Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(2):292-301. doi:10.7150/ijms.28047 This issue Cite
Review
Circular RNA: a novel biomarker and therapeutic target for human cancers
1. Department of Pathophysiology, Third Military Medical University, Chongqing 400038, China
2. Department of Microbiology and Immunology, Shanxi Medical University, Taiyuan 030001, China
3. State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing 400715, China
Received 2018-6-21; Accepted 2018-12-4; Published 2019-1-1
Abstract
Circular (circ)RNAs, a newly recognized class of noncoding RNA, have been implicated in the occurrence and development of several diseases, including neurological and cardiovascular diseases. Studies of human tumors, including those of liver cancer, gastric cancer, lung cancer and colorectal cancer, have shown differential expression profiles of circRNAs, suggesting regulatory roles in cancer pathogenesis and metastasis. In this review, we discuss the most recent research into tumor-related circRNAs, providing a comprehensive summary of the expression or/and function of these circRNAs and proposing rational perspectives on the potential clinical application of circRNAs as helpful biomarkers or therapeutic targets in human tumors.
Keywords: noncoding RNA, circRNA, cancer, biomarker, therapy
Introduction
The human transcriptome is very complex and diverse. A considerable portion of the mammalian genome can be transcribed into noncoding (nc)RNAs, rather than coding RNAs [1]. ncRNAs represent two broad categories: housekeeper ncRNAs, which include the ribosomal (r)RNAs, transfer (t)RNAs, small nuclear (sn)RNAs and small nucleolar (sno)RNAs; and regulatory ncRNAs. The regulatory ncRNAs are further classified according to the length of the nucleotide fragment, with the small ncRNAs having transcript lengths of less than 200 nucleotides, such as microRNAs (miRNAs), piwi-interacting (pi)RNAs and small interfering (si)RNAs, and the long noncoding (lnc)RNAs having transcript lengths of more than 200 nucleotides [2].
Among the lncRNAs, circular (circ)RNAs are a group of naturally occurring endogenous ncRNAs having transcript lengths of hundreds to thousands of nucleotides. The first evidence of circRNAs was reported in 1976, of RNA virus viriods−the uncoated infectious RNA molecules pathogenic to certain higher plants that exist as single-stranded covalently closed circular RNA molecules [3]. Since then, circRNAs have been found in mice, rats, fungi and humans [4-9]. At that time, however, these transcripts, detected at low abundance, were considered merely splicing errors [3]. Nevertheless, the recent robust development of second-generation sequencing techniques and bioinformatics has allowed researchers to confirm that there are many types of circRNAs with high stability in humans and to begin detailed investigations into their various functions [2].
Today, the majority of discovered circRNAs have been shown to originate from exons in the coding region of a gene, with others originating from the 5'- or 3'-untranslated regions (5'-UTRs or 3'-UTRs), introns and intergenic regions, as well as from antisense RNAs [10] . Accordingly, the circRNAs have been classified into four categories: exonic circRNAs, circular RNAs from introns, exon-intron circRNA, and intergenic circRNAs (Figure 1). In contrast to linear RNAs, circRNAs form a special circular covalently bonded structure without the 5'-terminal cap structure and 3'-terminal poly A, which renders a stronger tolerance to exonucleases and consequent stability in the cytoplasm, leading to relatively high abundance and prompting more research interest [2].
Biogenesis of circRNA. (A) Canonical splicing to form mRNA. (B) Lariat-driven circularization. First, a pre-mRNA is spliced, causing the 3'-hydroxyl of the upstream exon to covalently bind to the 5'-phosphate of the downstream exon. At the same time, the sequence between the exons becomes an RNA lariat, containing several exons and introns. Second, in the RNA lariat, the 2'-hydroxyl of the 5'-intron reacts with the 5'-phosphate of the 3'-intron, followed by the 3'-hydroxyl of the 3'-exon reacting with the 5'-phosphate of the 5'-exon. As a result, an RNA double lariat and a circular RNA are produced. Finally, some introns of the circular RNA are removed, producing an ecirRNA, EIciRNA, or ciRNA. (C) Intron pairing-driven circularization. The circular structure can be generated through direct base-pairing of the introns flanking inverted repeats or complementary sequences. The introns are removed or retained to form ecirRNA or EIciRNA. (D) RNA binding proteins (RBPs)-driven circularization. In this case, RBPs bind the upstream and downstream introns. The RBPs are attracted to each other, and form a bridge between the introns. The 2'-hydroxyl of the upstream intron then reacts with the 5'-phosphate of the downstream intron, which is followed by the 3'-hydroxyl of the 3'-exon reacting with the 5'-phosphate of the 5'-exon. Some introns of the circRNA are ultimately removed, producing an ecircRNA or EIcirRNA. CiRNAs, intronic circRNAs; ecirRNAs, exonic circRNAs; EIciRNAs, exon-intron circRNAs.
