Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(7):922-930. doi:10.7150/ijms.34322 This issue Cite
Research Paper
The molecular landscape of histone lysine methyltransferases and demethylases in non-small cell lung cancer
1. Department of Cardiothoracic Surgery, Yijishan Hospital, Wannan Medical College, Wuhu, 241001, Anhui, PR China
2. Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, 646000, Sichuan, PR China
3. South Sichuan Institution for Translational Medicine, Luzhou, 646000, Sichuan, PR China
4. The People's Hospital of Weiyuan, Neijiang, Sichuan, PR China
* These authors contribute equally to this work
Received 2019-2-22; Accepted 2019-4-22; Published 2019-6-2
Abstract
Background: Lung cancer is one of the most common malignant tumors. Histone methylation was reported to regulate the expression of a variety of genes in cancer. However, comprehensive understanding of the expression profiles of histone methyltransferases and demethylases in lung cancer is still lacking.
Methods: We analyzed the expression profile of methyltransferases and demethylases in non-small cell lung cancer (NSCLC) using TCGA and cBioportal databases. The mutation, expression level, association with survival and clinical parameters of histone methyltransferases and demethylases were determined.
Results: We found overall upregulation of histone regulators in NSCLC. Mutation and copy number alteration of histone methylation related genes both exist in NSCLC. The expression of certain histone methylation related genes were significantly associated with overall survival and clinical attributes.
Conclusions: Our result suggests that alteration of histone methylation is strongly involved in NSCLC. Some histone methylation related genes might serve as potential prognosis predictor or therapeutic target for NSCLC. The significance of some histone methylation related genes was contrary to the literature and awaits further validation.
Keywords: histone methylation, lung cancer, methyltransferases, demethylases, mutation, survival
Introduction
Lung cancer is the leading cause of cancer-related mortality in men and the second leading cause in women in the United States [1]. Approximately 85% to 90% lung cancer patients have non-small cell lung cancer (NSCLC). However, the survival of NSCLC patients has not significantly improved in over 30 years. The exploration of epigenetic modification as a therapeutic target for lung cancer has never stopped. Epigenetic modifications include DNA methylation, histone modification and noncoding RNA expression [2]. DNA methylation participates in carcinogenesis both at the transcriptional and post-transcriptional levels [3]. Histone modification represents one of the most critical epigenetic events in DNA function regulation in eukaryotic organisms and it includes methylation, acetylation, phosphorylation and ubiquitination [4]. More and more evidence suggest that histone modifications (such as methylation and acetylation) can serve as a binding platform to attract other protein complexes to chromatin [5-7].
Histone methylation usually occurs on the N-terminal histone tail of lysine (K) and arginine (R) residues [8]. Depending on the location and methylation level of amino acid residues, it can promote or inhibit the transcription of different genes and play a very complex role in cancer. In eukaryotic cells, the basic subunit of a chromatin is the nucleosome. Genomic DNA is wrapped around a protein octamer which contains four core histones (H2A, H2B, H3, H4), forming the structure of the nucleosome [9-11]. There are five lysines in histone H3 (K4, K9, K27, K36, K79) that have been shown to be modulated by methylation. In addition, a lysine in histone H4 (K20) could be methylated by the specific histone lysine methyltransferase. The methylation of H3K4 and H3K36 can active gene transcription while the methyltion at H3K9, H3K27, H3K79 and H4K20 can repress gene transcription [12]. Changes in histone methylation have been proved to be closely related to various malignant tumors.
Histone methylation is a dynamic process controlled by methylases and demethylases. Histone lysine methyltransferases (KMTs) add methyl groups, and they function as 'writers' of the histone code. Histone lysine demethylases (KDMs) are known as 'erasers' of methyl groups [13]. Methylation is catalyzed by methyltransferase, which can be modified by monovalent, divalent and trivalent methylation, and the latter is called “over” methylation modification (Hypermethylation) [14]. For example, EZH2, which acts as a histone lysine methyltransferase, mediates trimethylation of lysine 27 on histone H3 (H3K27me3), leading to chromatin condensation and the transcriptional repression of target genes, including tumor suppressor genes [15]. Methylation 'erasers' and 'writers' by removing or adding specific methyl groups fundamentally influence gene expression, genomic stability and cell fate [16, 17]. In addition, several inhibitors targeting histone methylation have entered clinical trials [18]. It has been reported that SMYD3 plays a pivotal role in the regulation of oncogenic Ras signaling in pancreatic ductal adenocarcinoma (PDAC) and lung cancer [19]. However, the molecular profiles of histone demethylases and methyltransferases have not been systematically studied. In this study, we comprehensively analyzed the gene alteration, mRNA expression and the relevance with clinical data of histone methyltransferases and demethylases in NSCLC.
