Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(12):1557-1563. doi:10.7150/ijms.39068 This issue Cite
Research Paper
Critical Roles of E2F3 in Growth and Musculo-skeletal Phenotype in Mice
1. Laboratory Animal Resource Center, Korea Research Institute of Bioscience and Biotechnology, Yeonjudanji-ro 30, Chungbuk 28116, Korea
2. Department of Animal Science and Technology, Chung-Ang University, Seodong-daero 4726, Gyeonggi 17546, Korea
*These authors are contributed equally to this study.
Received 2019-8-6; Accepted 2019-9-11; Published 2019-10-21
Abstract
E2F3, a member of the E2F family, plays a critical role in cell cycle and proliferation by targeting downstream, retinoblastoma (RB) a tumor suppressor family protein. The purpose of this study, was to investigate the role and function of E2F3 in vivo. We examined phenotypic abnormalities, by deletion of the E2f3 gene in mice. Complete ablation of the E2F3 was fully penetrant, in the pure C57BL/6N background. The E2f3+/- mouse embryo developed normally without fatal disorder. However, they exhibited reduced body weight, growth retardation, skeletal imperfection, and poor grip strength ability. Findings suggest that E2F3 has a pivotal role in muscle and bone development, and affect normal mouse growth.
Keywords: E2f3+/- Knockout, Development, Growth retardation, skeletal imperfection, Mouse phenotype
Introduction
E2F transcription factors (E2Fs) are found in most cell types of the body and contribute to cell cycle progression, cell proliferation, differentiation, and apoptosis processes [1]. These E2F transcription factors are potent regulators of a variety of genes, including cell-cycle checkpoints controlling genes in mammalian cells [2]. Until now, eight members have been characterized, E2F1 to 8, and they are generally classified into transcriptional activators (E2F1-3) and repressors (E2F4-8) [3, 4]. Some of these E2F family members have also a key role, in myeloid development and cardiac neovascularization [5, 6].
For transcriptional regulation, dimerization partner (DP) family and retinoblastoma (RB) family collaborate with E2Fs, by binding to E2F proteins. DP proteins identified DP1-3 form a heterodimer with an E2F, and allow it to bind to target promoters [7, 8]. RB family (pRB, p107, and p130) can bind to activation domains on the E2F1-5, and formed E2F/RB complex [1]. This complex on the promoters gives rise to repression of target genes in a cell cycle dependent manner, through epigenetic modification as well as by physical blocking [9-11]. Not all functions of the E2Fs are Rb-dependent. E2F3 protein also has functions, unrelated to Rb proteins [12, 13].
The E2F3 is expressed ubiquitously, like other E2F family members [14]. However, unlike other E2F family members, the E2F3 locus encodes two gene products, E2F3a and E2F3b, originating from two different promoters. They share the most coding sequence, such as the DNA binding domain and activation domain, as they possess a unique first exon [15]. Two types of the E2F3 isoform are different in expression, because of distinct promoters. E2F3a protein, the long one, is produced like E2F1 and E2F2. E2F3a promoter is expressed highly at G1/S transition, and is regulated by E2F-mediated negative feedback and Myc protein. In contrast, E2F3b promoter is not affected by E2F/Myc-mediated regulation mechanisms, and remains active throughout the cell cycle [15, 16].
E2F3 protein is a key factor in overall biological functions, as it regulates cell cycle progression. Additionally, E2F3 has played a key role in diverse biological processes such as lens development, cardiac neovascularization, DNA damage responses, neuronal migration, and myogenesis [6, 13, 17-21]. Dysregulation of E2F3 is closely related to carcinogenesis, and recent studies confirmed that overexpression of miRNAs targeting E2F3, inhibits cell migration and proliferation in many tumors [22-28]. E2F3 is considered a promising prognostic marker in specific carcinomas [27, 29, 30].
