3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(11):2438-2448. doi:10.7150/ijms.56457 This issue Cite
Review
Circular RNAs in stem cell differentiation: a sponge-like role for miRNAs
1. Department of Orthopedics, The Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, P. R. China.
2. Department of Orthopaedic Surgery, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, 250012, P. R. China.
3. Cheeloo College of Medicine, Shandong University, Jinan, Shandong, 250012, P. R. China.
4. Xiangya School of Medicine, Central South University, Changsha, Hunan, 410013, P. R. China.
5. Department of Dermatology, Xiangya Hospital, Central South University, Changsha, Hunan, 410008, P. R. China
*These authors contributed equally to this work.
Received 2020-11-26; Accepted 2021-4-9; Published 2021-4-22
Abstract
Circular RNAs (circRNAs) are novel endogenous non-coding RNAs that play a critical role during cellular signal transduction, gene transcription and translation. With the rapid advancement of bioinformatics analysis tools and high-throughput RNA sequencing, numerous circRNAs with important biological features have been identified. They function as competing endogenous RNAs (ceRNAs) of microRNAs and as such exhibit the potential to act as biomarkers for stem cell differentiation. In the recent past, several studies have shown the involvement of circRNAs in stem cells differentiation. The present review summarizes the molecular characteristics, biogenesis and mechanisms of newly identified circRNAs in the differentiation of stem cells. In conclusion, circRNAs regulate the stem cells differentiation via their ambient binding efficacy to modulate miRNA expression, as well as related gene translation. We believe that this review will provide reference guidance for future studies on stem cell differentiation.
Keywords: CircRNA, stem cell differentiation, miRNA, sponges, ceRNA
Introduction
Circular RNAs (CircRNAs) used to be misjudged as irrelevant during DNA transcription. However, with the development of RNA sequencing technology, circRNAs received increased attention [1-3]. Studies have shown that circRNAs are stable, abundant, cell-type specific and evolutionarily conserved [4, 5]. Some circRNAs can increase the complexity of RNA regulatory networks by acting as microRNA (miRNA) sponges, thereby suppressing the function of miRNAs in eukaryotic cells [3, 6-8]. There are other functions of circRNAs which include: 1) protein sponges that bind some proteins preventing them from binding to other targets [9]; 2) protein scaffolding which harbors binding sites for enzymes and substrates to facilitate their contact [10]; 3) splicing and transcription supporter that function in the nucleus to promote transcription or alternative splicing [1], and 4) translated into protein [11]. Among these functions mentioned above, the function of circRNAs as miRNA sponges is the most universal and most valued. The miRNAs regulate a battery of biological processes by binding to the 3'-untranslated regions (3'-UTR) of target messenger RNAs (mRNAs) to arrest their translation [12]. This form is mainly presented by promoting mRNA degradation via polyA tail shortening [13]. To inhibit the functions of miRNAs, antisense oligonucleotides or antagomirs were commonly used in vitro or in vivo [14, 15]. Recently, the theoretic hypothesis proposes that circRNAs bind to miRNAs to serve as competitive inhibitors and are thus referred as “miRNA sponges” that interrupt subsequent biological functions [2, 5]. The binding sites of circRNAs for miRNA exist either in the 3′-UTR or in the non-coding transcript of a specific gene [16]. Thousands of circRNAs have been predicted and identified by bioinformatics analysis and high throughput RNA sequencing in human genome. These studies have also indicated that circRNAs may serve as important post-transcriptional regulators [17, 18]. Recently, circRNAs downregulating the miRNA expression acting as miRNA sponges have been investigated in several different stem cells [19-22]. Cell differentiation causes the cells to change from a non-specific (non-specialized) state to a morphologically and functionally specific (specialized) state. This leads to the development of different types of cells with different phenotypic structures, and phenotypes which consequently form different tissues, organs, and systems to perform the complex functions of the body. During cell differentiation, totipotent or pluripotent stem cells gradually differentiate into mature cells and subsequently acquire a specific function, in which fluctuating expression of different genes play a vital role. In recent years, circRNAs have been more and more valued for their regulatory function on gene expression. A subset of circRNAs has been found to be enhanced in naive human ESCs (hESCs), for instance, circBIRC6 which acts as a sponge both for miR-34a and miR-145, involved in curbing the hESCs differentiation [22]. Additionally, during the differentiation of stem cells into mature cells, such as myocardial differentiation, epithelial differentiation, neuronal differentiation and osteogenic differentiation, the expression of some of the circRNAs have been found to remarkably elevated and many of these were derived from developmental genes [22-25].
Collectively, a large number of studies have revealed a sponge-like role for circRNAs, which prevents the expression of target miRNA consequently regulating the activity of downstream genes [23, 24]. Herein, we review the biogenesis, characterization and classification of circRNA and explore the relationship between miRNA expression and circRNA in stem cells differentiation.
Biogenesis of circRNAs
CircRNAs differ from other RNA species in that the 3' and 5' ends are covalently linked. Typical splicing signals are located on both sides of the end junctions of circRNAs indicative of the involvement of the splicing bodies in the generation of circRNAs and in constitutive and selective linear RNA splicing [25]. CircRNAs are formed by back-splicing [26]. According to a previous study, there are 6 models that explain the formation of circRNAs; A. cyclization driven by tRNA splicing [27], B. direct cyclization of lariat introns [26], C. cyclization mediated by RNA-binding proteins (RBPs) [28], D. cyclization driven by lariat mechanism [26], E. cyclization mediated by intron pairing [1, 26], F. cyclization driven by rRNA splicing [29].