Materials and Methods
Data acquisition
A total of 925 samples were employed for lung cancer genomic analysis, including 93 normal patients and 832 tumor samples. Preprocessed expression profiles of histone methylation related genes and patient clinical parameters were manually extracted from TCGA database (https://cancergenome.nih.gov/) and processed via automated pipelines (TCGAbiolinks [20]) in an attempt to accelerate analysis. Illumina HiSeq expression raw data was normalized based on Fragments per Kilobase of transcript per Million fragments mapped (FPKM) within the MATLAB software (www.mathworks.com). The Copy number variation (Amplification and Deep deletion) and somatic mutation data (Truncating mutation and Missense mutation) of lung cancer was downloaded from TCGA through cBioPortal and GISTIC.
Genomic and protein structure alteration analysis
We conducted analysis of histone methylation related regulators in lung cancer in TCGA using the oncoprint (http://cbioportal.org). The primary search included alterations, such as amplification, deep deletion, missense mutations, and truncating mutations, from GISTIC and TCGA data with the default setting. The diagram order was ranged according to alteration frequency of each cancer patients. Lollipops of each protein structure change of lung cancer were linked to COSMIC. The detailed mutation annotations from OncoKB, CIViC and Hotspot in different genes were displayed in different regions of the protein structure.
Differential expression and association with clinical parameters
Gene expression levels were evaluated across the tumor and normal samples using the median. The standard deviation of the gene expression level for each gene was computed with normalized FPKM. We reconstructed the diagram through computational bioinformatics method within the R version 3.5.0. In order to identify gene expression pattern of lung cancer samples across different clinical parameters, matching of the clinical data with expression data was performed using TCGA “hybridization” identifier. Eventually, 831 patients with gene expression data from 15 genes were included in the final analysis. All the genes with P<0.05 were displayed within smoking year, tumor status and pathologic stage.
Co-expression analysis
Co-expression between methylases and demethylases in mRNA level was analyzed using the liner regression. The 95% Confidence intervals were presented by dot lines.
Statistical analysis
GraphPad Prism 6 software was used for statistical analysis, data were presented as mean ± SD. Student's t test was used to compare two groups and one-way ANOVA was used to compare multiple groups. The correlation of mRNA expression was analyzed by Pearson test. Overall survival was shown as Kaplan-Meier curve with P values calculated using the log-rank test. P value<0.05 was considered statistically significant, and all P values are two-sided.
Results
Mutation of histone methyltransferases and demethylases in lung cancer
An oncoprint image of histone methyltransferases and demethylases gene alteration in lung adenocarcinoma and squamous cell carcinoma was generated in cBioportal based on TCGA data (Fig. 1). Gene amplification, deep deletion and missense mutation were frequently found in different histone methyltransferases and demethylases gene. Among H3K4 methyltransferases and demethylases, PRDM9 had the highest frequency of copy number amplification and missense mutation. The SETD1A, SMYD3, KDM5A and KDM5B genes were also amplified to different extent, among which KDM5A and KDM5B were also H3K9 demethylases. Moreover, H3K9 methyltransferase SETDB1 and H3K36 methyltransferase SMYD2 genes were frequently amplified in lung cancer. Mutation in other genes was less prevalent than that of the above mentioned genes.
The genetic alteration of histone methyltransferase and demethylases gene in NSCLC. Data represents various types of alterations including gene amplification, mutation, and deletion. Data was generated using TCGA datasets from cBioportal.
