Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(4):576-582. doi:10.7150/ijms.32773 This issue Cite
Research Paper
Decreased DNA methyltransferases expression is associated with coronary artery lesion formation in Kawasaki disease
1. Department of Pediatrics, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung, Taiwan.
2. Kawasaki Disease Center, Kaohsiung Chang Gung Memorial Hospital, Kaohsiung, Taiwan.
3. Institute for Translational Research in Biomedicine, Liver Transplantation Center and Department of Surgery, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung, Taiwan.
4. Baoan Maternity and Child Health Hospital, Shenzhen, Guangdong Province, China. 518100.
Received 2019-1-3; Accepted 2019-3-23; Published 2019-4-25
Abstract
Background: Kawasaki disease (KD) is the most common acute coronary vasculitis to occur in children. Although we have uncovered global DNA hypomethylation in KD, its underlying cause remains uncertain. In this study, we performed a survey of transcript levels of DNA methyltransferases and demethylases in KD patients.
Materials and Methods: We recruited 145 participants for this study. The chip studies consisted of 18 KD patients that were analyzed before undergoing intravenous immunoglobulin (IVIG) treatment and at least 3 weeks after IVIG treatment, as well as 36 control subjects, using Affymetrix GeneChip® Human Transcriptome Array 2.0. An additional study of 91 subjects was performed in order to validate real-time quantitative PCR.
Results: In our microarray study, the mRNA levels of DNMT1 and DNMT3A were significantly lower while TET2 was higher in acute-stage KD patients compared to the healthy controls. Through PCR validation, we observed that the expression of DNMT1 and TET2 are consistent with the Transcriptome Array 2.0 results. Furthermore, we observed significantly lower DMNT1 mRNA levels following IVIG treatment between those who developed CAL and those who did not.
Conclusion: Our findings provide an evidence of DNA methyltransferases and demethylases changes and are among the first report that transient DNA hypomethylation is induced during acute inflammatory phase of Kawasaki disease.
Introduction
Kawasaki disease (KD) is an acute vasculitis syndrome that covers multiple systems, has an unknown etiology, and primarily occurs in children under the age of 5 years old. In 1974, Tomisaku Kawasaki first published 50 cases of KD in the English language [1]. KD is characterized by prolonged fever, conjunctivitis, diffuse mucosal inflammation, polymorphous skin rashes, indurative edema of the hands and feet associated with peeling of finger tips, and nonsuppurative lymphadenopathy [2]. Vascular involvement in KD occurs in small and medium-sized blood vessels, particularly the coronary arteries. The most serious complication of KD is coronary artery lesions (CAL), including myocardial infarction and coronary artery aneurysms. A sequela of vasculitis, coronary artery aneurysms are developed in 20% of untreated children [3]. A U.S. multicenter study group established that a single high-dose of 2 g/kg intravenous immunoglobulin (IVIG) plus aspirin could lower the incidence of aneurysm from 20%-25% to 3-5% [4].
Epigenetic lesions result in changes to both the chromatin structure and the DNA methylation and acetylation pattern of the genome [5]. In general, the DNA methylation alteration of CpG sites is a powerful transcription inhibitor [6]. DNA methylation status is established by DNA methyltransferases (DNMTs) [6] and the Ten-eleven translocation (TET) family [7]. The three active DNA methyltransferases are DNMT1, DNMT3A, and DNMT3B, and three DNA demethylase, TET1-3, have been identified in mammals [7, 8]. We have previously shown considerably increased mRNA expressions in toll-like receptors [9], hepcidin [10, 11], matrix metalloproteinases [12], inflammasome sensors of NOD-like receptors [13], and hypomethylation at the gene promoters of these genes, as well as that IVIG treatment can drastically alter these methylation patterns in the WBC cells of KD patients [9-14]. Consistently, we have demonstrated that 87.8% of the most of the significant CpG markers between KD patients and controls are hypo-methylation of CpG markers by genome-wide screening on DNA methylation patterns with Illumina HumanMethylation450 (M450K) Bead-Chip microarray assay [15].