Given the significance of the E2F3 in key biological processes, we showed its effects on mouse phenotype using in vivo study. In this study, we investigated phenotypes of E2f3 null mutant (E2f3-/-) mouse in the C57BL/6N background. All E2f3-/- embryos were dead in the uterus or soon after birth, and we only obtained hetero mutant (E2f3+/-) mice. Also, we observed growth retardation and musculo-skeletal imperfection in E2f3+/- mice, supporting the key role of E2F3 after birth.
Materials and Methods
Generation of E2f3+/- mice
E2f3 gene knockout mouse was produced by Korea Mouse Phenotyping Center (KMPC) using a mouse embryonic stem cell clone with mutant E2f3 gene (E2f3tm1a(KOMP)Wisi) (EPD0034_2_C11, KOMP repository, CA, USA) obtained from the Mouse Biology Program at UC Davis. We produced the tm1b mutant mouse from the tm1a mutant mouse using HTN-Cre protein (HTN-Cre, Excellgen, USA) as described in the previous paper [31]. The tm1b genotype mice, were expanded for all studies. The care and use of all mice used in this study, were in accordance with the IACUC at KRIBB (KRIBB-AEC-17050).
Body weight curve of wild type and E2f3+/- mice
All mice obtained by mating female and male E2f3+/- mice were genotyped at 3 weeks of age. After grouping according to their sex and genotype, weekly body weight measurements were conducted for all the animals from 4 to 16 weeks of aged.
Dual energy X-ray absorptiometry (DEXA) and X-ray
DEXA and X-ray analysis were conducted at age 14 weeks in mice. The mice were anesthetized with intraperitoneal injection of 1.2% avertin solution, before these analyses. In the Dexa, Lunar Piximus II (GE Medical Systems, Wisconsin, USA) was used and all body composition parameters were obtained with whole body excluding the skull area. For X-ray analysis, Faxitron X-ray system (Model MX-20, Wheeling, IL, USA) was used and exposure X-ray dose per mouse was 300µSv. Six X-ray images per mouse were analyzed: Dorso-ventral (whole body), Lateral (whole body), Dorso-ventral (upper body), Lateral (head), Dorso-ventral (head), and Dorsoventral (lower body).
Grip strength test
Forelimb strength and combined forelimb/hind limb strength were measured, using the GRS meter apparatus (Chatillon, HMGU plate, USA) with 9-week-old animals. Three trials were conducted in succession, to measure forelimb strength. After a five-minute interval, three successive trials for measuring combined forelimb and hind limb grip strength were conducted.
Hematological assessment
At age 16 weeks, all the mice were autopsied after anesthesia with intraperitoneal injection of avertin. Before abdominal opening, blood was collected through a heart puncture with EDTA-treated tube (BD Microtainer, NJ, USA). Blood was used for hematology, blood chemistry. Hematological analysis was performed with a Cell-Dyn 3700 hematology analyzer (Abbott Laboratories, IL, USA). Blood biochemistry was conducted with a Hitachi 7020 automatic analyzer.
Fluorescence assisted cell sorting (FACS) analysis
Splenocytes were obtained from the spleen obtained from autopsy at age 16 weeks, and were stained with fluorescence labeled cell surface antibodies to analyze immune cell subsets. Cells which were surface stained, were analyzes with FACSAria III (BD Bioscience, San Jose, CA, USA). Cell surface staining was performed with antibodies as follow: Anti-mouse CD4 PE-Cyanine5 (#15-0042), Anti-mouse CD5 APC-eFluor® 780 (#47-0051), Anti-mouse CD8a FITC (#11-0081), Anti-mouse Ly-6G (Gr-1) FITC (#11-5931), Anti-mouse Ly-6C APC (#17-5932), Anti-mouse CD11b APC (#17-0112), Anti-mouse CD11c FITC (#11-0114), Anti-mouse CD11c PE-Cyanine5 (#15-0114), Anti-mouse CD11c Alexa Fluor® 700 (#56-0114), Anti-mouse CD19 PE-Cyanine5 (#15-0193), Anti-mouse CD19 PE-eFluor® 610 (#61-0193), Anti-mouse CD19 Alexa Fluor® 700 (#56-0193), Anti-mouse NK1.1 PE-eFluor® 610 (#61-5941) and Anti-mouse MHC Class II (I-A/I-E) PE-Cyanine7 (#25-5321), all from eBioscience (San Diego, CA, USA).