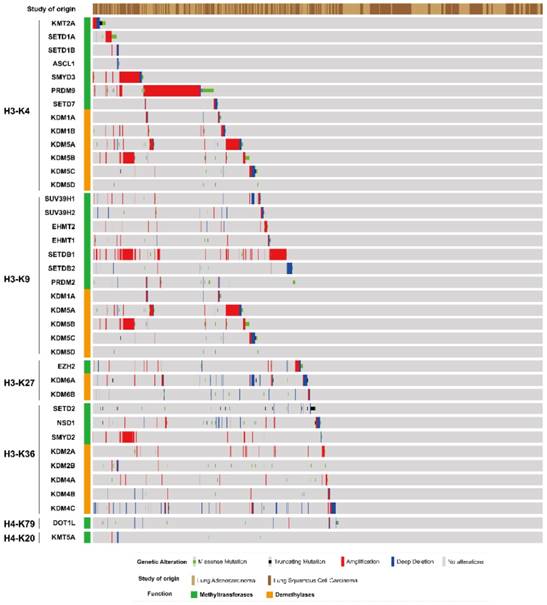
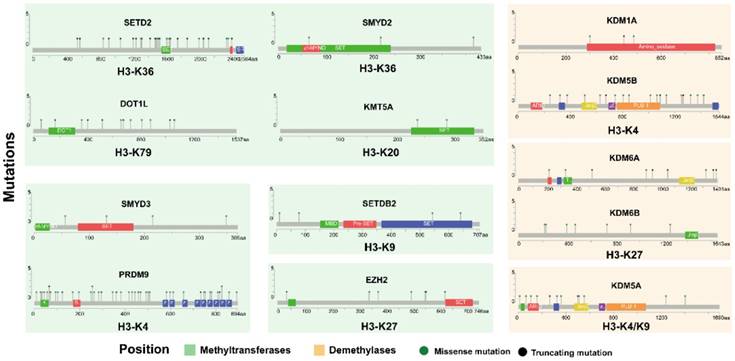
We further analyzed the influence of somatic mutation on protein structure in lung adenocarcinoma and squamous cell carcinoma in cBioportal based on OncoKB data (Fig. 2). We selected several genes with high frequency of mutation from previous result (Fig. 1), i.e. PRDM9, SETDB2, SETD2, KDM6A/B, DOT1L, etc. A variety of mutation points were found across the protein for SETD2, DOT1L, PRDM9 and KDM5A/B. Fewer mutation position was found for other proteins. Moreover, mutations were found in the catalytic SET domain for methyltransferases SETD2, SMYD2, SMYD3, KMT5A, SETDB2 and PRDM9 and Jmjc domain for demethylases KDM5A/B.
Expression level of histone methyltransferases and demethylases in lung cancer
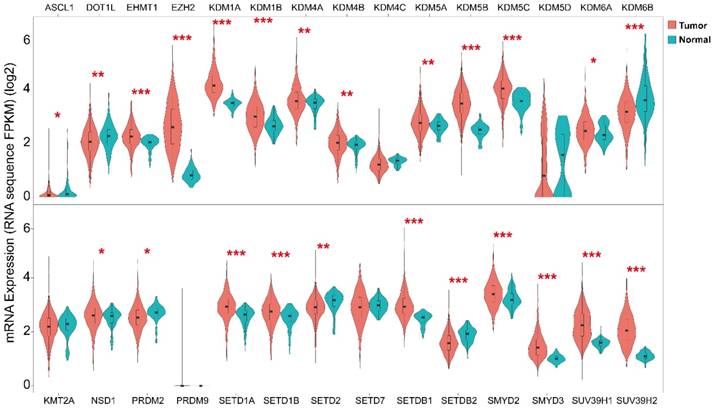
Next we analyzed the mRNA expression level of histone methylation related genes employing 946 patients (592 Lung adenocarcinoma and 354 Lung squamous cell carcinoma) RNA sequencing data from TCGA for comparison between tumor and normal patient samples (Fig. 3). The results indicated that most of the histone methylation related genes (methyltransferases and demethylases) were up-regulated in lung cancer compared to normal tissue (18/29). Noteworthy, H3K27 methyltransferase EZH2 was significantly up-regulated while H3K27 demethylase KDM6B was significantly down-regulated in lung cancer. Moreover, methyltransferases STDB2 and PRDM2 also revealed an opposite expression tendency with demethylases KDM1A and KDM5A/B/C in lung cancer in the current dataset.
The mutation in protein structure for key histone methyltransferases and demethylases. Mutation was analyzed in cBioportal based on OncoKB data.
Expression level of histone methyltransferases and demethylases in tumor versus normal tissue. The expression was determined in 946 patients using RNA sequencing data from TCGA. *P < 0.05, **P < 0.01 and ***P < 0.001.
Impact of histone methyltransferases and demethylases expression on overall survival. Kaplan-Meier analysis was performed using TCGA data.