Chen et al. reported that, of the 3193 CpG methylation regions with a methylation difference ≥ 20% between KD and controls, 3096 CpG loci revealed hypomethylation (97%) and only 3% hypermethylation [16], which indicates that more than 97% of genes in KD patients have a hypomethylation status, as well as a potential increase in gene expression levels. KD is a specific disease with an activated status of most genes, most of which have the condition of overexpression, including T helper 1 (Th1), Th2, Th17, innate immunity, adaptive immunity, inflammatory cytokines, chemokines, etc. Like the etiology, the reason why most genes are activated during the acute stage of KD is still unknown. Regulation of DNA methylation by DNA methyltransferases and TET may be key factors of this condition. This study is the first to evaluate the change of DNA methyltransferases and TET in KD and subsequent disease outcome.
Materials and Methods
Patients
We recruited 145 participants for this study (Table 1). The recruited KD patients met the American Heart Association diagnosis criteria of KD, which is characterized by fever for more than 5 days, oral mucosal inflammation with fissure lips or strawberry tongue, bilateral non-exudative conjunctivitis, non- suppurative lymphadenopathy over the neck, polymorphous skin rashes over the body surface, and indurative edema of the hands and feet associated with peeling of the finger tips [17, 18], and were treated with high-dose IVIG treatment (2 g/kg) over 12 hours at our hospital. In this study, we quantified and compared the gene expressions of DNA methylation status established by DNA methyltransferases (DNMTs) and the Ten-eleven translocation (TET) family in 18 KD patients (both before and at least 3 weeks after IVIG treatment), as well as in 18 healthy and 18 febrile controls using Affymetrix GeneChip® Human Transcriptome Array 2.0. Then, we validated the mRNA levels of genes in 39 KD patients and 52 controls using real-time quantitative PCR. The patients in the fever control group were diagnosed with acute tonsillitis, bronchitis, otitis media, bronchopneumonia, enterovirus, or urinary tract infection. We also used peripheral blood samples from KD patients before they underwent IVIG treatment (pre-IVIG) and then at least 3 days or 3 weeks after completing the IVIG treatment, as previously described in one of our previous studies [19]. CAL was identified through echocardiography and defined as a coronary artery with an internal diameter of at least 3 mm (4 mm if the patient was more than 5 years old), a segment with an internal diameter at least 1.5 times larger than that of an adjacent segment, as [20, 21], or a Z score ≧ 2.5, and the severity of the coronary was classified using Z scores according to the 2017 AHA statement [22, 23]. This study received approval from the Chang Gung Memorial Hospital's Institutional Review Board, and we also obtained written informed consent from the parents or guardians of all subjects. All of the methods used herein complied with the relevant guidelines established. The enrolled children were allowed to withdraw at any time during the study period, and all experimental results were anonymized before analysis.
Basal characteristics of patients with KD and controls
| Healthy controls | Febrile controls | Patients with KD | |
|---|---|---|---|
| Characteristic | (HTA 2.0=18 / qRT-PCR = 17) | (HTA 2.0 =18 /qRT-PCR = 35) | (HTA 2.0 = 18 /qRT-PCR = 39) |
| Male gender, n(%) | 9(50) / 11(64.7) | 8(44.4) / 22(62.9) | 9(50) / 32(82.1) |
| Mean (SEM), age (y) | 3.5±0.6 / 6.9±1.3 | 2.0±0.3 / 3.1±0.3 | 1.9±0.3 / 2.1±0.5 |
| Age range (y) | 1-10 / 1-16 | 0-4 / 0-12 | 1-5 / 0-18 |
| CAL formation | 6(33.3%) / 22(56.4%) | ||
| IVIG resistance | 1(6%) / 3(8%) |
CAL, coronary artery lesion; IVIG, intravenous immunoglobulin; KD, Kawasaki disease.
Experiment design
For this study, we collected whole blood samples from the subjects and submitted them to white blood cell (WBC) enrichment, as we have previously described in other studies [11, 14].
Gene expression profiling with microarray
To obtain unbiased results, we created pooled RNA libraries by evenly pooling six RNA samples, which resulted in three pooled healthy control, three fever control, three pre-IVIG, and three post-IVIG libraries, as previous described [9]. We performed microarray assay on the pooled RNA samples to establish the gene expression profiles and then further performed profiling with GeneChip® Human Transcriptome Array 2.0 (HTA 2.0, Affymetrix, Santa Clara). We used the WT PLUS Reagent kit to prepare the RNA samples and carry out hybridization on the HTA 2.0 microarray chips. Adhering to the Affymetrix instruction manual, we subjected the HTA 2.0 chips' raw data to quality control examination, as previously described in another study [9, 15].