Statistical analysis
Data were presented as the mean and standard deviation. All statistical differences between groups were analyzed by Student's t-test using a statistical program (GraphPad Prism 7.04, San Diego, CA). P<0.05 was considered as significant.
Results
E2F3 is critical for embryo survival and E2f3-/- neonates died in early development stage
To validate the role of E2F3 in mouse development, we mated the E2f3 heterozygous mutant C57BL/6N mice. However, we couldn't observe viable E2f3-/- neonates, only E2f3+/+ and E2f3+/-animals were obtained (Table 1). In the previous study that investigated viability of the E2f3 mutant mouse, all the E2F3-/- 129/Sv embryos were dead. But C57BL/6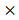 129/Sv mixed mice survived partially, suggesting that E2f3 is an essential gene for embryonic viability in a strain-specific manner [19]. Our data also indicate that deletion of E2f3 affect embryo survival in C57BL/6N background and that the heterozygosity of E2F3 gene is enough for the survival in the mice.
129/Sv mixed mice survived partially, suggesting that E2f3 is an essential gene for embryonic viability in a strain-specific manner [19]. Our data also indicate that deletion of E2f3 affect embryo survival in C57BL/6N background and that the heterozygosity of E2F3 gene is enough for the survival in the mice.
Analysis of progeny, arising from mating of E2F3+/- mice in the C57BL/6N background.
| E2f3+/+ | E2f3+/- | E2f3-/- | |
|---|---|---|---|
| Number of pups | 41 | 77 | 0 |
| Expected ratio | 1 | 2 | 1 |
| Observed ratio | 1 | 1.88 | 0 |
E2f3+/- mice have lower body weight than wild type mice, suggesting its role in development
Although E2f3+/- animals were born ordinarily, they showed immature phenotype. We conducted various phenotypic analysis for E2f3+/- animals, to investigate effects of partial E2f3 defects after birth. Weekly body weight measurements were conducted, after weaning until age 16 weeks. Our data indicates that female and male E2f3+/- mutant mice had shown significantly lower body weight than wild mice throughout the measurement period, suggesting E2f3 is responsible for development as well as maturation in mice (Fig. 1).
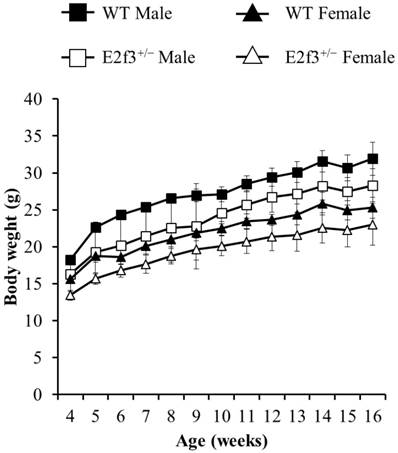
Also, body composition of these mice measured by DEXA indicated that E2f3+/- mutant mice had lower bone mineral density (BMD) and content (BMC) and lower lean mass, indicating poor bone and muscle development in E2f3+/- mutant mice. Three skeletal parameters (BMD, BMC and bone area) of all E2f3+/- mutant mice were significantly lower (P<0.01) than wild littermate animals, but there were no morphological abnormalities in the bone when observed in X-ray records (Fig 2 A-B and data not shown). Consistent with reduced body weight and skeletal parameters, E2f3+/- mice showed significantly (P<0.01) poor lean mass (male: 21.4±0.8g, female: 17.5±0.9g) than the control groups (male: 23.6±0.7g, female: 19.9±0.8g). However, fat mass did not correlate with E3f3 mutation (Fig. 3).