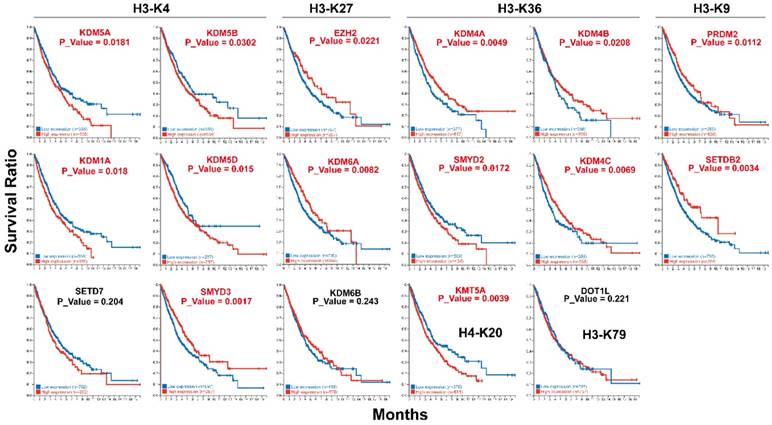
Impact of histone methyltransferases and demethylases on patient survival
Kaplan-Meier analysis using TCGA data revealed that the expression level of several histone methyltransferases and demethylases were significantly associated with overall patient survival (Fig. 4). Our results indicated that patients with higher expression of H3K4 histone demethylases (KDM1A, KDM5A, KDM5B and KDM5D) had a significantly worse prognosis (Fig. 4). Meanwhile, low expression of H3K4 histone methyltransferases SMYD3 was also associated with poor overall survival. High expression of H3K27 histone methylation regulators EZH2 and KDM6A both predicts poor overall survival. For H3K36 histone methylation regulators, high expression of methyltransferase SMYD2 (P=0.0017) and demethylase KDM4C (P=0.0069) and low expression of demethylases KDM4A (P=0.0049), KDM4B (P=0.0208) were significantly associated with poor overall patient survival. Among H3K9 regulators, low expression of SETDB2 (P=0.0003) and high expression of PRDM2 (P=0.0112) were associated with poor overall survival. The high expression of H4K20 histone methyltransferases KMT5A also predicts poor survival.
Association of histone methylation regulators with clinical parameters
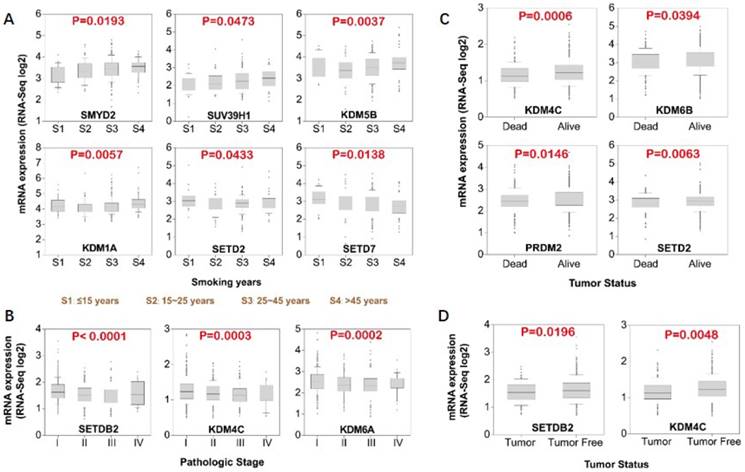
Since we have shown that differential expression of several histone methylation regulators significantly influence patient overall survival, we further studied the association of the expression of these regulators with clinical parameters (Fig. 5). Result of the expression of histone methylation regulators and smoking history revealed that the level of SMYD2 (P=0.0193), SUV39H1 (P=0.0473), KDM5B (P=0.0037) and KDM1A (P=0.0057) were significantly elevated in patients with longer smoking history (Fig. 5A). In contrast, SETD7 level decreased with smoking years. In different tumor pathologic stages, we found that SETDB2 (P<0.0001), KDM4C (P=0.0003) and KDM6A (P=0.0002) expression levels significantly downregulated from stage I to III (Fig. 5B). Moreover, the levels of KDM4C (P=0.0006), KDM6B (P=0.0394), SETD2 (P=0.0063) and PRDM2 (P=0.0146) were lower in dead patients compared with live patient (Fig. 5C). The level of SETDB2 (P=0.0196) and KDM4C (P=0.0048) were also significantly lower in tumor compared with tumor free samples, although marginally (Fig. 5D). Taken together, the data suggests a tumor suppressive role for SETDB2 and KDM4C.