RNA isolation and real-time quantitative RT-PCR
To quantify the mRNA levels of DNMT1, DNMT3A, DNMT3B, and TET1-3, we adopted the LightCycler® 480 Real-Time PCR System (Roche Molecular Systems, Inc., IN, USA) to perform real- time quantitative PCR. We separated the total mRNA from the WBC using an isolation kit (mirVana™ miRNA Isolation Kit, Catalog number: AM1560, Life Technologies, Carlsbad, CA) and then calculated both the quality (RIN value) and quantity of the RNA samples using Bioanalyzer (ABI) and Qubit (Thermo) in accordance with the manufacturer's instructions. All RNA samples passed the criterion of RIN≧7. We performed PCR using a SYBR Green PCR Master Mix containing 10 μM of specific forward and reverse primers. We performed the relative quantification of gene expression based on the comparative threshold cycle (CT) method, which allowed us to determine the target amount as 2-(ΔCT target - Δ CT calibrator) or 2-ΔΔCT [24]. Primers were designed to amplify the target genes, as demonstrated in Table 2.
Statistical Analysis
All data are presented as mean ± standard error. Once chips passed the quality control criteria, we evaluated them with Partek (Partek, St. Louis), commercial software specifically designed to analyze microarray data. We adopted one-way ANOVA or Student's t-test as necessary to evaluate the quantitative data, while we used the paired sample t-test to evaluate any data changes before and after IVIG treatment [9]. We carried out all statistical analyses with SPSS version 12.0 for Windows XP (SPSS, Inc., Chicago, USA), and we considered a two-sided p-value less than 0.05 statistically significant.
Primers list
| Gene symbol | Accession number | Hybridization | Primers (5' to 3') |
|---|---|---|---|
| RNA18S5 | NR_003286.2 | forward | GTAACCCGTTGAACCCCATT |
| reverse | CCATCCAATCGGTAGTAGCG | ||
| DNMT1 | NM_001130823 | forward | CCAAAGAACCAACACCCAAAC |
| reverse | CTCATCTTTCTCGTCTCCATCTTC | ||
| DNMT3A | NM_175630 | forward | ACGATTGCTAGACTGGGATAATG |
| reverse | AGTAAGCAGGCCAGGTAGA | ||
| DNMT3B | NM_175850 | forward | GGAGCCACGACGTAACAAATA |
| reverse | GTAAACTCTAGGCATCCGTCATC | ||
| TET1 | NM_030625 | forward | GGTCCTAGCAAATCAGACAGAG |
| reverse | GTCGGTAGCAAAGTGGTATAGG | ||
| TET2 | NM_017628 | forward | CTTCCTCACTTAGCTCGTCATATC |
| reverse | TAACCCTACAGTGGCCTCTAA | ||
| TET3 | NM_001287491.1 | forward | TTGGTTCCACACCTGTCTTC |
| reverse | CCTGGCTATGAGAATGCCTATC |
Results
Significantly altered expressions of DNMTs and TETs' mRNA levels in KD patients and controls and changes following IVIG treatment
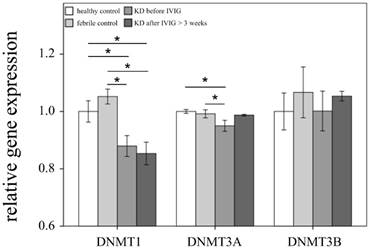
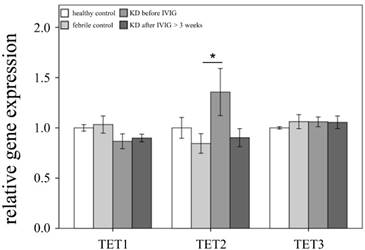
This study included 145 participants. At the beginning of this study, we used Affymetrix GeneChip® Human Transcriptome Array 2.0 to identify the expression profiling of DNMTs and TETs in both the KD patients and the control subjects. As shown in Figures 1 and 2, we observed differential expressions of DNMT1, DNMT3A, and TET2 in KD patients when compared to both the febrile and healthy control subjects. The mRNA levels of both DNMT1 and DNMT3A were significantly lower, while TET2 was higher, in acute-stage KD patients compared to the healthy controls (p=0.047, 0.022, 0.176, respectively) and febrile controls (p = 0.011, 0.045, 0.044, respectively). Furthermore, DNMT1 expression values were significantly lower in KD patients after they underwent IVIG treatment (p<0.05).
