Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(5):654-659. doi:10.7150/ijms.31288 This issue Cite
Research Paper
HSDL2 Promotes Bladder Cancer Growth In Vitro and In Vivo
1. Graduate Faculty, Jiangxi Medical College, Nanchang University, Nanchang 330006;
2. Department of Urology, Jiangxi Provincial People's Hospital Affiliated to Nanchang University, Nanchang 330006;
3. Departments of Gerontology, The First Affiliated Hospital of Nanchang University, Nanchang 330006;
4. Division of Nephrology, The Fifth People's Hospital of Shanghai, Fudan University, Shanghai 200240;
5. Department of Pathology, The First Affiliated Hospital of Nanchang University, Nanchang 330006;
6. Department of Respiratory Medicine, Jiangxi Provincial People's Hospital Affiliated to Nanchang University, Nanchang 330006.
Received 2018-11-6; Accepted 2019-3-27; Published 2019-5-7
Abstract
Bladder cancer is a common malignant urinary tumor, and patients with bladder cancer have poor prognosis. Abnormal lipid metabolism in peroxisomes is involved in tumor progression. Hydroxysteroid dehydrogenase-like 2 (HSDL2) localized in peroxisomes regulates fatty acid synthesis. In the present study, we reported that HSDL2 was upregulated in two human bladder cancer cell lines 5637 and T24 compared to normal human urothelial cells. Furthermore, lentiviral-mediated HSDL2 knockdown inhibited the proliferation and colony formation while promoted the apoptosis of human bladder cancer T24 cells in vitro. In nude mice HSDL2 knockdown inhibited the growth of T24 derived xenografts in vivo. In conclusion, our results suggest that HSDL2 plays an oncogenic role in bladder cancer and might serve as a potential target for bladder cancer therapy.
Keywords: bladder cancer, HSDL2, shRNA, cell proliferation, apoptosis
Introduction
Great efforts have been taken to understand the malignant phenotypes of bladder cancer (BCa), the ninth most common cancer globally [1,2]. Currently, the most common approach to BCa treatment is surgery, although chemotherapy has shown some efficacy [3]. However, about 60-70% of cases with metastatic BCa relapse in the first year due to chemoresistance [4]. Therefore, it is urgent to understand the mechanisms of BCa progression.
Abnormal lipid metabolism is an important hallmark of cancer, and tumor cells have significantly increased level of ether lipids [5,6]. ADHAPS, ether lipid synthesis enzyme, was upregulated in various cancer cells and tissues such as breast cancers and melanomas [7]. Peroxisome is an exclusive organelle required for ether lipid production [8]. Hydroxysteroid dehydrogenase-like 2 (HSDL2) is widely expressed in human tissues and localized in peroxisomes [9,10]. HSDL2 plays an important role in fatty acid metabolism [11]. Previous study has shown that HSDL2 is involved in glioma development [11]. However, the role of HSDL2 in BCa remains elusive.
This study aims to investigate the role of HSDL2 in BCa progression. First, we investigated the expression of HSDL2 in human BCa cells. Next, we employed lentivirus mediated small hairpin (shRNA) to knockdown HSDL2 and examined the effects on BCa cell phenotypes in vitro, and tumor growth in vivo.
Materials and Methods
Cell culture
Two human BCa cell lines 5637 and T24 were provided by Cell Bank at Chinese Academy of Sciences (Shanghai, China), and cultured at 37°C in DMEM medium (Gibco, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml streptomycin/penicillin (Sangon, Shanghai, China) in a humid incubator with 5% CO2. Normal human urothelial cells (NHUCs) were provided by Oligene (Berlin, Germany) and cultured in urothelial cell medium (Oligene).
Lentivirus construction and infection
The human HSDL2-specific targeting sequence (5′-CCA GAA GCA GTT AGC AAG AAA-3′) and a scrambled shRNA (5′-TTC TCC GAA CGT GTC ACG T-3′) were designed at GeneChem (Shanghai, China). HSDL2 or scrambled hairpin oligonucleotides were subcloned into pGCSIL-GFP lentiviral vector (GeneChem) and named as shHSDL2 and shCtrl, respectively. T24 and 5637 cells were seeded in 6-well plates and cultured for 48 h. Then, the cells were incubated with lentivirus shHSDL2 or shCtrl (MOI = 5) for 72 h, the efficiency of infection was calculated by evaluating GFP expression under fluorescence microscope (XI71, Olympus, Tokyo, Japan), and cells were collected for further analysis.
PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen). cDNA was synthesized using oligo dT primers and M-MLV reverse transcriptase (Promega, Shanghai, China). The level of HSDL2 mRNA was determined by PCR with SYBR master mixture (Takara Biotech, Dalian, China) as follows: denaturation at 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 40 s. The primers were synthesized by Sangon Biotech with the following sequences: HSDL2 forward: 5′-AAG CCA CTC AAG CAA TCT ATC TG-3′; HSDL2 reverse: 5′-GCT CTC CAT ATC CGA CAT TCC C-3′. GAPDH forward: 5′-TGA CTT CAA CAG CGA CAC CCA-3′; GAPDH reverse: 5′-CAC CCT GTT GCT GTA GCC AAA-3′. Relative HSDL2 mRNA level was normalized to GAPDH and calculated with delta-delta CT method.
Western blot analysis
T24 cells were lysed in lysis buffer (100 mM Tris-HCl, pH 7.4, 0.15 M NaCl, 1% Triton X-100, 5 mM EDTA, and 5 mM DTT) supplemented with protease inhibitors. Total 20 μg proteins were separated by 12.5% SDS-PAGE and transferred onto the membranes (Sangon Biotech), which were blocked in 5% milk dissolved in TBST for 1 h at room temperature and incubated with antibodies to HSDL2 and GAPDH (Santa Cruz Biotech, Santa Cruz, CA, USA) overnight at 4°C. The membranes were then washed three times with TBST and incubated with goat anti-mouse IgG coupled to HRP (Santa Cruz Biotech), and the blots were examined by using ECL-Plus kit (Sangon Biotech).
MTT assay
T24 and 5637 cells at logarithmic phase were collected and seeded in 96-well plates at 2,000 cells/well in triplicate. Cells were incubated at 37°C in a humid incubator with 5% CO2 for 5 days. Each day, 20 μL 5 mg/mL MTT solution was added into each well, 4 h later the supernatant was removed and 150 μL DMSO was added. The plates were shaken gently for 10 min, and the absorbance at 490 nm was quantified using a microplate reader.
Annexin V-APC assay
The apoptosis was examined using apoptosis detection kit (eBioscience, San Diego, CA, USA). T24 cells were washed with PBS and suspended at a density of 1 ×106/ml. 100 μl cell suspensions were incubated with 5 μl annexin V-APC for 10 min in the dark, and the stained cells were immediately used for cytometric analysis on a FACS Calibur (Becton- Dickinson, San Jose, CA, USA).
Colony formation assay
After infection with ShHSDL2 or shCtrl, T24 cells were seeded into 6-well plates (800 cells/well) in triplicate, and incubated at 37°C with 5% CO2 for 14 days. Then the cells were washed with PBS and fixed in 4% paraformaldehyde for 1 h. Next, the cells were washed with PBS and stained in 500 μl Giemsa (Sigma) for 20 min, and the number of colony was counted under light microscope.
Xenograft on nude mice
Female BALB/c nude mice (4-week old) were injected with 1×105 T24 cells subcutaneously. Tumor volume was measured once every 2-3 days from 27 days after cell injection. Bioluminescent imaging was performed with the In Vivo Imaging Solutions (IVIS, PerkinElmer, Waltham, USA) as described previously [12]. The mice were anesthetized with isofluorance, injected with 10 µl/g D-Luciferin (Sigma) and imaged by IVIS. Images were analyzed using Living Image software v4.1 (PerkinElmer) as described previously [13].
Statistical analysis
Data are expressed as mean ± SD and statistical analysis was performed by using SPSS version 16.0 software (SPSS Inc, Chicago, IL, USA). The differences were comapred by Student's t test, and P value < 0.05 was considered to be statistically significant.