CircRNAs as sponges of miRNAs in stem cells
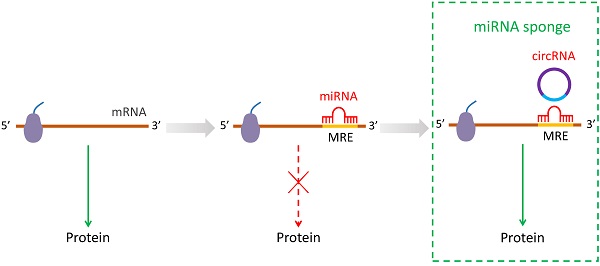
The most significant role of circRNAs is that circRNAs can repress the function of miRNAs by binding them as competing endogenous RNAs (ceRNAs) [16, 26, 30]. It has been shown that circRNAs have multiple miRNA response elements (MREs) [3]. A previous study has identified over 60 conserved seed match segments on miR-7 for circRNAs sponge suggestive of a very dense combination [3]. The interactions between circRNAs with miRNAs by sponges are recognized gradually during cellular living activities [23, 24]. The function of circRNAs is dependent on the cellular localization. CircRNAs turn into large RNA-protein complexes by binding to RBPs in the nucleus, which in turn may affect the transcription of mRNAs [31, 32]. Moreover, circRNAs also exhibit regulatory role in gene expression through linear pre-mRNA splicing competition. For example, circMbl sponges to muscle blind (MBL) by regulating the splicing of its own pre-mRNA into a translatable mRNA or circMbl [33]. It is known that circRNAs can participate in many biological processes, while many functions remain yet to be elucidated. Almost all known circRNAs display sponge activity that localize to the cytoplasm [16]. Hitherto, accumulating evidence suggest that circRNAs regulate the development and progression of stem cells differentiation by suppressing miRNA species (Figure 1).
Identified circRNAs in stem cells differentiation
Recent evidence has shown that circRNAs are associated with the differentiation of several types of stem cells, such as dental pulp stem cells (DPSCs), maxillary sinus membrane stem cells (MSMSCs), periodontal ligament stem cells (PDLSCs), embryonic stem cells (ESCs), bone marrow-derived mesenchymal stem cells (BMSCs), induced pluripotent stem cells (iPSCs), epidermal stem cells (EpSCs), germline stem cells (GSCs), intestinal stem cells (ISCs) and hematopoietic stem cells (HSCs).
CircRNAs in the differentiation of dental pulp stem cells
Dental pulp stem cells (DPSCs) are present in the dental pulp and exhibit higher proliferation rates, self-renewal and cloning potential due to its vested abilities. Recent studies have identified that circRNAs play a critical role in the involvement in the differentiation of DPSCs. From these investigations, circRNA124534 has been reported to promote osteogenic differentiation as a miR-496 sponge in human DPSCs. Additionally, circRNA124534/miR-496/β-catenin axis has been demonstrated as a covert curative clue for elevation of dental bone remodeling [34]. Moreover, circRNA hsa_circ_0026827 has also been reported to play a sponging role by targeting miR-188-3p via Beclin1 and RUNX1 signaling pathways that enhances the osteogenic differentiation in human DPSCs [35]. In yet another study, circRNA SIPA1L1 was found to regulate miR-617/Smad3 axis to potentiate osteogenic differentiation of DPSCs [20]. Furthermore, role of exosomes in osteogenic differentiation of DPSCs has been investigated and it was found that circLPAR1 adsorbs to hsa-miR-31 to avoid its inhibitory effects on osteogenesis of DPSCs [36]. A microarray analysis revealed 43 upregulated circRNAs and 144 downregulated circRNAs to be differentially expressed in human DPSCs during odontogenic differentiation process. Moreover, hsa_circRNA_104101 was highly assumed to enhance the odontogenic differentiation of human DPSCs [37]. Based on above, it is well established that the common mechanisms of circRNAs in DPSCs are competently binding with miRNAs to deliver a promoting effect on osteogenic differentiation (Figure 2A).
The miRNA sponging by circRNA during post-transcriptional process.
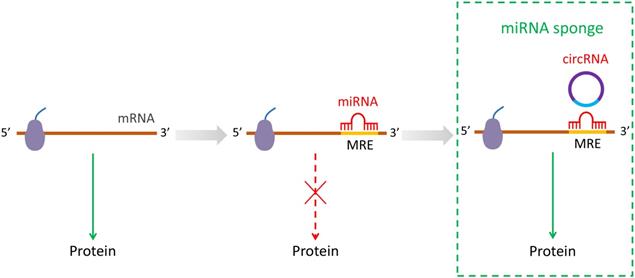
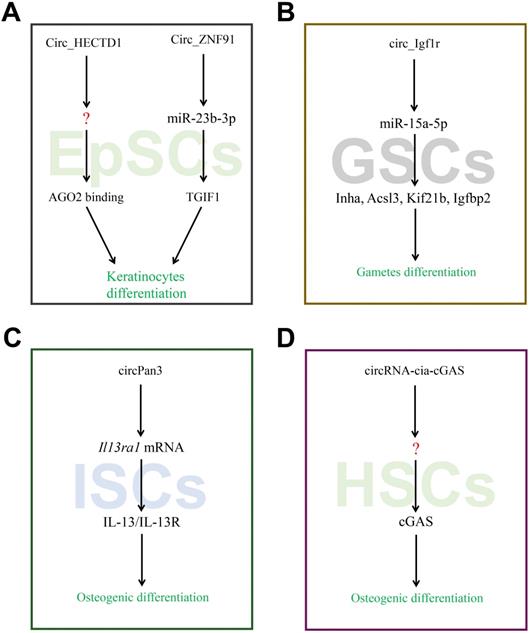
CircRNAs associated with stem cells differentiation. (A-D) CircRNAs affected its target miRNAs via miRNA sponge which reduce the function of translation in DPSCs, MSMSCs, PDLSCs and EMCs.