Significantly decreased DNMT1 and increased TET2 expressions in the WBCs of KD patients
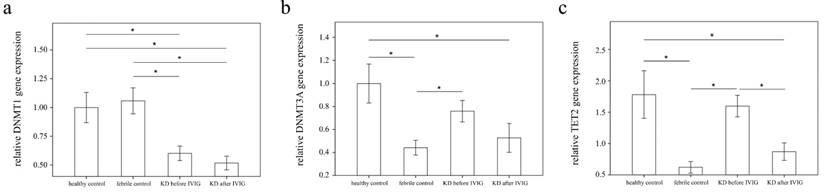
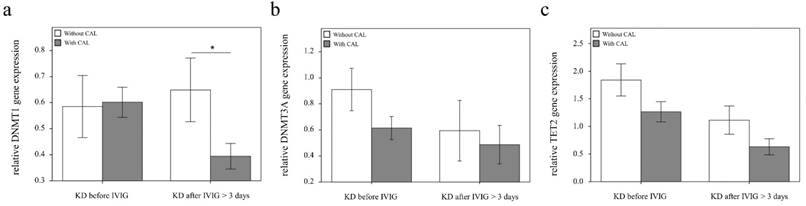
Using real-time PCR, we investigated the mRNA levels of DNMT1, DNMT3A, and TET2 in a separate cohort of 39 KD patients, 17 health and 35 febrile controls. We found decreased DNMT1 in the WBCs of KD patients compared to those of the health and febrile control subjects, as shown in Figure 3 (p =0.018, p =0.001, respectively) and increased TET2 mRNA levels in the KD patients compared to those of febrile controls (p <0.001). Both the DNMT1 and TET 2 findings were consistent with the Affymetrix GeneChip® Human Transcriptome Array 2.0 results. Furthermore, the mRNA level of TET2 decreased following IVIG treatment (p =0.001) (Figure 3). Notably, in KD patients who had already been treated with IVIG, we observed significantly lower DMNT1 mRNA levels between those who developed CAL and those who did not (p = 0.037) (Figure 4).
Discussion
Our particularly noteworthy observations include that KD patients demonstrated differential expressions of DNMT1, DNMT3A, and TET2 when compared to both the febrile and healthy control subjects. The mRNA levels of DNMT1 and DNMT3A were significantly lower, while TET2 was significantly higher, in the acute stage of KD patients than in the healthy controls. Of particular note, we observed significantly lower DMNT1 mRNA levels following IVIG treatment between those who developed CAL and those who did not.
Comparison of DNA methyltransferases (DNMTs) mRNA expressions by GeneChip® Human Transcriptome Array 2.0 between acute-stage Kawasaki disease (KD) patients and control subjects. * indicates significance (p < 0.05). Data are expressed as mean ± standard error for the three replications.
Comparison of Ten-eleven translocation (TET) family mRNA expressions by GeneChip® Human Transcriptome Array 2.0 between acute-stage Kawasaki disease (KD) patients and control subjects. * indicates significance (p < 0.05). Data are expressed as mean ± standard error for the three replications.
Analyses of DNA methyltransferases (DNMTs) and demethylases (TETs) mRNA in the peripheral white blood cells of 39 patients with KD before and after intravenous immunoglobin administration as well as 52 controls using a real-time quantitative polymerase chain reaction. Data are expressed as mean ±standard error. *indicates a p < 0.05 between the groups.
Comparison of DNMT1, 3A, and TET2 mRNA in KD patients with (n = 20) and without (n = 19) coronary artery lesion (CAL). Data are presented as mean ±standard error. *indicates a p < 0.05 between the groups
KD is a systemic vasculitis that primarily affects children under the age of 5 years old that can result in life-threatening complications. Vasculitis represents a group of systemic inflammatory diseases of the blood vessels. Despite recent progress with regard to understanding the genetic basis and the underlying pathogenic mechanisms of vasculitis, the etiology and pathogenesis of vasculitis, like the etiology of KD, remain unknown. Epigenetic dysregulation plays a crucial role in immune-mediated diseases, and the contribution of epigenetic aberrancies in vasculitis is being increasingly recognized [25]. Previous studies have revealed important epigenetic contributions to vasculitides, including KD, Behçet's disease, giant cell arteritis, and IgA vasculitis [26]. More recently, genome-wide epigenomic studies have been performed for several vasculitides [25]. Our results of a decreased expression of DMNT1 and an increased expression of TET2 are consistent with our previous reports of hypomethylation of promotor of β-catenin [16], NOD-like receptors [13], matrix metalloproteinases [9], toll-like receptors [9] and HAMP [11] in KD patients when compared to age-matched controls that presented with fever/without fever and no history of KD. Global genomic hypomethylation in PBMCs has been observed not only in our recent studies of Kawasaki disease, but also in a number of inflammatory and autoimmune diseases, such as systemic lupus erythematous (SLE), rheumatoid arthritis (RA), etc., where it also correlates with aberrant gene expression that likely contributes to pathogenesis [27-30]. However, many aspects of DNA hypomethylation in their pathology are still lacking research.