Weight curve of wild-type and E2f3+/- male/female mice during 16 weeks. Body weight was measured once a week from age four-16 weeks. During the whole observation periods, body weight of E2f3+/- Male (open squares, n = 8), was significantly lower than that of WT Male (filled squares, n = 8). Likewise, body weight of E2f3+/- Female (open triangles, n = 8), was significantly lower than that of WT Female (filled triangles, n = 8).
Analysis of muscle strength in male/female E2f3+/- and wild mice
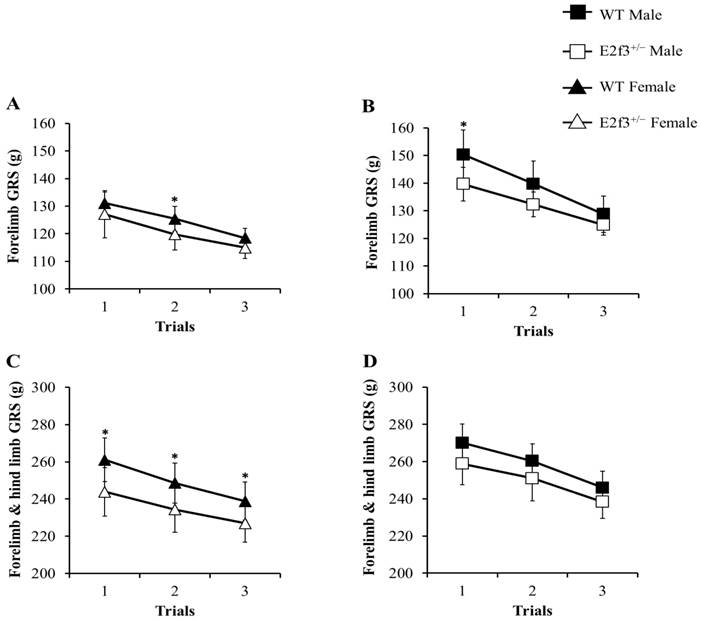
The grip strength (GRS) test was performed, to measure neuromuscular function as maximal muscle strength. In all trials, E2f3+/- mice showed weak grip strength, compared to WT mice (Fig. 4). Forelimb grip strength of the male E2f3+/- mice was significantly lower, than that of the wild mice at trial 1 and on average. Similarly, female E2f3+/- mice showed weaker strength, than the wild group at trial 2. In the case of combined forelimb/hind limb strength, female E2f3+/- mice exhibited significantly lower strength than the wild type group in all three trials. However, in the male, the values were also lower but not statistically significant.
E2f3+/- male and female mice showed reduced BMD and BMC, than WT. (A) BMD was significantly reduced in E2f3+/- Male (dark gray bar, n = 7) and Female (white bar, n = 8) mice, compared to their control groups (black bar for WT Male; n = 8, light gray bar for WT Female; n = 8). (B) BMC was similarly reduced, in both genders of E2f3+/- mice. **P<0.01.
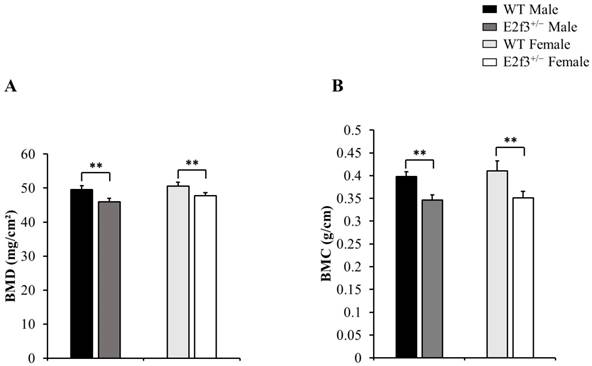
Body composition of 14-week-old E2f3+/- mice (both genders) has lower body weight, lean mass than WT strain. Body weight (A) and lean mass (B) were significantly lower in E2f3+/- Male (dark gray bar, n = 7) and Female (white bar, n = 8) mice, than their control groups (black bar for WT Male; n = 8, light gray bar for WT Female; n = 8). (C) However, fat mass between WT and E2f3+/- was not significantly different in both genders. *P<0.05 and **P<0.01.