CircRNA in maxillary sinus membrane stem cells differentiation
Maxillary sinus membrane stem cells (MSMSCs) exhibit osteogenic potential and may find their use in bone repair of maxilla. By using microarray analysis, Peng et al. revealed that the novel pathway circRNA_33287/miR-214-3p/Runx3 plays a role in the regulation of the osteoblastic differentiation in MSMSCs. Additionally, BMP2 may also induce the osteogenic differentiation by enhancing the expression of circRNA_33287 [38]. However, only a few studies have been carried out on this aspect of circRNAs in MSMSCs and therefore the underlying mechanisms remain largely elusive (Figure 2B).
CircRNAs in periodontal ligament stem cells osteogenic differentiation
Gu et al. indicated that in the phase of periodontal ligament stem cells (PDLSCs) osteogenic differentiation, recognition and disposal of circRNAs plays an imperative role in ceRNAs. They used RNA sequencing and found 1456 circRNAs to be differentially expressed in normal and osteogenic inductive PDLSCs. Furthermore, 1382 circRNAs were found to combine with 148 common miRNAs, which were predicted to be involved in osteoblast differentiation [39]. By acting as miR-7 sponge, circRNA-CDR1as has been reported to mediate the inhibitory effect of LPS on proliferation of PDLSCs by activating the ERK signaling pathway [39]. Importantly, circRNA-CDR1as was also reported to regulate the osteoblastic differentiation of PDLSCs by triggering the activation of Smad and p38 MAPK signaling pathway, as well as upregulation of GDF5 [40]. The circRNA-CDR1as/miR-7 axis may be involved in PDLSCs upon periodontitis burst. Moreover, Zheng et al. ascertained that hsa_circ_0003489 is located at cyclin-dependent kinase 8 (CDK8) gene so they referred it as circCDK8 [41]. Meanwhile, the overexpression of circCDK8 was found to induce autophagy and apoptosis, whereas the silencing of circCDK8 reversed the inhibitory effects of cobalt chloride (CoCl2) on osteogenic differentiation of PDLSCs. Hereof, Zheng et al, pointed out that circCDK8 inhibits the osteogenic differentiation of PDLSCs in a hypoxic environment [42]. In addition, the mechanical force may induce the changes in the expression of circRNAs in PDLSCs with regard to miRNAs transformation [42]. A previous study showed that miRNA146a exerted its unique role by post-transcriptionally modulating the expression levels of the target genes to regulate the PDLSC osteogenic differentiation [43]. Moreover, in another study Chen et al. [44] showed that miRNA-34a inhibits the differentiation of osteoblasts. Interfering with miRNA34a and miRNA146a as we inferred, circRNA BANP coupled with circRNA ITCH could modulate PDLSC osteogenic differentiation through MAPK pathway. DUSP1, FAS and RAC1 are targeted by miRNA34a, while as PDGFRA, TGFBR2 and MYC are targeted by miRNA146a. Kovacić et al. showed that Fas/Fas ligand system inhibits the differentiation of murine osteoblasts [45]. The inhibition of RAC1 has been reported to promote the BMP-2-induced osteoblastic differentiation [46]. Circ_BANP/circ_ITCH-miR-34a-FAS/RAC1 may be an axial regulator of osteoblast differentiation. Zheng et al examined the expression profile of circRNAs, miRNAs and mRNAs during the osteogenic differentiation of PDLSCs, and a network containing potential targeting relationships was constructed [47]. A previous report indicated that miR-204 inhibits the osteogenesis and promotes adipogenesis of mesenchymal stem cells by targeting Runx2 [48]. In the regulatory aspect of osteogenesis and chondrogenesis, RUNX2 has been regarded as a pivotal transcription factor [49]. Circ_2929 and circ_ARHGAP35-miR-204-5p-Runx2 may be potential axial regulators of osteoblast differentiation (Figure 2C).
CircRNAs in the differentiation of embryonic stem cells
Using RNA-sequencing analysis, a total of 3,894 circRNAs within the body of 2,097 known genes were identified expressed in murine embryonic stem cells (ESCs) derived motor neurons (MNs) [50]. Analysis from studies showed that most of them having a homologous trend like the annular structures, but the circ_5906 and circ_2193 were counter examples. According to Lorenzo Errichelli et al reported that the expression of circ_5906 upregulates during the mESCs differentiation, whereas circ_2193 was found to be downregulated [50]. Interestingly, the RNA-binding protein FUS-dependent linear correlations were highly presented in mESCs and down-regulated under differentiation. Both of them may play an important role in preventing the murine embryonic stem cell differentiation. Intriguingly, several candidate circRNAs were identified to play crucial roles in neuronal differentiation in mESCs and circZNF827 was reported to be negatively regulated in this process through nerve growth factor (NGF) signaling [51].
Human embryonic stem cells (hESCs) with the characteristics of unlimited self-renewal and pluripotency are studies on first priority for applicability [52]. It is known that miR-34a and miR-145 can promote in vitro differentiation of hESCs by repressing pluripotency-associated genes [22]. Yu et al. identified a subgroup of circRNAs that are enriched in undifferentiated hESCs [22]. Additionally, circBIRC6 is associated with the pluripotent state. Moreover, it was found that circBIRC6 interacts with miR-34a and miR-145 and is enriched in the AGO2 complex. Correspondingly, circBIRC6 suppresses hESCs differentiation by down-regulating the expression of miR-34a and miR-145. Furthermore, the biogenesis of circBIRC6 in hESCs was found to be regulated by the hESC-enriched splicing factors (SFs) ESRP1. The expression of ESRP1 was controlled by the core pluripotency-associated factors, NANOG and OCT4. CircRNAs can suppress the hESCs differentiation by serving as a microRNA “sponge” (Figure 2D).