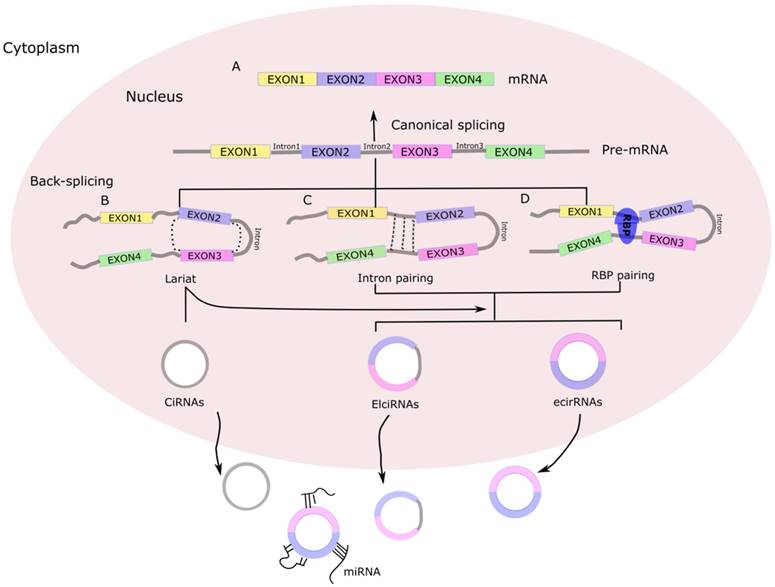
The rapid development of RNA sequencing technology and bioinformatics has led to a plethora of diverse circRNAs being investigated and characterized as regulators of physiological conditions and developmental stages [11]. The myriad functions recognized for the circRNAs now include sequestering proteins from their native subcellular localization, regulating parental gene expression, and RNA-protein interactions. In addition, a role as miRNA sponges has been discovered [12], a function by which the circRNAs may serve as competitive endogenous RNAs (ceRNAs) to affect gene expression by binding to and preventing target miRNAs from regulating their downstream target genes (Figure 2). In addition, many of the circRNAs have been implicated in pathogenic pathways of common diseases, such as atherosclerosis and nervous system disorders [13].
Recent studies have also revealed that circRNAs are differentially expressed in several human tumors and play indispensable roles in cancer pathogenesis, namely in carcinogenesis and metastasis [13-15]. As such, circRNAs have clinical potential for cancer risk assessment, diagnosis, prognosis and monitoring of treatment response, and may even serve as targets for cancer treatment. Herein, we review the most recently published circRNAs related to cancers, including gastric cancer, hepatocellular carcinoma, lung cancer, colorectal cancer and bladder cancer, providing evidence for the impact of circRNAs on various cancer types. The potential significance of these circRNAs in cancer diagnosis, prognosis and therapy is discussed in the context of the various molecular mechanisms underlying their regulatory roles in cancer pathogenesis.