E2f3+/- mice have abnormal heart weight than wild mice
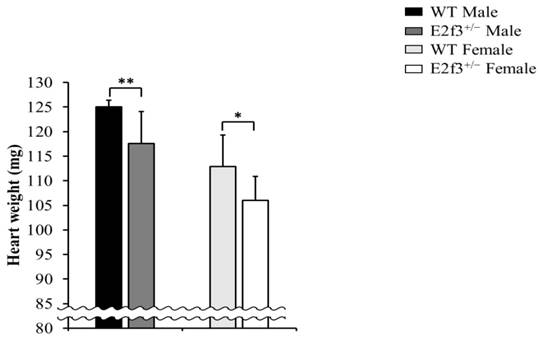
Full necropsy at age 16 weeks was performed. There was no obvious abnormal gross pathology except in heart weight. The E2f3+/- mice exhibited significantly lower heart weight (male: 117.59±6.42mg, female: 106.05±4.88mg) than wild mice (male: 125.01±1.36mg, female: 112.86±6.51mg), and it is parallel to light body weight and lean mass (Fig. 5).
FACS analysis and assessment of hematological parameters
We also performed hematological test and FACS analysis to assess hematological and immunological problems, but there were no abnormal findings in these analyses when compared with those in the wild type mice (Table 2 and data not shown).
Discussion
In this study, we explored the function of the E2F3 gene in normal development of mice. E2F3 that regulates cell cycle progression is required for development of most tissues and organs in the body. Because of their importance in development and other important processes, here we conducted several in vivo analyses to identify abnormalities in E2f3+/- mice, which pave the way for further research.
FACS data analysis, for major cell types.
| Parameters (%) | Male | Female | ||
|---|---|---|---|---|
| WT | E2f3+/- | WT | E2f3+/- | |
| T cells | 15.83±0.51 | 17.07±2.87 | 25.23±1.98 | 20.83±1.00* |
| CD4 T cells | 10.80±0.26 | 12.03±1.88 | 14.37±1.1 | 12.27±0.75* |
| CD8 T cells | 4.35±0.24 | 4.94±0.98 | 8.39±0.76 | 6.51±0.2* |
| B cells | 40.67±2.85 | 40.87±2.75 | 38.4±2.6 | 38.27±1.29 |
| NK cells | 2.31±0.05 | 2.39±0.07 | 1.88±0.24 | 2.17±0.26 |
| NKT cells | 0.59±0.05 | 0.76±0.11 | 0.98±0.11 | 0.95±0.03 |
| Neutrophils | 2.14±0.32 | 2.28±0.34 | 1.77±0.34 | 1.66±0.29 |
| Monocytes | 0.57±0.11 | 0.68±0.12 | 0.49±0.09 | 0.39±0.05 |
| Macrophages | 1.68±0.31 | 1.66±0.31 | 1.31±0.42 | 1.5±0.21 |
Herein, we showed that E2F3 is the pre-requisite factor for normal development in mice. In a previous study that investigated viability of the E2F3 mutant mouse, all E2f3-/- 129/Sv embryos were dead. However, E2f3-/- mice with C57BL/6 × 129/Sv mixed background survived partially, suggesting that E2f3 is an essential gene for embryonic viability in a strain-specific manner [19]. Our data also indicate that presence of E2F3 in C57BL/6N background is essential for embryo survival and that the amount of E2F3 is also important for the normal development of bones and muscles in mice. It might be possible that incidence of viability of E2F3 embryo depends on the genetic background of mice.