Correlation between methyltransferases and demethylases
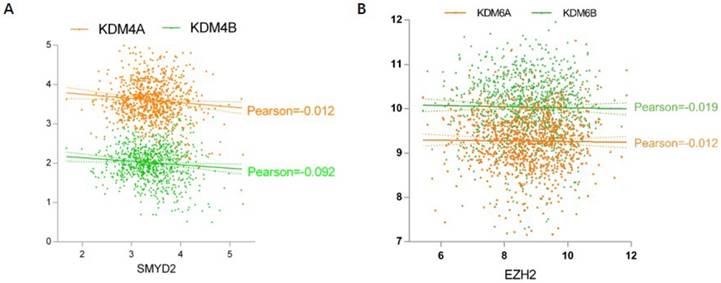
Next we analyzed the relationship of methyltransferase and demethylase expression in lung cancer (Fig. 6). Pearson's correlation analysis indicated that the expression of some methyltransferases and demethylases were negatively correlated. For H3K36, the methyltransferase SMYD2 showed a negative correlation with demethylases KDM4A and KDM4B expression (Fig. 6A). For H3K27, the methyltransferase EZH2 was negatively correlated with both KDM6A and KDM6B (Fig. 6B).
Association of histone methyltransferases and demethylases expression with clinical parameters. (A) Association of mRNA expression with smoking history. (B) Association of methyltransferase and demethylases mRNA expression with tumor stage. (C) Expression level of methyltransferases and demethylases in dead versus alive patients. (D) Expression level of methyltransferases and demethylases in tumor versus tumor free patients.
Correlation between methyltransferases and demethylases. (A) SMYD2 is negatively correlated with KDM4A and KDM4B. (B) EZH2 has a negative correlation with KDM6A and KDM6B
Discussion
The methylation of histones refers to the different degrees of methylation occurring at different sites in the H3 and H4 histone N terminal arginine or lysine residues, which are catalyzed by the histone methyltransferase containing the SET [Su(var)3-9, Enhancer-of-zeste, Trithorax] domain [9]. A growing body of evidence indicates that amplification, translocation or mutation of histone methyltransferases and demethylases is linked to the development of many human cancers [21]. We analyzed genetic alteration of histone methyltransferase and demethylases in lung adenocarcinoma and squamous cell carcinoma using TCGA data. Relatively high alteration rate was seen in histone methyltransferases (SETD1A, SMYD3, PRDM9, SETDB1, EZH2, SMYD2) and demethylases (KDM5A/B, KDM6A/B, KDM2A) (Fig. 1). PRDM9 (PR domain-containing protein 9) was highly active histone methyltransferase catalyzing mono-, di-, and trimethylation of the H3K4 mark [22]. PRDM9 has also been regarded as a meiosis-specific protein that methylates H3K4 and variation strongly influences recombination hot-spot activity and meiotic instability in human [23]. Recently, PRDM9 variability has been implicated in genome instability and having a potential role in the risk of acquiring genome rearrangements associated with childhood leukemogenesis [24]. KDM5A [25, 26] and KDM5B [27] were previously identified as DNA damage response proteins and critical regulators of genome stability and associated with tumor cell migration. Moreover, previous study also indicated that SETDB1 function as an oncogene in lung cancer [28, 29]. We further analyzed mutation position across the protein structure of histone methylation regulators in lung cancer patients (Fig. 2). Results revealed that some mutations were within the catalytic domain, which may influence the activity of histone methylation regulators.
The expression level of histone methyltransferases and demethylases was studied. Overall, most of the histone regulators were significantly overexpressed in lung cancer (18/29), whereas ASCL1, DOT1L, KDM6B, PRDM2, SETD2, SETDB2 were significantly downregulated (Fig. 3). Accordingly, KDM6B, PRDM2 and SETD2 have been shown to possess tumor suppressive function [30, 31] and PRDM2 was demonstrated as tumor suppressor in lung cancer [32]. We further analyzed the differential expression of histone methyltransferases and demethylases on patient overall survival (Fig. 4). In accordance with our finding, overexpression of KDM5B, KDM1A, EZH2 was reported to be associated with shorter overall survival in different cancers including lung cancer [33-38]. Among them, EZH2 might be involved in the progression and metastasis of lung cancer [39]. Increased activity of EZH2 has been reported in different cancers and it is a potential target in cancer therapy [40]. KDM6A (UTX) and KDM6B (JMJD3), antagonists to EZH2, were both tumor suppressors and the high expression of them was associated with better patient survival [31, 41, 42]. However, our result for KDM6A was in contrary to the literature and KDM6B was not a significant prognosis predictor. Though SMYD2 displayed a small amount of somatic mutation positions (Fig. 2), its high expression level in lung cancer was associated with poor prognosis (Fig. 4), which was in accordance with previous finding in hepatocellular carcinomas (HCC) [43], esophageal squamous cell carcinoma [44], gastric cancer [45], head and neck carcinomas [46] and acute lymphoblastic leukemia [47]. KMT5A is a potential oncogene [48, 49], however, its relationship with patient survival has not been explored. Consistent with its oncogenic function, high expression of KMT5A predicts poor survival from our result. In contrast to our finding, low expression of KDM5D was associated with poor overall survival in prostate cancer [50, 51]. Moreover, high expression of some oncogenes from the literature predicts good prognosis from our study including SMYD3, KDM4A, KDM4B and SETDB2. SMYD3 is a well-studied oncogene [19] and its overexpression is reported to be associated with poor survival [52]. Although SMYD3 was found overexpressed in our result, its association with patient survival is contradictory to the literature and needs further investigation in larger patient cohort. KDM4A, KDM4B and KDM4C were overexpressed in various cancers [53] and high expression of them has been associated with worse patient survival [54, 55]. Moreover, KDM4A appeared to have a significant role in the metastatic spread of lung cancer [56]. SETDB2 was reported as an oncogene [57] and its low expression was associated with shorter disease-free survival in clear cell renal cell carcinoma (ccRCC) [58]. Further enlargement of sample size and clarification of their function are needed in lung cancer.