CircRNAs in bone marrow-derived mesenchymal stem cells osteoblast differentiation
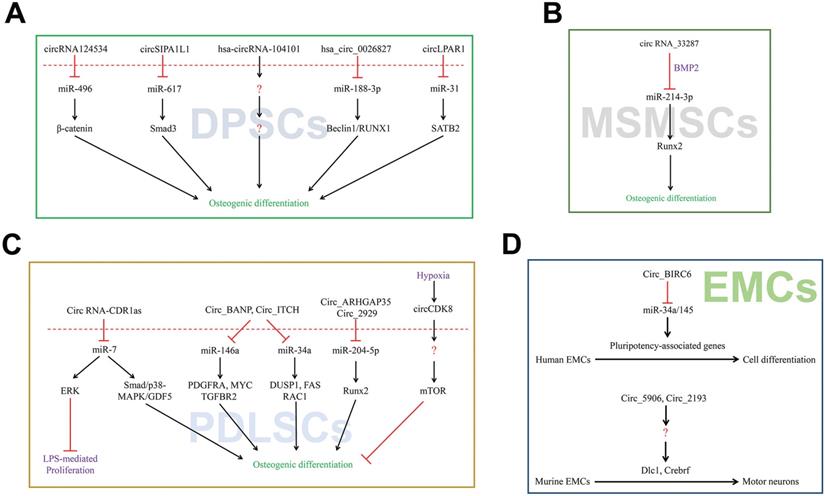
The proliferation and differentiation of bone marrow-derived mesenchymal stem cells (BMSCs) into osteoblasts is first stage for osteogenesis. In presence of calcitonin gene-related peptide (CGRP) stimulation, the expression of circRNA_003795 was significantly increased to promote proliferation and FOS like 2 AP-1 transcription factor subunit (FOSL2) expression, whereas slicing it dramatically up-regulated the miR-504-3p expression. These results indicated that circRNA_003795 indirectly regulates FOSL2 expression via sponging of miR-504-3p to promote BMSCs proliferation which prepared for further differentiation [53]. The milieu of BMSCs was also found to regulate its differentiation by influencing the expression of numerous circRNAs. As ceRNAs, circRNAs have been detected to exhibit variable expression in BMSCs on the different materials surface [54]. Recently, our study also discovered hsa_circ_0074834 to act as a ceRNA to regulate the expression of ZEB1 and VEGF via microRNA-942-5p to promote osteogenic differentiation of BMSCs which is advantageous to bone remodeling [55]. Opposite regulation of circRNAs with miRNAs was reported in the early osteogenic differentiation of BMSCs, and masked genes (FLBN1, MT2A, and BSCL2) were given rise to attention during this part [56]. The adipogenic or osteogenic differentiation of BMSCs has been reported to be associated with the pathogenesis of steroid-induced osteonecrosis in femoral head. A recently carried out study identified circRNA CDR1as promotes the development of adipogenic differentiation which dampens the osteogenic differentiation in BMSCs via CDR1as-miR-7-5p-WNT5B axis providing new insights into the treatment of related diseases (Figure 3A) [57].
Previous studies showed that ERβ plays an important role in the osteogenesis and trigger MC3T3-E1 cells to undergo osteogenic differentiation [58]. However, little is known about the relationship between circRNAs and bone metabolism. In a recent study, RNA-Seq was performed to detect the differentially expressed circRNAs between control and estrogen receptor beta (ERβ) deficient rat BMSCs. It was found that up to 146 cases of circRNAs were diversely expressed by the range of fold-change 2.0 (p ≤ 0.05). Moreover, a total of 68 circRNAs were down-regulated, while 78 were found to be up-regulated [59]. Li et al constructed circRNA-microRNA network to predict the miRNA sponges. This study showed that ERβ may regulate osteoblast differentiation via miR-328-5p-mRNA axis. Previous study indicated that miR-328 up-regulates C/EBPα expression to inhibit cell proliferation [59]. Taken together, circRNAs-miRNAs network plays an important role in the regulation of osteogenic differentiation and C/EBPα may be considered as a mediator during this process. RNA-seq was performed with the aim to explore differential expression of circRNAs in osteoblast differentiation using particular cells named MC3T3-E1 [60]. A total of 158 circRNAs were differentially expressed between BMP2-treated MC3T3-E1 cells and untreated control cells, and among them 74 were up-regulated while 84 were down-regulated. The expression of circRNA.5846, circRNA.19142 and circRNA.10042 was confirmed to be dramatically up-regulated in the BMP2-treated cells. BMP2 was found to enhance the bone formation and remodeling by regulating the transcription of osteogenic genes both in osteoblast and BMSCs. Bioinformatic analysis of circ_19142 and circ_5846 showed Ywhaz, Plcg1, Src, Thbs1, Ncor2, Rps6kb1, Pdk1, Xiap and Prkci are potential target genes related to FGF, EGF, PDGF and Wnt pathways during this process. The circ_19142/circ_5846-miR-7067-5p-mRNAs axis may participate in the regulation of osteoblast differentiation (Figure 3B).
CircRNAs in induced pluripotent stem cells differentiation
Induced pluripotent stem cells (iPSCs) are the reversion of somatic cells differentiation and cell reprogramming. Several circRNAs have been shown to participate in this process. Yu et al reported that circRNA BIRC6 reprograms the somatic cells to human iPSCs by directly acting as a sponge for miRNA involved in the maintenance of the pluripotent state [22]. Another circRNA, circFOXP1, has been shown to be downregulated in mesenchymal stem cells when reprogramming to pluripotency [61]. The circular RNA map for human iPSCs of fetal origin was also provided and it may highly benefit from clinical research [62]. Zhang et al demonstrated that circRNAs upregulate the expression levels of exogenous octamer-binding protein 4 (OCT4) by sponging regulatory microRNAs in iPSCs [63]. Overall, circRNAs act as ceRNA for miRNAs that play a critical role during cellular reprogramming.