A previous study demonstrated that E2F3, plays a pivotal role in normal cardiac development. A small fraction of E2f3-/- neonates showed normal heart. However, these animals ultimately died from defects in the cardiac muscle, and as a result of congestive cardiac failure [32]. Our observation that E2f3+/- mice had lower heart weight compared with that of littermate wild mice, may be associated with the partial deficit of E2F3 that may affect poor heart muscle development in E2f3+/- mice.
Grip strength (GRS) analysis of WT and E2f3+/- mice. Forelimb GRS was observed lower in female (A) and male (B) E2f3+/- mice (filled square for WT Male; n = 8, open square for E2f3+/- Male; n = 7, filled triangle for WT Female; n = 8 open triangle for E2f3+/- Male; n = 8). (C) Forelimb and hind limb combined GRS of female E2f3+/- mice was significantly low, compared to their control group in all trials. (D) Forelimb and hind limb combined GRS of male E2f3+/- mice was weaker, than their control group like in female, but was not statistically significant. *P<0.05.
Heart weight analysis of E2f3+/- mice, at necropsy. Heart weight was significantly reduced in E2f3+/- mice (dark gray bar for E2f3+/- Male; n = 7, white bar for E2f3+/- Female; n = 8), compared to WT animals (black bar for WT Male; n = 8, light gray bar for WT Female; n = 8). *P<0.05 and **P<0.01.
On the contrary heterozygous mutant, E2f3+/- mice had shown remarkable significant phenotypes. E2f3 is not an allelic exclusive gene, and is expressed more in E2f3+/+ cells than E2f3+/- cells [33]. Growth retardation with skeletal imperfection of E2f3+/- mice, suggest that E2f3 expression level is crucial for postnatal musculoskeletal development. E2F3b has been characterized as an essential player, in myogenic differentiation and development [13]. Reduced lean mass, grip strength, and heart weight of E2f3+/- mice, may be associated with specific function of E2f3b in myogenesis. Unlike muscle development, the specific role of E2f3 in osteogenesis is unknown. Small bone area and low BMD and BMC of E2f3+/- mice strongly suggest E2f3 has a unique function in bone development. Earlier studies have shown that activator E2F transcription factors share a binding motif, and that loss of E2F3 activity triggers compensation effects of other activator of E2Fs [34-37]. Our results imply that compensation effects are insufficient to make up for a unique function of E2F3 in muscle and bone development, and leads to growth retardation.
Previous studies have demonstrated the importance of the E2F family in hematopoiesis [5, 38, 39]. Individual knock-out of E2f1, E2f2, and E2f4 causes' hematopoietic impairment, and also, E2f1-3 triple knock-out leads to tremendous decline of bone marrow cellularity and CD11b+ myeloid cell count [5, 38-41]. However, individual Mx-Cre; E2f3fl/fl mice exhibited normal bone marrow cellularity [5]. In this study, E2f3+/- mice showed normal blood cell count/hematocrit level including CD11b+ cells, and is in contrast to musculoskeletal imperfection. It seems that E2f3 has functional redundancy with other E2Fs in hematopoietic development, and consequently E2f3 deletion does not cause hematopoietic defects.
Also, because E2F3 contributes to neural development, more explanation is needed as to if E2F3 mutation affects neural functions or not [18, 42].
In this study, we found that complete ablation of E2f3 was fully penetrant in the pure C57BL/6N background, and that E2f3+/- mouse embryo developed normally without fatal disorders. However, they exhibited reduced body weight, growth retardation, skeletal imperfection, and poor muscle condition, but no detectable hematopoietic defects. Findings suggest that E2F3 has a pivotal role in muscle and bone development. However, it remains to be elucidated how E2F3 works, in different tissues or environments.
