease. N Engl J Med. 2017;377:1765-76
9. Li A, Lee HY, Lin YC. The Effect of Ketoanalogues on Chronic Kidney Disease Deterioration: A Meta-Analysis. Nutrients. 2019;11:957
10. Chen HY, Sun CY, Lee CC, Wu IW, Chen YC, Lin YH. et al. Ketoanalogue supplements reduce mortality in patients with pre-dialysis advanced diabetic kidney disease: A nationwide population-based study. Clin Nutr. 2021
11. Hahn D, Hodson EM, Fouque D. Low protein diets for non-diabetic adults with chronic kidney disease. Cochrane Database Syst Rev. 2018;10:Cd001892
12. Menon V, Kopple JD, Wang X, Beck GJ, Collins AJ, Kusek JW. et al. Effect of a very low-protein diet on outcomes: long-term follow-up of the Modification of Diet in Renal Disease (MDRD) Study. Am J Kidney Dis. 2009;53:208-17
13. Wang YC, Juan SH, Chou CL, Hsieh TC, Wu JL, Fang TC. Long-Term Effects of Ketoanalogues on Mortality and Renal Outcomes in Advanced Chronic Kidney Disease Patients Receiving a Low-Protein Diet. Nutrients. 2020 12
14. Yamada S, Tokumoto M, Tatsumoto N, Tsuruya K, Kitazono T, Ooboshi H. Very low protein diet enhances inflammation, malnutrition, and vascular calcification in uremic rats. Life Sci. 2016;146:117-23
15. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776-80
16. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105-8
17. Black AP, Anjos JS, Cardozo L, Carmo FL, Dolenga CJ, Nakao LS. et al. Does Low-Protein Diet Influence the Uremic Toxin Serum Levels From the Gut Microbiota in Nondialysis Chronic Kidney Disease Patients? J Ren Nutr. 2018;28:208-14
18. Lai S, Molfino A, Testorio M, Perrotta AM, Currado A, Pintus G. et al. Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease. Nutrients. 2019 11
19. Jiang S, Wang B, Sha T, Li X. Changes in the Intestinal Microbiota in Patients with Stage 5 Chronic Kidney Disease on a Low-Protein Diet and the Effects of Human to Rat Fecal Microbiota Transplantation. Med Sci Monit. 2020;26:e921557
20. Di Iorio BR, Rocchetti MT, De Angelis M, Cosola C, Marzocco S, Di Micco L. et al. Nutritional Therapy Modulates Intestinal Microbiota and Reduces Serum Levels of Total and Free Indoxyl Sulfate and P-Cresyl Sulfate in Chronic Kidney Disease (Medika Study). J Clin Med. 2019 8
21. Wu IW, Lee CC, Hsu HJ, Sun CY, Chen YC, Yang KJ. et al. Compositional and Functional Adaptations of Intestinal Microbiota and Related Metabolites in CKD Patients Receiving Dietary Protein Restriction. Nutrients. 2020 12
22. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264-9 w64
23. Sterne JA, Hernán MA, Reeves BC, Savović J, Berkman ND, Viswanathan M. et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. Bmj. 2016;355:i4919
24. Chang LC, Lin HM, Sibille E, Tseng GC. Meta-analysis methods for combining multiple expression profiles: comparisons, statistical characterization and an application guideline. BMC Bioinformatics. 2013;14:368
25. Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012;40:3785-99
26. Crespo-Salgado J, Vehaskari VM, Stewart T, Ferris M, Zhang Q, Wang G. et al. Intestinal microbiota in pediatric patients with end stage renal disease: a Midwest Pediatric Nephrology Consortium study. Microbiome. 2016;4:50
27. Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ. et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308-15
28. Wong J, Piceno YM, DeSantis TZ, Pahl M, Andersen GL, Vaziri ND. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am J Nephrol. 2014;39:230-7
29. Wu IW, Gao SS, Chou HC, Yang HY, Chang LC, Kuo YL. et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics. 2020;10:5398-411
30. Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med. 2016;8:46
31. Keir M, Yi Y, Lu T, Ghilardi N. The role of IL-22 in intestinal health and disease. J Exp Med. 2020;217:e20192195
32. Wu IW, Hsu KH, Lee CC, Sun CY, Hsu HJ, Tsai CJ. et al. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol Dial Transplant. 2011;26:938-47
33. Lee TH, Park D, Kim YJ, Lee I, Kim S, Oh CT. et al. Lactobacillus salivarius BP121 prevents cisplatin-induced acute kidney injury by inhibition of uremic toxins such as indoxyl sulfate and p-cresol sulfate via alleviating dysbiosis. Int J Mol Med. 2020;45:1130-40
34. Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG. et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523-35
35. Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput Biol. 2016;12:e1004977
36. Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A. et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67:882-91
37. Kawasaki M, Ikeda Y, Ikeda E, Takahashi M, Tanaka D, Nakajima Y. et al. Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer. 2021;127:512-9
38. Li Y, Wang HF, Li X, Li HX, Zhang Q, Zhou HW. et al. Disordered intestinal microbes are associated with the activity of Systemic Lupus Erythematosus. Clin Sci (Lond). 2019;133:821-38
39. Sun M, Wu W, Chen L, Yang W, Huang X, Ma C. et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9:3555
40. Pluznick JL. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int. 2016;90:1191-8
41. Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56-64
42. Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2016;12:169-81
43. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E. et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585-8
44. Rebholz CM, Zheng Z, Grams ME, Appel LJ, Sarnak MJ, Inker LA. et al. Serum metabolites associated with dietary protein intake: results from the Modification of Diet in Renal Disease (MDRD) randomized clinical trial. Am J Clin Nutr. 2019;109:517-25
45. Lobel L, Cao YG, Fenn K, Glickman JN, Garrett WS. Diet posttranslationally modifies the mouse gut microbial proteome to modulate renal function. Science. 2020;369:1518-24
46. Rocchetti MT, Di Iorio BR, Vacca M, Cosola C, Marzocco S, di Bari I. et al. Ketoanalogs' Effects on Intestinal Microbiota Modulation and Uremic Toxins Serum Levels in Chronic Kidney Disease (Medika2 Study). J Clin Med. 2021 10
47. Klingbeil E, de La Serre CB. Microbiota modulation by eating patterns and diet composition: impact on food intake. Am J Physiol Regul Integr Comp Physiol. 2018;315:R1254-r60
Author contact
![]() Corresponding author: I-Wen Wu, MD., Department of Nephrology, Chang Gung Memorial Hospital, Keelung, Taiwan. 222, Mai-Chin Road, Keelung 20401, Taiwan. Phone: 886-2-24313131-6211; Fax: +886-2-27191623; E-mail: fliawucom.
Corresponding author: I-Wen Wu, MD., Department of Nephrology, Chang Gung Memorial Hospital, Keelung, Taiwan. 222, Mai-Chin Road, Keelung 20401, Taiwan. Phone: 886-2-24313131-6211; Fax: +886-2-27191623; E-mail: fliawucom.

 Global reach, higher impact
Global reach, higher impact