CircRNAs associated with stem cells differentiation. (A-C) CircRNAs affected its target miRNAs via miRNA sponge which reduce the function of translation in BMSCs, MC3T3-E1 and iPSCs.
Previous studies have revealed a close linkage between alternations in ribosome and proliferation, cell development and differentiation. Siede et al used RNA-seq to detect 320 differentially expressed circRNAs in human dilated cardiomyopathy (DCM) and control patients [64]. The same dynamics of circRNAs from ATXN10, CHD7, DNAJC6 and SLC8A1 were also validated. The rodent homologs of circ_MYOD, circ_SLC8A1, circ_ATXN7 and circ_PHF21A interact with the ribosome during human induced pluripotent stem cell derived cardiomyocytes (hiPSC-CMs) differentiation, which are highly relevant in cardiac dysfunction. Functional studies are required to further clarify the role of these circRNAs refer to their host genes in pathophysiology (Figure 3C).
CircRNAs in epidermal stem cells differentiation
The RNA-seq was used to detect 624 unique circRNAs in the epidermal stem cells (EpSCs) and differentiated keratinocytes [65]. It was found that the differentiated cells exhibit higher expression of circRNAs than the undifferentiated cells, many of which were derived from developmental genes. The discovered changes in circRNA expression were largely different from the expression of host gene. On the other hand, compared to stably expressed circRNAs, circRNAs which were up-regulated upon differentiation have stronger tendency to AGO2 binding and more promising miRNA combination sites. In particular, up--regulated circRNAs from the ZNF91 and HECTD1 genes have predicted miRNA target sites and exceptionally high numbers of AGO2 binding sites. Moreover, circ_ZNF91 contains 24 target sites for miR-23b-3p. It has been reported that circZNF91 functions as a miRNA sponge in human cells [65]. Barbollat-Boutrand L et al. showed that mi-RNA-23b-3p regulates human keratinocyte differentiation via activation of the TGF-β-SMAD2 signaling pathway and repression of TGIF1 [66]. In yet another study, Circ_ZNF91-miR-23b-3p-TGIF1 axis has been reported to regulate the differentiation of EpSC (Figure 4A).
CircRNAs associated with stem cells differentiation. (A-D) CircRNAs affected its target miRNAs via miRNA sponge which reduce the function of translation in EpSCs, GSCs, ISCs and HSCs.
CircRNA in germline stem cells differentiation
A study used high-throughput sequencing to identify the circRNAs in germline stem cells and the expression was further confirmed using RT-PCR [21, 67]. Among them, 921 circRNAs showed sex-biased expression, which may reveal a vast amount of circRNA involved in diverse genetic imprinting between female and male germline stem cells. The circ_Igf1r can increase target gene Kif21b, Acsl3, Igfbp2 and Inha expressions by binding competitively with miRNA-15a-5p. A previous study indicated that circ_Igf1r acts as the ceRNAs of miR-15a-5p that targets the Kif21b, Acsl3, Igfbp2 and Inha mRNAs. Liu et al. showed that miRNA-15a-5p regulates VEGFA during endometrial mesenchymal stem cell differentiation. Kageyama A et al. identified ACSL3 as mediator of PA-induced osteoblastic differentiation [68]. In another study, Xi et al. found that IGFBP-2 can directly stimulate osteoblast differentiation [69]. Collectively, Circ_Igf1r-miRNA-15a-5p-Inha, Acsl3, Kif21b, and Igfbp2 may act as axis regulators of germline stem cells differentiation (Figure 4B).
CircRNA in intestinal stem cells differentiation
Intestinal stem cells (ISCs) play a critical role in the repair of intestinal mucosal barrier by precise self-renewal and differentiation under the normal circumstances. Recent study showed that circRNA circPan3 is highly expressed in mouse and human ISCs. It binds to Il13ra1 mRNA in ISCs to promote their stability, and is necessary for the self-renewal and differentiation of ISCs via IL-13/IL-13R-mediated signaling pathway [70]. However, it is still unknown whether the circRNAs/miRNAs interactions exist during this process (Figure 4C).
CircRNA in hematopoietic stem cells differentiation
Hematopoietic stem cells (HSCs) located in the bone marrow are adult stem cells of the blood system which exhibit the ability of long-term self-renewal and the potential to differentiate into various mature blood cells. Its aberrant form has been linked to the occurrence of hematological cancers. Benoit P Nicolet et al examined the expression pattern of circRNAs in human HSCs and found that circRNAs expression is cell-type specific as well as circRNAs splicing variants, and increase when cell maturates [71]. Previously, Xia et al reported that a circRNA cia-cGAS to be highly expressed in the nucleus of long-term HSCs which binds to cGAS and subsequently inactivates the enzymatic activity of cGAS to protect dormant long-term HSCs from cGAS-mediated recognition, autoimmune response and exhaustion [72]. Overall, the miraculous expression pattern of circRNAs in HSCs may manifest the vital role during hematopoiesis (Figure 4D).
Discussion
Recently, circRNAs and stem cell have emerged as a new filed of research. Previous studies reported various methods to investigate the functions of circRNAs. Gene knockdown strategies can be used for functional study. We can identify target circRNAs via RNA sequencing and microarray. Quantitative real-time PCR, western blot, FISH and northern blot can be used to detect circRNA expression. Bioinformatics, immunohistochemical, and over-expression studies can be used to decipher the mechanisms. Furthermore, there are several databases or online services have been developed to provide the prediction or basic information about circRNAs and their potential regulatory networks (Table 1) [6, 73-80].