Acknowledgements
This research was supported by a grant from KRIBB Research Initiative Program and Korea Mouse Phenotyping Project (NRF-2017M3A9D5A01072797) of the Korean Ministry of Science and ICT through the National Research Foundation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245-62
2. Lavia P, Jansen-Durr P. E2F target genes and cell-cycle checkpoint control. Bioessays. 1999;21:221-30
3. Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658-62
4. Logan N, Graham A, Zhao X, Fisher R, Maiti B, Leone G. et al. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene. 2005;24:5000-4
5. Trikha P, Sharma N, Opavsky R, Reyes A, Pena C, Ostrowski MC. et al. E2f1-3 are critical for myeloid development. J Biol Chem. 2011;286:4783-95
6. Zhou J, Wu M, Xu S, Cheng M, Ding C, Liu Y. et al. Contrasting roles of E2F2 and E2F3 in cardiac neovascularization. PLoS One. 2013;8:e65755
7. Rubin SM, Gall AL, Zheng N, Pavletich NP. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093-106
8. Ormondroyd E, de la Luna S, La Thangue NB. A new member of the DP family, DP-3, with distinct protein products suggests a regulatory role for alternative splicing in the cell cycle transcription factor DRTF1/E2F. Oncogene. 1995;11:1437-46
9. Macaluso M, Montanari M, Noto PB, Gregorio V, Bronner C, Giordano A. Epigenetic modulation of estrogen receptor-alpha by pRb family proteins: a novel mechanism in breast cancer. Cancer Res. 2007;67:7731-7
10. Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX. et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79-89
11. Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E. et al. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813-25
12. Liao Y, Du W. Rb-independent E2F3 promotes cell proliferation and alters expression of genes involved in metabolism and inflammation. FEBS Open Bio. 2017;7:1611-21
13. Asp P, Acosta-Alvear D, Tsikitis M, van Oevelen C, Dynlacht BD. E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes Dev. 2009;23:37-53
14. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397-406
15. Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L. et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626-32
16. Adams MR, Sears R, Nuckolls F, Leone G, Nevins JR. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol Cell Biol. 2000;20:3633-9
17. Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G. et al. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 2010;30:524-36
18. Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L. et al. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol Cell Biol. 2009;29:414-24
19. Cloud JE, Rogers C, Reza TL, Ziebold U, Stone JR, Picard MH. et al. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol Cell Biol. 2002;22:2663-72
20. Wenzel PL, Chong JL, Saenz-Robles MT, Ferrey A, Hagan JP, Gomez YM. et al. Cell proliferation in the absence of E2F1-3. Dev Biol. 2011;351:35-45
21. McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D. et al. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27:4825-43
22. Sun FB, Lin Y, Li SJ, Gao J, Han B, Zhang CS. MiR-210 knockdown promotes the development of pancreatic cancer via upregulating E2F3 expression. Eur Rev Med Pharmacol Sci. 2018;22:8640-8
23. Wang Y, Sun G, Wang C, Guo W, Tang Q, Wang M. MiR-194-5p inhibits cell migration and invasion in bladder cancer by targeting E2F3. J BUON. 2018;23:1492-9
24. Liu J, Si L, Tian H. MicroRNA-148a inhibits cell proliferation and cell cycle progression in lung adenocarcinoma via directly targeting transcription factor E2F3. Exp Ther Med. 2018;16:5400-9
25. Guo Y, Qi Y, Guo A, Du C, Zhang R, Chu X. miR-564 is downregulated in gastric carcinoma and targets E2F3. Oncol Lett. 2017;13:4155-60