We also determined the association of histone methyltransferases and demethylases with clinicopathological parameters (Fig. 5). Increasing levels of SMYD2, SUV39H1, KDM5B, and KDM1A and decreasing levels of SETD7 were associated with longer smoking years (Fig. 5A). Consistent with previous result, high expression of SMYD2, KDM5B, and KDM1A predicts poor patient survival (Fig. 4). KDM1A (also called LSD1), the first reported histone demethylase is upregulated in many cancers [59] and is associated with undifferentiated, malignant phenotype of neuroblastoma [60]. SETDB2 and KDM4C were decreased from tumor stage I to III (Fig. 5B) and their expression were higher in tumor free versus tumor samples (Fig. 5D). Moreover, KDM4C expression was higher in alive compared with dead patients (Fig. 5C). Altogether, the data indicated that SETDB2 and KDM4C might be tumor suppressors in lung cancer, which is contradictory to the literature. Thus, the expression and function of them needs to be further confirmed in lung cancer. Furthermore, we tried to explore the relationship of histone methylation regulators expression. Correlation analysis revealed a negative correlation between methyltransferase and demethylases for H3K27 and H3K36 (Fig. 6).
In summary, our research findings demonstrated the molecular landscape of histone lysine methyltransferases and demethylases in lung cancer and identified some potential prognosis and/or therapeutic targets for lung cancer. Further study is warranted to confirm their expression and function.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant nos. 81770562, 81503093, 81602166, and 81672444), the Joint Funds of the Southwest Medical University & Luzhou (2016LZXNYD-T01, 2017LZXNYD-Z05 and 2017LZXNYD-J09) and the University Natural Science Research Project of Anhui Province (KJ2017A272).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Alberg AJ, Brock MV, Ford JG, Samet JM, Spivack SD. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e1S-e29S
2. Darilmaz Yuce G, Ortac Ersoy E. [Lung cancer and epigenetic modifications]. Tuberk Toraks. 2016;64(2):163-170
3. Li B, Lu Q, Song ZG, Yang L, Jin H, Li ZG. et al. Functional analysis of DNA methylation in lung cancer. Eur Rev Med Pharmacol Sci. 2013;17(9):1191-1197
4. Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1362-1372
5. Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57-85
6. Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015;16(11):1467-1481
7. Karki R, Zhang Y, Igwe OJ. Activation of c-Src: a hub for exogenous pro-oxidant-mediated activation of Toll-like receptor 4 signaling. Free Radic Biol Med. 2014;71:256-269
8. Muller MM, Muir TW. Histones: at the crossroads of peptide and protein chemistry. Chem Rev. 2015;115(6):2296-2349
9. Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491-507
10. da Silva IT, de Oliveira PS, Santos GM. Featuring the nucleosome surface as a therapeutic target. Trends Pharmacol Sci. 2015;36(5):263-269
11. Zhang Y, Karki R, Igwe OJ. Toll-like receptor 4 signaling: A common pathway for interactions between prooxidants and extracellular disulfide high mobility group box 1 (HMGB1) protein-coupled activation. Biochem Pharmacol. 2015;98(1):132-143
12. Yi X, Jiang XJ, Li XY, Jiang DS. Histone methyltransferases: novel targets for tumor and developmental defects. Am J Transl Res. 2015;7(11):2159-2175
13. D'Oto A, Tian QW, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1(2):34-40
14. Van Rechem C, Whetstine JR. Examining the impact of gene variants on histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1463-1476
15. Takashina T, Kinoshita I, Kikuchi J, Shimizu Y, Sakakibara-Konishi J, Oizumi S. et al. Combined inhibition of EZH2 and histone deacetylases as a potential epigenetic therapy for non-small-cell lung cancer cells. Cancer Sci. 2016;107(7):955-962
16. Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521