However, the research of circRNAs in stem cells differentiation is still in the infancy. CircRNAs used to be considered as errors in RNA splicing. However, they are now regarded as stable, abundant, active and ubiquitously expressed RNA molecules. CircRNAs play a remarkable role in translation, act as sponge and regulatory molecules and also may be utilized as reliable biomarkers. Meanwhile, circRNAs usually regulate miRNA expression via harboring MREs [81]. The interaction of circRNA-miRNA can act through down-regulation transcriptional activity of miRNA target genes. Among them many miRNA target genes have been reported to play a significant role in stem cells differentiation, such as miR-34a and miR-146a. The miR-34a is the target gene of different circRNAs and plays an important role in the differentiation of hESCs and PDLSCs. In future, we can explore more connections between circRNAs and differentiation via those miRNAs. Gene silencing for this type of miRNAs may enable the achievement of the target and cause strong side effects in clinical applications. Nonetheless, the sponge effect of miRNAs on the target gene can be better targeted by specific circRNA. This point may provide a new way for cell experiments and animal models. The importance of circRNA dysregulation has been shown to be associated with the differentiation of different stem cells. In the present article we reviewed a novel mechanism by which circRNAs regulate miRNAs at the post-transcriptional level and we believe that it will provide insights into various processes associated with the stem cell differentiation. Additionally, the studies on miRNAs and circRNAs may enable the development of reliable diagnostic biomarkers and potential therapeutic targets for nonunion patients.
Online services of circRNA
| Name | Website | Description |
|---|---|---|
| StarBase v2.0 | http://starbase.sysu.edu.cn/ | To predict miRNA-circRNA interactions |
| CircBase | http://www.circbase.org | To explore public circRNA datasets |
| Tissue-Specific CircRNA Database | http://gb.whu.edu.cn/TSCD/ | To provide a global view for tissue-specific circRNA in main tissues of human and mouse |
| MiOncoCirc | https://mioncocirc.github.io/ | Query the expression of a certain circRNA in different cancer clinical samples |
| ExoRBase | http://www.exoRBase.org | To trigger new circulating biomarker discovery and functional implication for human diseases. |
| CirclncRNAnet | http://app.cgu.edu.tw/circlnc/ | To view the expression information of database samples according to gene names. |
| TRCirc | http://www.licpathway.net/TRCirc/view/index | To search and browse TFBSs of circRNAs, and other related information, for specific TFs, cell lines or circRNAs of interest |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | To collect circRNA databases that are potentially associated with human diseases or traits |
| CircRNADisease | http://cgga.org.cn:9091/circRNADisease/ | To explore circRNA and disease associations |
| DeepBase v2.0 | http://rna.sysu.edu.cn/deepBase/ | To annotate and discover small (microRNAs), lncRNAs and circRNAs from next generation sequencing data |
| CircRNADb | http://reprod.njmu.edu.cn/circrnadb | A database for annotating exonic circRNAs, and can be a valuable resource for large-scale studies of circRNA |
| RegRNA 2.0 | http://regrna2.mbc.nctu.edu.tw/index.html | To identify functional RNA motifs in an input RNA sequence |
| MiRWalk 3.0 | http://mirwalk.umm.uni-heidelberg.de/ | To predict and experimentally verify miRNA-target interactions with various novel and unique features |
| CircNet | http://circnet.mbc.nctu.edu.tw/ | Identification of new circRNAs and integration of circRNA-miRNA-mRNA interaction network. |
| Cirbank | http://www.circbank.cn/help.html | To analyze the coding potential of circular RNA proteins |
| CircFunBase | http://bis.zju.edu.cn/CircFunBaseBlast/ | To quickly query the name and function introduction of circleRNA |
| CircAtlas | http://circatlas.biols.ac.cn/ | To use GO and KEGG databases to predict the potential functions of these circRNAs |
| BIOINF | http://www.bioinf.com.cn/ | CircRNA primer design |
| Cancer-specific circRNA database | http://gb.whu.edu.cn/CSCD/ | To find circRNA based on tumor cells |
| CIRCpedia | http://www.picb.ac.cn/rnomics/circpedia | To annotate alternative back-splicing and alternative splicing in circRNAs across different cell lines |
| Circular RNA Interactome | https://circinteractome.nia.nih.gov/ | To predict and map the binding sites for RBPs and miRNAs on reported circRNAs |
Conclusions and perspectives
Taken together, circRNAs are confirmed to play a critical role in stem cell differentiation by regulation the miRNA functions via their sponging activity. The interactions between circRNAs and miRNAs may activate downstream signaling to promote stem cell differentiation. We hypothesize that whether circRNAs with potential sponging function is related to existence of each miRNAs. Furthermore, we conclude that the role of osteogenic promotion by circRNAs is the major part during the differentiation of several stem cells, such as DPSCs, MSMSCs, PDLSCs and BMSCs. Interventions targeting relevant circRNAs may have potential to regulate stem cells differentiation. Ultimately, we propose several regulatory axes such as circRNA_33287/miR-214-3p/Runx3, Circ_2929-circ_ARHGAP35/miR-204-5p/Runx2, circ_BANP/circ_ITCH-miR-34a-FAS/RAC1, and circ_19142/circ_5846-miR-7067-5p-mRNAs as vital players during stem cells osteogenic differentiation.