26. Feng Z, Peng C, Li D, Zhang D, Li X, Cui F. et al. E2F3 promotes cancer growth and is overexpressed through copy number variation in human melanoma. Onco Targets Ther. 2018;11:5303-13
27. Liu ZL, Bi XW, Liu PP, Lei DX, Wang Y, Li ZM. et al. Expressions and prognostic values of the E2F transcription factors in human breast carcinoma. Cancer Manag Res. 2018;10:3521-32
28. Ma C, Han J, Dong D, Wang N. MicroRNA-152 Suppresses Human Osteosarcoma Cell Proliferation and Invasion by Targeting E2F Transcription Factor 3. Oncol Res. 2018;26:765-73
29. Al Ahmed HA, Nada O. E2F3 transcription factor: A promising biomarker in lung cancer. Cancer Biomark. 2017;19:21-6
30. Zeng X, Yin F, Liu X, Xu J, Xu Y, Huang J. et al. Upregulation of E2F transcription factor 3 is associated with poor prognosis in hepatocellular carcinoma. Oncol Rep. 2014;31:1139-46
31. Ryder E, Doe B, Gleeson D, Houghton R, Dalvi P, Grau E. et al. Rapid conversion of EUCOMM/KOMP-CSD alleles in mouse embryos using a cell-permeable Cre recombinase. Transgenic Res. 2014;23:177-85
32. King JC, Moskowitz IP, Burgon PG, Ahmad F, Stone JR, Seidman JG. et al. E2F3 plays an essential role in cardiac development and function. Cell Cycle. 2008;7:3775-80
33. Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690-703
34. Kong LJ, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321-7
35. To B, Andrechek ER. Transcription factor compensation during mammary gland development in E2F knockout mice. PLoS One. 2018;13:e0194937
36. Freedman JA, Chang JT, Jakoi L, Nevins JR. A combinatorial mechanism for determining the specificity of E2F activation and repression. Oncogene. 2009;28:2873-81
37. Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18:1763-77
38. Humbert PO, Rogers C, Ganiatsas S, Landsberg RL, Trimarchi JM, Dandapani S. et al. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol Cell. 2000;6:281-91
39. Rempel RE, Saenz-Robles MT, Storms R, Morham S, Ishida S, Engel A. et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell. 2000;6:293-306
40. Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG Jr, Livingston DM. et al. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549-61
41. Murga M, Fernandez-Capetillo O, Field SJ, Moreno B, Borlado LR, Fujiwara Y. et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959-70
42. Santerre M, Bagashev A, Gorecki L, Lysek KZ, Wang Y, Shrestha J. et al. HIV-1 Tat protein promotes neuronal dysregulation by inhibiting E2F transcription factor 3 (E2F3). J Biol Chem. 2019;10:3618-33
Author contact
![]() Corresponding authors: Ki-Hoan Nam, D.V.M., Ph.D.; Laboratory Animal Resource Center, Korea Research Institute of Bioscience and Biotechnology, Yeongudanji-ro 30, Ochang-eup, Cheongwon-gu, Cheongju, Chungbuk 28116, Republic of Korea; Phone: +82-43-240-6561; E-mail: namkre.kr. Hyoung-Chin Kim, D.V.M., Ph.D.; Laboratory Animal Resource Center, Korea Research Institute of Bioscience and Biotechnology, Yeongudanji-ro 30, Ochang-eup, Cheongwon-gu, Cheongju, Chungbuk 28116, Republic of Korea; Phone: +82-43-240-6560; E-mail: hckimre.kr
Corresponding authors: Ki-Hoan Nam, D.V.M., Ph.D.; Laboratory Animal Resource Center, Korea Research Institute of Bioscience and Biotechnology, Yeongudanji-ro 30, Ochang-eup, Cheongwon-gu, Cheongju, Chungbuk 28116, Republic of Korea; Phone: +82-43-240-6561; E-mail: namkre.kr. Hyoung-Chin Kim, D.V.M., Ph.D.; Laboratory Animal Resource Center, Korea Research Institute of Bioscience and Biotechnology, Yeongudanji-ro 30, Ochang-eup, Cheongwon-gu, Cheongju, Chungbuk 28116, Republic of Korea; Phone: +82-43-240-6560; E-mail: hckimre.kr

 Global reach, higher impact
Global reach, higher impact