17. Li XY, Li Y, Zhang Y, Wang K, Yuan X, Jin J. et al. A novel bisindolymaleimide derivative (WK234) inhibits proliferation and induces apoptosis through the protein kinase Cbeta pathway, in chronic myelogenous leukemia K562 cells. Leuk Lymphoma. 2011;52(7):1312-1320
18. McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol Ther. 2015;150:1-22
19. Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S. et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510(7504):283-287
20. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71
21. Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34(2):229-241
22. Eram MS, Bustos SP, Lima-Fernandes E, Siarheyeva A, Senisterra G, Hajian T. et al. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J Biol Chem. 2014;289(17):12177-12188
23. Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA. et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42(10):859-863
24. Hussin J, Sinnett D, Casals F, Idaghdour Y, Bruat V, Saillour V. et al. Rare allelic forms of PRDM9 associated with childhood leukemogenesis. Genome Res. 2013;23(3):419-430
25. Gale M, Sayegh J, Cao J, Norcia M, Gareiss P, Hoyer D. et al. Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance. Oncotarget. 2016;7(26):39931-39944
26. Hou J, Wu J, Dombkowski A, Zhang K, Holowatyj A, Boerner JL. et al. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am J Transl Res. 2012;4(3):247-256
27. Li X, Liu L, Yang S, Song N, Zhou X, Gao J. et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci U S A. 2014;111(19):7096-7101
28. Rodriguez-Paredes M, Martinez de Paz A, Simo-Riudalbas L, Sayols S, Moutinho C, Moran S. et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33(21):2807-2813
29. Wu PC, Lu JW, Yang JY, Lin IH, Ou DL, Lin YH. et al. H3K9 histone methyltransferase, KMT1E/SETDB1, cooperates with the SMAD2/3 pathway to suppress lung cancer metastasis. Cancer Res. 2014;74(24):7333-7343
30. Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7(31):50719-50734
31. Tokunaga R, Sakamoto Y, Nakagawa S, Miyake K, Izumi D, Kosumi K. et al. The Prognostic Significance of Histone Lysine Demethylase JMJD3/KDM6B in Colorectal Cancer. Ann Surg Oncol. 2016;23(2):678-685
32. Tan SX, Hu RC, Liu JJ, Tan YL, Liu WE. Methylation of PRDM2, PRDM5 and PRDM16 genes in lung cancer cells. Int J Clin Exp Pathol. 2014;7(5):2305-2311
33. Kuo KT, Huang WC, Bamodu OA, Lee WH, Wang CH, Hsiao M. et al. Histone demethylase JARID1B/KDM5B promotes aggressiveness of non-small cell lung cancer and serves as a good prognostic predictor. Clin Epigenetics. 2018;10(1):107
34. Huang D, Qiu Y, Li G, Liu C, She L, Zhang D. et al. KDM5B overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck. J Cancer. 2018;9(1):198-204
35. Dai B, Hu Z, Huang H, Zhu G, Xiao Z, Wan W. et al. Overexpressed KDM5B is associated with the progression of glioma and promotes glioma cell growth via downregulating p21. Biochem Biophys Res Commun. 2014;454(1):221-227
36. Kong L, Zhang P, Li W, Yang Y, Tian Y, Wang X. et al. KDM1A promotes tumor cell invasion by silencing TIMP3 in non-small cell lung cancer cells. Oncotarget. 2016;7(19):27959-27974
37. Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P. et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051
38. Yu S, Jia L, Zhang Y, Wu D, Xu Z, Ng CF. et al. Increased expression of activated endothelial nitric oxide synthase contributes to antiandrogen resistance in prostate cancer cells by suppressing androgen receptor transactivation. Cancer Lett. 2013;328(1):83-94
39. Wan L, Li X, Shen H, Bai X. Quantitative analysis of EZH2 expression and its correlations with lung cancer patients' clinical pathological characteristics. Clin Transl Oncol. 2013;15(2):132-138
40. Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li F. et al. Oncogenic Deregulation of EZH2 as an Opportunity for Targeted Therapy in Lung Cancer. Cancer Discov. 2016;6(9):1006-1021
41. Li SH, Lu HI, Huang WT, Tien WY, Lan YC, Lin WC. et al. The Prognostic Significance of Histone Demethylase UTX in Esophageal Squamous Cell Carcinoma. Int J Mol Sci. 2018:19 (1)