Results
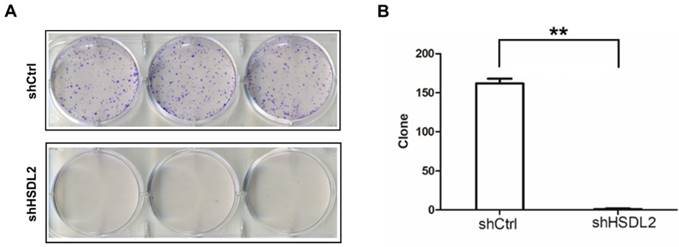
Lentivirus-based shRNA strategy to knockdown HSDL2
To explore the functional role of HSDL2 in BCa, first we need to establish the cell model. We compared HSDL2 mRNA expression in two human BCa cell lines and NHUCs and found significant higher expression of HSDL2 in BCa cells, especially in T24 cells (Fig. 1A). Next we employed shRNA lentivirus to knockdown HSDL2 in T24 cells. qPCR analysis of HSDL2 mRNA showed that knockdown efficiency of shHSDL2 was approximately 82% (Fig. 1B). Furthermore, Western blot analysis showed that shHSDL2 efficiently inhibited HSDL2 protein expression in T24 cells (Fig. 1C, D).
HSDL2 knockdown inhibited the proliferation of T24 cells
Next we evaluated the proliferation of human BCa cells after HSDL2 knockdown. MTT assay showed that the proliferation of both T24 and 5637 cells was inhibited significantly after transduction of shHSDL2 lentivirus (Fig. 2A, B). These data indicate that HSDL2 could promote BCa cell proliferation.
HSDL2 knockdown in BCa cells. A. HSDL2 expression at mRNA level in two human BCa cell lines 5637 and T24, and normal human urothelial cells (NHUCs). *P< 0.05 vs. NHUCs. B. HSDL2 expression at mRNA level in T24 cells after infection with shRNA lentivirus. **P< 0.01. C. Western blot analysis of HSDL2 protein level in T24 cells after infection with shRNA lentivirus. D. Densitometry analysis of HSDL2 protein level in T24 cells after infection with shRNA lentivirus. **P< 0.01.
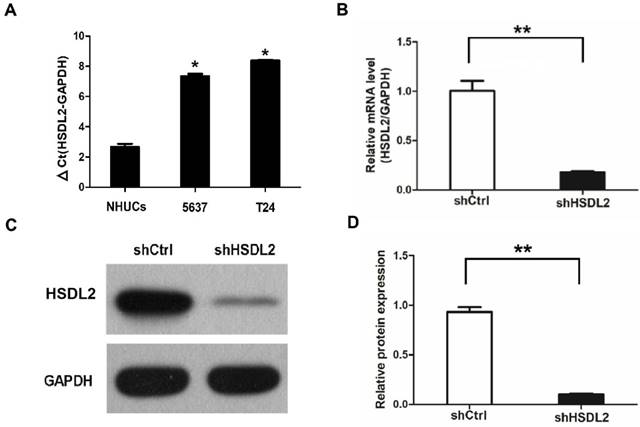
HSDL2 knockdown inhibited the proliferation of BCa cells. Cell proliferation was analyzed by MTT assay for continuous 5 days. Cell proliferation is shown as fold change compared to absorbance at OD490 on day 1. A. T24 cells transduced with shRNA lentivirus. B. 5637 cells transduced with shRNA lentivirus. The results are presented as the mean ± SD of three separate experiments.
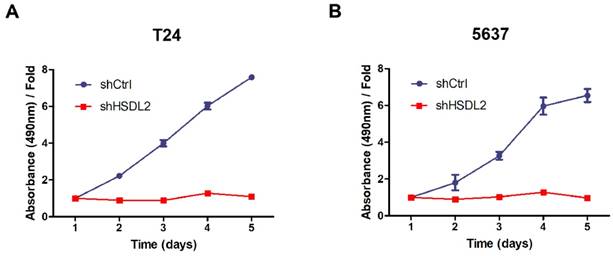
HSDL2 knockdown augmented the apoptosis of T24 cells. A. Representative images of apoptosis analysis of T24 cells infected with lentivirus shCtrl or shHSDL2. B. Quantitative analysis of apoptosis percentage in T24 cells infected with lentivirus shCtrl or shHSDL2. Data shown are the mean ± SD from three separate experiments. **P < 0.01.