Abbreviations
CircRNAs: Circular RNAs; DPSCs: dental pulp stem cells; ESCs: embryonic stem cells; BMSCs: bone marrow-derived mesenchymal stem cells; PDLSCs: periodontal ligament stem cells; PSCs: pluripotent stem cells; EpSCs: epidermal stem cells; GSCs: germline stem cells; ADSCs: adipose-derived mesenchymal stem cells; MSMSCs: maxillary sinus membrane stem cells; ISCs: intestinal stem cells; HSCs: hematopoietic stem cells; miRNAs: microRNAs; 3'-UTR: 3ʹ-untranslated regions; mRNAs: messenger RNAs; RBPs: RNA-binding proteins; ceRNAs: competing endogenous RNAs; MREs: miRNA response elements; RPCs: RNA-protein complexes; MNs: motor neurons; NGF: nerve growth factor; SFs: splicing factors; CGRP: calcitonin gene-related peptide; FOSL2: FOS like 2 AP-1 transcription factor subunit; ERβ: estrogen receptor beta; CDK8: cyclin-dependent kinase 8; HIF-1α: hypoxia-inducible factor-1α; CoCl2: cobalt chloride; RNA-seq: RNA sequencing; DCM: dilated cardiomyopathy; hiPSC-CMs: human induced pluripotent stem cell derived cardiomyocytes.
Acknowledgements
The authors would like to thank all the reviewers who participated in the review and MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities of Central South University (Grant No. 2018zzts930), the Scientific Research Project of Hunan Health Committee (Grant No. 20200357), the Central South University Sports Medicine Scholarship, the National College Students' Innovation and Entrepreneurship Training Program (Grant No. 201710422116) and the National Natural Science Foundation of China (Grant No. 81000821 and 81672176), the Natural Science Foundation of Hunan Province, China (Grant No. 2018JJ2565) and the National Science Foundation for Post-doctoral Scientists of China (Grant No. 2017M622601).
Authors' contributions
JZ, CQ and ZHF drafted the manuscript. TYL and TL provided suggestions and edited the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
2. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
3. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
4. Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T. et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932-9
5. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-52
6. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666-70
7. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777
8. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838-47
9. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
10. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z. et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357-70
11. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37.e9
12. Izaurralde E. GENE REGULATION. Breakers and blockers—miRNAs at work. Science. 2015;349:380-2
13. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51
14. Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR. et al. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:5898-903
15. Patutina OA, Bichenkova EV, Miroshnichenko SK, Mironova NL, Trivoluzzi LT, Burusco KK. et al. miRNases: Novel peptide-oligonucleotide bioconjugates that silence miR-21 in lymphosarcoma cells. Biomaterials. 2017;122:163-78
16. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42-51
17. Li H, Hao X, Wang H, Liu Z, He Y, Pu M. et al. Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cell Physiol Biochem. 2016;40:1334-44
18. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
19. Chia W, Liu J, Huang YG, Zhang C. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020;11:372
20. Ge X, Li Z, Zhou Z, Xia Y, Bian M, Yu J. Circular RNA SIPA1L1 promotes osteogenesis via regulating the miR-617/Smad3 axis in dental pulp stem cells. Stem Cell Res Ther. 2020;11:364
21. Li X, Tian GG, Zhao Y, Wu J. Genome-wide identification and characterization of long noncoding and circular RNAs in germline stem cells. Sci Data. 2019;6:8
22. Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY. et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149
23. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
24. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-31
25. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14:1035-45
26. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
27. Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14:978-84
28. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
29. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829-42
30. Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J. et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics. 2021;11:2755-69
31. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440-1
32. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
33. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
34. Ji F, Pan J, Shen Z, Yang Z, Wang J, Bai X. et al. The Circular RNA circRNA124534 Promotes Osteogenic Differentiation of Human Dental Pulp Stem Cells Through Modulation of the miR-496/β-Catenin Pathway. Front Cell Dev Biol. 2020;8:230
35. Ji F, Zhu L, Pan J, Shen Z, Yang Z, Wang J. et al. hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells Through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p. Front Cell Dev Biol. 2020;8:470
36. Xie L, Guan Z, Zhang M, Lyu S, Thuaksuban N, Kamolmattayakul S. et al. Exosomal circLPAR1 Promoted Osteogenic Differentiation of Homotypic Dental Pulp Stem Cells by Competitively Binding to hsa-miR-31. Biomed Res Int. 2020;2020:6319395
37. Chen M, Yang Y, Zeng J, Deng Z, Wu B. circRNA Expression Profile in Dental Pulp Stem Cells during Odontogenic Differentiation. Stem Cells Int. 2020;2020:5405931
38. Peng W, Zhu S, Chen J, Wang J, Rong Q, Chen S. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother. 2019;109:1709-17
39. Wang F, Chen X, Han Y, Xi S, Wu G. circRNA CDR1as Regulated the Proliferation of Human Periodontal Ligament Stem Cells under a Lipopolysaccharide-Induced Inflammatory Condition. Mediators Inflamm. 2019;2019:1625381
40. Li X, Zheng Y, Zheng Y, Huang Y, Zhang Y, Jia L. et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9:232
41. Zheng J, Zhu X, He Y, Hou S, Liu T, Zhi K. et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci. 2020
42. Wang H, Feng C, Jin Y, Tan W, Wei F. Identification and characterization of circular RNAs involved in mechanical force-induced periodontal ligament stem cells. J Cell Physiol. 2019;234:10166-77