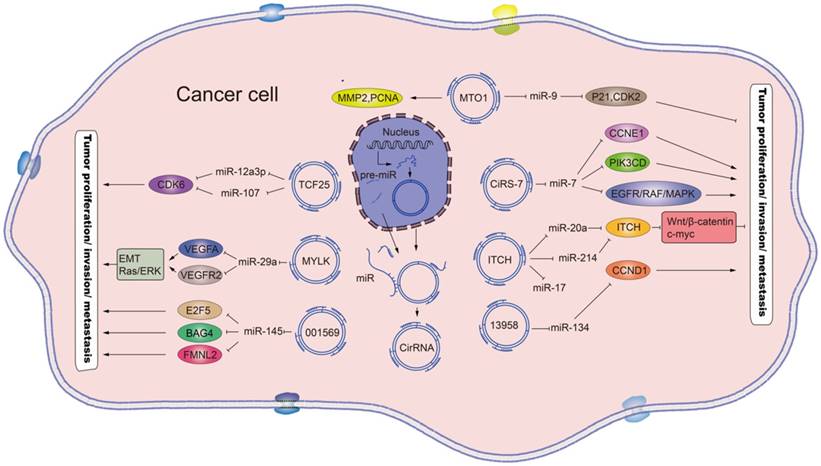
Schematic diagram of the effects and the underlying mechanisms of circRNAs. Mature circRNAs, such as the circ_MTO1, circ_TCF25, circ_MYLK, circ_001569, circ_ciRs-7, circ_ITCH and circ_13958, are released from the nucleus and can function as sponges for the indicated miRNAs, which regulate the respective target genes to promote or inhibit tumor proliferation and metastasis. Standard-shaped arrow, stimulation; T-shaped arrow, inhibition.
CircRNAs in Gastric Cancer
Gastric cancer is the fourth most common malignant tumor and the third leading cause of cancer death worldwide [16]. Development of extensive radical surgery has increased the overall survival rate of gastric cancer patients; unfortunately, the diagnosis of many gastric cancer patients occurs in the advanced disease state, after the best opportunity for radical surgery has passed [17]. Thus, foremost aims of gastric cancer research currently are improving survival through earlier diagnosis and effective targeted therapy in all stages. The evidenced involvement of circRNAs in the development of gastric cancer has led to their classification as candidate diagnostic markers or therapeutic targets.
Valuable markers for diagnosis and prognosis of gastric cancer
Recently, Li et al [13] found that hsa_circ_002059 was markedly down-regulated in gastric cancer tissues and plasma, in a study of 101 gastric cancer tissues with paired adjacent nontumorous tissues and 36 paired plasma samples from preoperative and postoperative patients. The lower expression level of the circRNA in gastric cancer tissues was further found to be significantly correlated with distant metastasis, tumor-node-metastasis (TNM) stage, sex and age by one-way analysis of variance (ANOVA) (Table 1), suggesting the potential of hsa_circ_002059 as a new stable diagnostic biomarker for gastric cancer [13]. In a later study by another group, Li et al [18] found that low expression of hsa_circ_104916 in gastric cancer tissues was also associated with higher tumor stage and more frequent lymph node metastasis in patients with gastric cancer.
Another research group demonstrated that hsa_circ_0001649 expression was significantly down-regulated in gastric cancer tissues, which however was significantly up-regulated after the therapeutic surgery [19]. Further analysis demonstrated the diagnostic value of this marker for the early detection of gastric cancer [19]. Similarly, Shao et al [20] found that circRNA hsa_circ_0014717 was significantly down-regulated in 77.2% of gastric cancer tissues and that its expression level in gastric cancer tissue is negatively correlated to tumor stage, distant metastasis and tissue expression levels of the routinely used tumor markers carcinoembryonic antigen and carbohydrate antigen 19-9. In addition, hsa_circ_0014717 was also detected in human body fluid [20], suggesting its potential for development as a convenient biomarker for gastric cancer screening (Table 1).