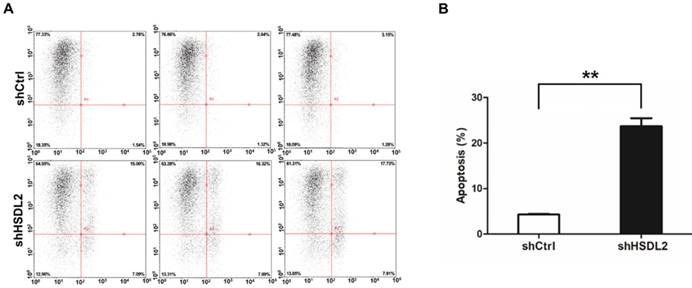
HSDL2 knockdown inhibited T24 cell colony formation. A. Photomicrographs of Giemsa-stained T24 colonies in 6-well plates 10 days post seeding. B. Quantitative analysis of colonies formed in T24 cells infected with lentivirus shCtrl or shHSDL2. Data shown are the mean ± SD from three separate experiments. **P < 0.01.
HSDL2 knockdown induced the apoptosis of T24 cells
To determine how HSDL2 could promote BCa cell proliferation, we examined the apoptosis of T24 cells by Annexin V-APC assay. As shown in Fig. 3, 4.3% of cells infected with shCtrl underwent apoptosis, but 23.7% of cells infected with shHSDL2 underwent apoptosis (P<0.05). These data indicate that HSDL2 may promote BCa cell proliferation by inhibiting apoptosis.
HSDL2 knockdown repressed colony formation of T24 cells
Next, we investigated the colony formation capacity of T24 cells infected by shHSDL2 or shCtrl lentivirus. As shown in Fig. 4, the number of colonies was significantly less in HSDL2 knockdown group compared to shCtrl group.
HSDL2 knockdown inhibited BCa in vivo
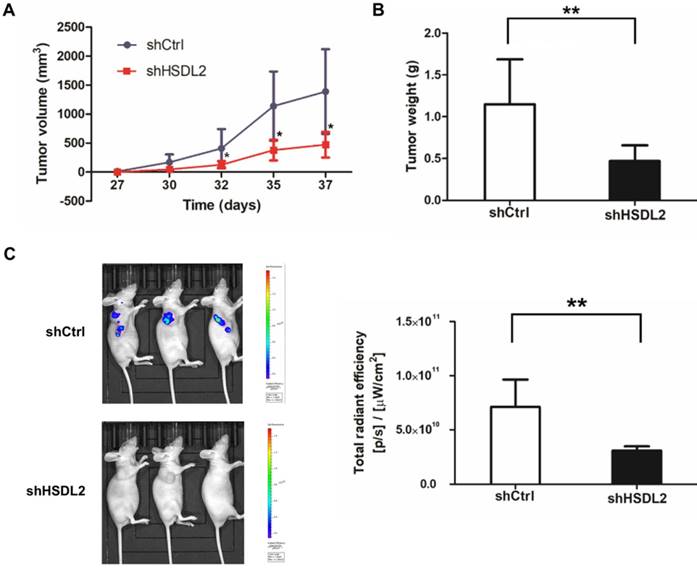
Finally, to determine in vivo effects of HSDL2 knockdown on BCa, we established nude mice xenografted with T24 cells. More than 5 weeks following injection, tumors in group were smaller compared to shCtrl group (Fig. 5A, B). Moreover, bioluminescent imaging showed that all mice injected with shHSDL2-infected T24 cells had rarely detectable tumors 37 days after implantation (Fig. 5C). Collectively, these results suggest that HSDL2 promotes the progression of BCa in vivo.
Discussion
In this study, we found that HSDL2 was highly expressed in two BCa cell lines. We further showed that T24 cell proliferation and colony formation were significantly decreased after shRNA lentivirus mediated HSDL2 knockdown, accompanied by the accumulation of apoptosis. Furthermore, our in vivo data in nude mice demonstrated that HSDL2 knockdown significantly inhibited BCa growth.
HSDL2 knockdown inhibited tumor growth in nude mice. A. Reduced tumor volume of xenografts generated by T24 cells infected with lentivirus shHSDL2. *P < 0.05, compared to T24 cells infected with control lentivirus. B. Reduced tumor weight of xenografts generated by T24 cells infected with lentivirus shHSDL2. **P < 0.01. C. Representative bioluminescent imaging of T24 implant and luminescent units on day 37 after cell injection. Data shown are the mean ± SD (n=10). **P < 0.01.
HSDL2 protein is localized in the peroxisomes and mitochondria and plays a crucial role in fatty acid metabolism [14,15]. However, up to date, only one study reported abnormal HSDL2expression in glioma. Moreover, knockdown of HSDL2 inhibited the proliferation and induced the apoptosis of glioma cells [11]. Consistent with previous study, our results confirmed that HSDL2 promoted BCa progression because HSDL2 knockdown suppressed the proliferation and accumulated the apoptosis in human BCa T24 cells. Furthermore, HSDL2 knockdown led to less colony formation in vitro and reduced tumour growth in vivo.