The DNMT family consists of a conserved set of DNA-modifying cytosine methylases that have a vital role in epigenetic regulation [6]. DNMT activity is highly regulated in humans. Key genetic regulatory mechanisms include molecular interactions, post- translational modifications, alternative splicing, and gene duplication or gene loss [31]. DNMTs have an important role in the epigenetic alteration of immune cells, as well as potentially in the pathogenesis of disease through gene expression dysregulation [32] . DNMT1 was the most significantly and consistently decreased DNA methyltransferase in comparison to both healthy and febrile controls in this study. Since KD has generally been considered an autoimmune- like systemic vasculitis, this result also agrees with observations made in several inflammatory and autoimmune diseases [27-30]. For example, impaired DNMT1 expression contributes to global DNA hypomethylation, and autoimmunity has been best studied in drug-induced SLE [29]. Furthermore, global DNA hypomethylation was specifically observed in T cells and monocytes of RA patients, together with a lower expression of DNMT1 [27]. The global DNA hypomethylation in the PBMCs of KD patients was primarily observed in our previous study using HumanMethylation27 BeadChip assay, in which we identified an increase of FCGR2A associated with its hypomethylation and a susceptibility to IVIG resistance [33]. We further illustrated a more comprehensive study using HumanMethylation27 BeadChip assay, which showed that 97% of CpG regions with a methylation difference ≥ 20% between KD and controls were hypomethylated [16]. We determined that a significant decrease of β-catenin was associated with its hypomethylation in the promoter, as well as in the pathogenesis and cause of coronary arterial lesions in KD. Interestingly, global DNA hypomethylation can relapse after IVIG treatment, which indicates that a dynamic balance in enzymatic regulation for DNA methylation may still exist (data not shown). In our present study, the down-regulation of DNMT1 seems to be a major factor in DNA hypomethylation. However, the decrease of DNMT1 was not affected by IVIG treatment in KD patients. The expression level of DNMT3A was even reduced in KD patients 3 days after receiving IVIG treatment. Therefore, the significant increase of demethylation enzyme TET2 and its concomitant decrease after KD patients undergo IVIG treatment may also be responsible for and participate in the dynamic regulation of global DNA methylation in KD patients. The TET family of enzymes has recently been discovered to oxidize 5mC to hydroxymethyl cytosine (5hmC) and subsequently trigger passive, DNA replication-dependent DNA demethylation and contribute to the dynamics of DNA methylation [34-36]. One previous study has suggested that TET proteins play a protective role against abnormal methylation caused by oxidative stress by interacting with DNMTs in a Yin-Yang relationship toward targeted transcription events [37]. However, little is known regarding the mechanisms with which both methyltransferases and demethylation enzymes were dysregulated, contribute to global DNA hypomethylation in PBMCs, and are associated with disease progression in KD.
Conclusion
This report is the first to provide an epigenetic and genetic study of the changes of DNA methyltransferases and demethylases and among the first to suggest transient DNA hypomethylation during KD's acute inflammatory phase.
Abbreviations
CAL: coronary artery lesions; DNMT: DNA methyltransferases; IVIG: intravenous immunoglobulin; KD: Kawasaki disease; PCR: Polymerase chain reaction; TET: Ten-eleven translocation; WBC: white blood cell.
Acknowledgements
This study received funding from the following grants: MOST 105-2314-B-182-050-MY3 and MOST 103-2410-H-264-004, provided by the Ministry of Science and Technology of Taiwan, and CMRPG8F1931, 8E0212, and CORPG8F0012, provided by Chang Gung Memorial Hospital in Taiwan. Although these organizations provided financial support, they had no influence on the manner in which we collected, analyzed, or interpreted the data or prepared this manuscript.