42. Wang J, Liu L, Xi W, Long Q, Wang Y, Bai Q. et al. Prognostic value of UTX expression in patients with clear cell renal cell carcinoma. Urol Oncol. 2016;34(8):338 e319-327
43. Zuo SR, Zuo XC, He Y, Fang WJ, Wang CJ, Zou H. et al. Positive Expression of SMYD2 is Associated with Poor Prognosis in Patients with Primary Hepatocellular Carcinoma. J Cancer. 2018;9(2):321-330
44. Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y. et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30(7):1139-1146
45. Komatsu S, Ichikawa D, Hirajima S, Nagata H, Nishimura Y, Kawaguchi T. et al. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer. 2015;112(2):357-364
46. Ohtomo-Oda R, Komatsu S, Mori T, Sekine S, Hirajima S, Yoshimoto S. et al. SMYD2 overexpression is associated with tumor cell proliferation and a worse outcome in human papillomavirus-unrelated nonmultiple head and neck carcinomas. Hum Pathol. 2016;49:145-155
47. Sakamoto LH, Andrade RV, Felipe MS, Motoyama AB, Pittella Silva F. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk Res. 2014;38(4):496-502
48. Veschi V, Liu Z, Voss TC, Ozbun L, Gryder B, Yan C. et al. Epigenetic siRNA and Chemical Screens Identify SETD8 Inhibition as a Therapeutic Strategy for p53 Activation in High-Risk Neuroblastoma. Cancer Cell. 2017;31(1):50-63
49. Liao T, Wang YJ, Hu JQ, Wang Y, Han LT, Ma B. et al. Histone methyltransferase KMT5A gene modulates oncogenesis and lipid metabolism of papillary thyroid cancer in vitro. Oncol Rep. 2018;39(5):2185-2192
50. Li N, Dhar SS, Chen TY, Kan PY, Wei Y, Kim JH. et al. JARID1D Is a Suppressor and Prognostic Marker of Prostate Cancer Invasion and Metastasis. Cancer Res. 2016;76(4):831-843
51. Komura K, Jeong SH, Hinohara K, Qu F, Wang X, Hiraki M. et al. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc Natl Acad Sci U S A. 2016;113(22):6259-6264
52. Liu Y, Luo X, Deng J, Pan Y, Zhang L, Liang H. SMYD3 overexpression was a risk factor in the biological behavior and prognosis of gastric carcinoma. Tumour Biol. 2015;36(4):2685-2694
53. Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73(10):2936-2942
54. Jin X, Xu H, Wu X, Li T, Li J, Zhou Y. et al. KDM4A as a prognostic marker of oral squamous cell carcinoma: Evidence from tissue microarray studies in a multicenter cohort. Oncotarget. 2017;8(46):80348-80357
55. Yuan X, Kong J, Ma Z, Li N, Jia R, Liu Y. et al. KDM4C, a H3K9me3 Histone Demethylase, is Involved in the Maintenance of Human ESCC-Initiating Cells by Epigenetically Enhancing SOX2 Expression. Neoplasia. 2016;18(10):594-609
56. Soini Y, Kosma VM, Pirinen R. KDM4A, KDM4B and KDM4C in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(10):12922-12928
57. Nishikawaji T, Akiyama Y, Shimada S, Kojima K, Kawano T, Eishi Y. et al. Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget. 2016;7(41):67251-67265
58. Ferreira MJ, Pires-Luis AS, Vieira-Coimbra M, Costa-Pinheiro P, Antunes L, Dias PC. et al. SETDB2 and RIOX2 are differentially expressed among renal cell tumor subtypes, associating with prognosis and metastization. Epigenetics. 2017;12(12):1057-1064
59. Maiques-Diaz A, Somervaille TC. LSD1: biologic roles and therapeutic targeting. Epigenomics. 2016;8(8):1103-1116
60. Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R. et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69(5):2065-2071
Author contact
![]() Corresponding authors: Boying Ding, Department of Cardiothoracic Surgery, Yijishan Hospital, Wannan Medical College, Wuhu, 241001, Anhui, PR China, E-mail: dby0067com and Zhangang Xiao, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, 646000, Sichuan, PR China; Email: xzg555898com, Tel: (0086)18308330263
Corresponding authors: Boying Ding, Department of Cardiothoracic Surgery, Yijishan Hospital, Wannan Medical College, Wuhu, 241001, Anhui, PR China, E-mail: dby0067com and Zhangang Xiao, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, 646000, Sichuan, PR China; Email: xzg555898com, Tel: (0086)18308330263

 Global reach, higher impact
Global reach, higher impact