Peroxisomes are pivotal for lipid production, in particular for ether lipids production. Abnormality in ether lipids in various cancers has been reported recently [16,17]. Further studies are needed to elucidate the mechanism how abnormal HSDL2 expression and ether lipid synthesis regulate BCa cell proliferation and invasion. Recently, gene chip analysis has been used to reveal the mode of actions of anti-cancer reagents [18]. We will employ similar approach to elucidate molecular mechanism by which abnormal HSDL2 expression regulates BCa progression. In conclusion, our study suggests that HSDL2 is a potential therapeutic target for BCa.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Salehi S, Mansoori B, Mohammadi A. et al. An analysis of suppressing migratory effect on human urinary bladder cancer cell line by silencing of snail-1. Biomed Pharmacother. 2017;96:545-550
2. Liu M, Zhang X. An integrated analysis of mRNA-miRNA transcriptome data revealed hub regulatory networks in three genitourinary cancers. Biocell. 2017;41:19-26
3. Rosenberg JE. Current status of neoadjuvant and adjuvant chemotherapy for muscle-invasive bladder cancer. Expert Rev Anticancer Ther 7: 1729-1736. 2007
4. Sun L, Lu J, Niu Z. et al. A Potent Chemotherapeutic Strategy with Eg5 Inhibitor against Gemcitabine Resistant Bladder Cancer. PloS One 10: e0144484. 2015
5. Albert DH, Anderson CE. Ether-linked glycerolipids in human brain tumors. Lipids 12: 188-192. 1977
6. Roos DS, Choppin PW. Tumorigenicity of cell lines with altered lipid composition. Proc Natl Acad Sci USA 81: 7622-7626. 1984
7. Benjamin DI, Cozzo A, Ji X. et al. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci U S A 110: 14912-14917. 2013
8. Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab 19: 380-392. 2014
9. Dai J, Xie Y, Wu Q. et al. Molecular cloning and characterization of a novel human hydroxysteroid dehydrogenase-like 2 (HSDL2) cDNA from fetal brain. Biochem Genet 41: 165-174. 2003
10. Gronemeyer T, Wiese S, Ofman R. et al. The proteome of human liver peroxisomes: identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PloS One 8: e57395. 2013
11. Ruokun C, Yake X, Fengdong Y, Xinting W, Laijun S, Xianzhi L. Lentivirus-mediated silencing of HSDL2 suppresses cell proliferation in human gliomas. Tumour Biol 37: 15065-15077. 2016
12. Toyoshima M, Tanaka Y, Matumoto M. et al. Generation of a syngeneic mouse model to study the intraperitoneal dissemination of ovarian cancer with in vivo luciferase imaging. Luminescence 24: 324-331. 2009
13. Neff BA, Voss SG, Allen C. et al. Bioluminescent imaging of intracranial vestibular schwannoma xenografts in NOD/SCID mice. Otol Neurotol 30: 105-111. 2009
14. Brites P, Motley AM, Gressens P. et al. Impaired neuronal migration and endochondral ossification in Pex7 knockout mice: a model for rhizomelic chondrodysplasia punctata. Hum Mol Genet 12: 2255-2267. 2003
15. Kowalik D, Haller F, Adamski J, Moeller G. In search for function of two human orphan SDR enzymes: hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O). J Steroid Biochem Mol Biol 117: 117-124. 2009
16. Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380-92
17. Dean JM, Lodhi IJ. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196-206
18. Zhong J, Deng L, Jiang Y, Zou L, Yuan H, Tan S. Gene expression profiling of HepG2 cells after treatment with black tea polyphenols. Biocell. 2018;42:99-104
Author contact
![]() Corresponding author: Dr. Qiu-Gen Li, Department of Respiratory Medicine, Jiangxi Provincial People's Hospital Affiliated to Nanchang University, Nanchang 330006, P.R. China. E-mail: Drlqg66com
Corresponding author: Dr. Qiu-Gen Li, Department of Respiratory Medicine, Jiangxi Provincial People's Hospital Affiliated to Nanchang University, Nanchang 330006, P.R. China. E-mail: Drlqg66com

 Global reach, higher impact
Global reach, higher impact