43. Hung PS, Chen FC, Kuang SH, Kao SY, Lin SC, Chang KW. miR-146a induces differentiation of periodontal ligament cells. J Dent Res. 2010;89:252-7
44. Chen L, Holmstrøm K, Qiu W, Ditzel N, Shi K, Hokland L. et al. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. 2014;32:902-12
45. Kovacić N, Lukić IK, Grcević D, Katavić V, Croucher P, Marusić A. The Fas/Fas ligand system inhibits differentiation of murine osteoblasts but has a limited role in osteoblast and osteoclast apoptosis. J Immunol. 2007;178:3379-89
46. Kim JH, Kim K, Kim I, Seong S, Nam KI, Kim KK. et al. Adaptor protein CrkII negatively regulates osteoblast differentiation and function through JNK phosphorylation. Exp Mol Med. 2019;51:1-10
47. Zheng Y, Li X, Huang Y, Jia L, Li W. The Circular RNA Landscape of Periodontal Ligament Stem Cells During Osteogenesis. J Periodontol. 2017;88:906-14
48. Jiang X, Zhang Z, Peng T, Wang G, Xu Q, Li G. miR-204 inhibits the osteogenic differentiation of mesenchymal stem cells by targeting bone morphogenetic protein 2. Mol Med Rep. 2020;21:43-50
49. Su YW, Wong DSK, Fan J, Chung R, Wang L, Chen Y. et al. Enhanced BMP signalling causes growth plate cartilage dysrepair in rats. Bone. 2021;145:115874
50. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D. et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741
51. Hollensen AK, Thomsen HS, Lloret-Llinares M, Kamstrup AB, Jensen JM, Luckmann M. et al. circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. Elife. 2020 9
52. Piao J, Zabierowski S, Dubose BN, Hill EJ, Navare M, Claros N. et al. Preclinical Efficacy and Safety of a Human Embryonic Stem Cell-Derived Midbrain Dopamine Progenitor Product, MSK-DA01. Cell Stem Cell. 2021;28:217-29.e7
53. Ren W, Yang L, Deng T, Wu C, Li Y, Wu J. et al. Calcitonin gene-related peptide regulates FOSL2 expression and cell proliferation of BMSCs via mmu_circRNA_003795. Mol Med Rep. 2019;19:3732-42
54. Zhu S, Zhu Y, Wang Z, Liang C, Cao N, Yan M. et al. Bioinformatics analysis and identification of circular RNAs promoting the osteogenic differentiation of human bone marrow mesenchymal stem cells on titanium treated by surface mechanical attrition. PeerJ. 2020;8:e9292
55. Ouyang Z, Tan T, Zhang X, Wan J, Zhou Y, Jiang G. et al. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019;10:932
56. Della Bella E, Menzel U, Basoli V, Tourbier C, Alini M, Stoddart MJ. Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells. 2020 9
57. Chen G, Wang Q, Li Z, Yang Q, Liu Y, Du Z. et al. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone. 2020;133:115258
58. Yin X, Wang X, Hu X, Chen Y, Zeng K, Zhang H. ERβ induces the differentiation of cultured osteoblasts by both Wnt/β-catenin signaling pathway and estrogen signaling pathways. Exp Cell Res. 2015;335:107-14
59. Cao JX, Wen F, Wang H, Liao P, Li JJ. [miR-328 Inhibits K562 Cell Proliferation by Up-regulation of C/EBPα Expression]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2015;23:350-5
60. Qian DY, Yan GB, Bai B, Chen Y, Zhang SJ, Yao YC. et al. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed Pharmacother. 2017;90:492-9
61. Cherubini A, Barilani M, Rossi RL, Jalal MMK, Rusconi F, Buono G. et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019;47:5325-40
62. Barilani M, Cherubini A, Peli V, Polveraccio F, Bollati V, Guffanti F. et al. A circular RNA map for human induced pluripotent stem cells of foetal origin. EBioMedicine. 2020;57:102848
63. Zhang X, Zhang J, Zheng K, Zhang H, Pei X, Yin Z. et al. Long noncoding RNAs sustain high expression levels of exogenous octamer-binding protein 4 by sponging regulatory microRNAs during cellular reprogramming. J Biol Chem. 2019;294:17863-74
64. Siede D, Rapti K, Gorska AA, Katus HA, Altmüller J, Boeckel JN. et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. 2017;109:48-56
65. Kristensen LS, Okholm TLH, Venø MT, Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280-91
66. Barbollat-Boutrand L, Joly-Tonetti N, Dos Santos M, Metral E, Boher A, Masse I. et al. MicroRNA-23b-3p regulates human keratinocyte differentiation through repression of TGIF1 and activation of the TGF-ß-SMAD2 signalling pathway. Exp Dermatol. 2017;26:51-7
67. Li X, Ao J, Wu J. Systematic identification and comparison of expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in mouse germline stem cells. Oncotarget. 2017;8:26573-90
68. Kageyama A, Matsui H, Ohta M, Sambuichi K, Kawano H, Notsu T. et al. Palmitic acid induces osteoblastic differentiation in vascular smooth muscle cells through ACSL3 and NF-κB, novel targets of eicosapentaenoic acid. PLoS One. 2013;8:e68197
69. Xi G, Wai C, DeMambro V, Rosen CJ, Clemmons DR. IGFBP-2 directly stimulates osteoblast differentiation. J Bone Miner Res. 2014;29:2427-38
70. Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y. et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20:183-94
71. Nicolet BP, Engels S, Aglialoro F, van den Akker E, von Lindern M, Wolkers MC. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018;46:8168-80
72. Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z. et al. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity. 2018;48:688-701.e7
73. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277-87
74. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984-92
75. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34-42
76. Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL. et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196-202
77. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-7
78. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985
79. Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4
80. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839-47
81. Han C, Seebacher NA, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622-37
Author contact
![]() Corresponding author: Tang Liu, Department of Orthopedics, The Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, China. E-mail: liutang1204edu.cn.
Corresponding author: Tang Liu, Department of Orthopedics, The Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, China. E-mail: liutang1204edu.cn.

 Global reach, higher impact
Global reach, higher impact