Authors' contributions
Ying-Hsien Huang, Wei-Dong Huang and Ho- Chang Kuo conceptualized and designed the study, drafted the initial manuscript, critically reviewed the manuscript, and approved the final manuscript as submitted.
Mao-Hung Lo, Xin-Yuan Cai, Ling-Sai Chang and Kuang-Den Chen designed the data collection instruments, coordinated, supervised data collection and approved the final manuscript as submitted.
Ethics approval and consent to participate
This study received approval from the Chang Gung Memorial Hospital's Institutional Review Board, and we also obtained written informed consent from the parents or guardians of all subjects. All of the methods used herein complied with the relevant guidelines established. The enrolled children were allowed to withdraw at any time during the study period, and all experimental results were anonymized before analysis.
Availability of data and material
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kawasaki T, Kosaki F, Okawa S, Shigematsu I, Yanagawa H. A new infantile acute febrile mucocutaneous lymph node syndrome (MLNS) prevailing in Japan. Pediatrics. 1974;54:271-6
2. Wang CL, Wu YT, Liu CA, Kuo HC, Yang KD. Kawasaki disease: infection, immunity and genetics. Pediatr Infect Dis J. 2005;24:998-1004
3. Newburger JW, Takahashi M, Burns JC, Beiser AS, Chung KJ, Duffy CE. et al. The treatment of Kawasaki syndrome with intravenous gamma globulin. N Engl J Med. 1986;315:341-7
4. Newburger JW, Takahashi M, Beiser AS, Burns JC, Bastian J, Chung KJ. et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N Engl J Med. 1991;324:1633-9
5. Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138-62
6. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annual review of biochemistry. 2006;75:243-69
7. Liu Y, Peng W, Qu K, Lin X, Zeng Z, Chen J. et al. TET2: A Novel Epigenetic Regulator and Potential Intervention Target for Atherosclerosis. DNA Cell Biol. 2018;37:517-23
8. Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem: a European journal of chemical biology. 2002;3:274-93
9. Huang YH, Li SC, Huang LH, Chen PC, Lin YY, Lin CC. et al. Identifying genetic hypomethylation and upregulation of Toll-like receptors in Kawasaki disease. Oncotarget. 2017;8:11249-58
10. Huang YH, Yang KD, Hsu YW, Lu HF, Wong HS, Yu HR. et al. Correlation of HAMP gene polymorphisms and expression with the susceptibility and length of hospital stays in Taiwanese children with Kawasaki disease. Oncotarget. 2017;8:51859-68
11. Huang YH, Kuo HC, Li SC, Cai XY, Liu SF, Kuo HC. HAMP promoter hypomethylation and increased hepcidin levels as biomarkers for Kawasaki disease. J Mol Cell Cardiol. 2018;117:82-7
12. Kuo HC, Li SC, Huang LH, Huang YH. Epigenetic hypomethylation and upregulation of matrix metalloproteinase 9 in Kawasaki disease. Oncotarget. 2017;8:60875-91
13. Huang YH, Lo MH, Cai XY, Kuo HC. Epigenetic hypomethylation and upregulation of NLRC4 and NLRP12 in Kawasaki disease. Oncotarget. 2018;9:18939-48
14. Li SC, Chan WC, Huang YH, Guo MM, Yu HR, Huang FC. et al. Major methylation alterations on the CpG markers of inflammatory immune associated genes after IVIG treatment in Kawasaki disease. BMC Med Genomics. 2016;9(Suppl 1):37
15. Huang LH, Kuo HC, Pan CT, Lin YS, Huang YH, Li SC. Multiomics analyses identified epigenetic modulation of the S100A gene family in Kawasaki disease and their significant involvement in neutrophil transendothelial migration. Clin Epigenetics. 2018;10:135
16. Chen KD, Huang YH, Ming-Huey Guo M, Lin TY, Weng WT, Yang HJ. et al. The human blood DNA methylome identifies crucial role of beta-catenin in the pathogenesis of Kawasaki disease. Oncotarget. 2018;9:28337-50
17. Newburger JW, Takahashi M, Gerber MA, Gewitz MH, Tani LY, Burns JC. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 2004;110:2747-71
18. Kuo HC, Lo MH, Hsieh KS, Guo MM, Huang YH. High-Dose Aspirin is Associated with Anemia and Does Not Confer Benefit to Disease Outcomes in Kawasaki Disease. PLoS One. 2015;10:e0144603
19. Kuo HC, Wang CL, Yang KD, Lo MH, Hsieh KS, Li SC. et al. Plasma Prostaglandin E2 Levels Correlated with the Prevention of Intravenous Immunoglobulin Resistance and Coronary Artery Lesions Formation via CD40L in Kawasaki Disease. PLoS One. 2016;11:e0161265
20. Kuo HC, Wang CL, Liang CD, Yu HR, Huang CF, Wang L. et al. Association of lower eosinophil-related T helper 2 (Th2) cytokines with coronary artery lesions in Kawasaki disease. Pediatr Allergy Immunol. 2009;20:266-72
21. Kuo HC, Yang KD, Liang CD, Bong CN, Yu HR, Wang L. et al. The relationship of eosinophilia to intravenous immunoglobulin treatment failure in Kawasaki disease. Pediatr Allergy Immunol. 2007;18:354-9
22. Liu YC, Lin MT, Wang JK, Wu MH. State-of-the-art acute phase management of Kawasaki disease after 2017 scientific statement from the American Heart Association. Pediatr Neonatol. 2018;59:543-52
23. McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M. et al. Diagnosis, Treatment, and Long-Term Management of Kawasaki Disease: A Scientific Statement for Health Professionals From the American Heart Association. Circulation. 2017;135:e927-e99
24. Yang YL, Wang FS, Li SC, Tiao MM, Huang YH. MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. Int J Mol Sci. 2017:18
25. Renauer P, Coit P, Sawalha AH. Epigenetics and Vasculitis: a Comprehensive Review. Clin Rev Allergy Immunol. 2016;50:357-66
26. Coit P, Direskeneli H, Sawalha AH. An update on the role of epigenetics in systemic vasculitis. Curr Opin Rheumatol. 2018;30:4-15
27. de Andres MC, Perez-Pampin E, Calaza M, Santaclara FJ, Ortea I, Gomez-Reino JJ. et al. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis research & therapy. 2015;17:233
28. Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. Journal of autoimmunity. 2013;41:92-9
29. Meda F, Folci M, Baccarelli A, Selmi C. The epigenetics of autoimmunity. Cellular & molecular immunology. 2011;8:226-36
30. Javierre BM, Hernando H, Ballestar E. Environmental triggers and epigenetic deregulation in autoimmune disease. Discovery medicine. 2011;12:535-45
31. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81-92
32. Hedrich CM, Mabert K, Rauen T, Tsokos GC. DNA methylation in systemic lupus erythematosus. Epigenomics. 2017;9:505-25
33. Kuo HC, Chang JC, Yu HR, Wang CL, Lee CP, Huang LT. et al. Identification of an association between genomic hypomethylation of FCGR2A and susceptibility to Kawasaki disease and intravenous immunoglobulin resistance by DNA methylation array. Arthritis Rheumatol. 2015;67:828-36
34. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300-3
35. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-5
36. Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45-68
37. Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H. et al. Acetylation Enhances TET2 Function in Protecting against Abnormal DNA Methylation during Oxidative Stress. Molecular cell. 2017;65:323-35
Author contact
![]() Corresponding author: Ho-Chang Kuo, MD, PhD, FAAAAI, Kawasaki Disease Center and Department of Pediatrics, Kaohsiung Chang Gung Memorial Hospital, Taiwan. #123 Da-Pei Road, Niaosong District, Kaohsiung 83301, Taiwan. Tel.: +8867-7317123 ext. 8795; Fax: +886-7-7338009; E-mail: erickuo48com.tw or dr.hckuocom or Wei-Dong Huang, MD, Baoan Maternity and Child Health Hospital, Shenzhen, Guangdong Province, China. 518100. E-mail: wdhuang126com
Corresponding author: Ho-Chang Kuo, MD, PhD, FAAAAI, Kawasaki Disease Center and Department of Pediatrics, Kaohsiung Chang Gung Memorial Hospital, Taiwan. #123 Da-Pei Road, Niaosong District, Kaohsiung 83301, Taiwan. Tel.: +8867-7317123 ext. 8795; Fax: +886-7-7338009; E-mail: erickuo48com.tw or dr.hckuocom or Wei-Dong Huang, MD, Baoan Maternity and Child Health Hospital, Shenzhen, Guangdong Province, China. 518100. E-mail: wdhuang126com

 Global reach, higher impact
Global reach, higher impact