Expression and function of circRNAs in different cancers
| Tumor type | CircRNA | Research model | Expression change | Function | Mechanism | Ref |
|---|---|---|---|---|---|---|
| GC | Hsa_circ_002059 | GC tissue and plasma | ↓ | Biomarker | Hsa_circ_002059 levels were negatively related to TNM stage (p = 0.042), distal metastasis (p = 0.036), gender (p = 0.002), and age (p = 0.022); ROC curve was 0.73, sensitivity and specificity were 0.81 and 0.62, respectively. | [13] |
| Hsa_circ_0001649 | GC tissue and serum | ↓ | Biomarker | Hsa_circ_0001649 levels were negatively correlated with pathological differentiation (p = 0.039); ROC curve was 0.834, sensitivity and specificity were 0.711 and 0.816, respectively. | [19] | |
| Hsa_circ_0014717 | GC tissue and gastric juice | ↓ | Biomarker | Hsa_circ_0014717 levels were negatively correlated with tumor stage (p = 0.037), distal metastasis (p = 0.048), tissue carcinoembryonic antigen (p = 0.001), and carbohydrate antigen 19-9 expression (p = 0.021). | [20] | |
| Hsa_circ_104916 | GC tissue and cell lines | ↓ | Tumor suppressor | Over-expression suppresses the migration and invasion of GC cells through alteration of the EMT process. | [18] | |
| Hsa_circ_100269 | GC tissue and cell lines | ↓ | Tumor suppressor | Over-expression suppresses tumor cell growth by targeting miR-630. | [21] | |
| HCC | ciRS-7(cdr1as) | HCC tissue | ↑ | Biomarker | ciRS-7 levels were positively correlated with age <40 years (p = 0.02), serum AFP ≥400 ng/µL (p < 0.01), and hepatic microvascular invasion (MVI) (p = 0.03); Median disease-free survival (DFS) time in the low ciRS-7 group was longer than that in the high ciRS-7 group (25 vs. 18 months). | [22] |
| Hsa_circ_0005075 | HCC tissue | ↑ | Biomarker | Hsa_circ_0005075 levels were positively correlated with HCC tumor size (p = 0.042); ROC curve was 0.94, sensitivity and specificity were 0.833 and 0.900, respectively. | [4] | |
| Circ-ITCH | HCC tissue | ↓ | Biomarker | High expression of circ-ITCH was positively associated with favorable survival of HCC (HR = 0.45, 95% CI = 0.29-0.68; p value < 0.001). | [23] | |
| Hsa_circ_0005986 | HCC tissue and cell lines | ↓ | Regulate cell cycle and proliferation | Promote the G0/G1 to G2 phase transition in HCC cell line. | [24] | |
| circ_MTO1 | HCC tissue and cell lines | ↓ | Tumor suppressor | Act as the sponge of oncogenic miR-9 to promote p21 expression. | [25] | |
| Hsa_circ_0004018 | HCC tissue and cell lines | ↓ | Biomarker; Tumor suppressor | Hsa_circ_0004018 level was negatively correlated with serum AFP level (p = 0.027), tumor diameter (p = 0.045), differentiation (p = 0.006), BCLC stage (p = 0.040), and TNM stage (0.029); ROC curve was 0.848, sensitivity and specificity were 0.716 and 0.815, respectively. | [26] | |
| Hsa_circ_0001649 | HCC tissue and cell lines | ↓ | Promote HCC tumorigenesis and metastasis | Up-regulate several matrix metallopeptidases (MMP9, MMP10, and MMP13). | [15] | |
| ciRS-7 | HCC tissue and cell lines | ↑ | Promote HCC proliferation and invasion | ciRS-7 acts as an oncogene through targeting miR-7 and promoting HCC cell proliferation and invasion by regulating expression of the CCNE1 and PIK3CD genes. | [27] | |
| Lung cancer | Hsa_circ_100876 | NSCLC tissue | ↑ | Biomarker | Hsa_circ_100876 level was positively correlated with lymph node metastasis (p = 0.001) and tumor staging (p = 0.001) in NSCLC; Overall survival time of NSCLC patients with high hsa_circ_100876 expression was significantly shorter than for those patients with low hsa_circ_100876 expression (p = 0.000). | [28] |
| Hsa_circ_0013958 | LAC tissue LAC cell lines | ↑ | Promote cell proliferation and invasion and inhibit apoptosis of LAC cell lines | Act as the sponge of miR-134 and subsequently up-regulate CCND1 protein. | [29] | |
| CircRNA-ITCH | Lung cancer tissue and cell lines | ↓ | Suppress lung cancer cell proliferation | Act as sponge of oncogenic miR-7 and miR-214 to enhance ITCH expression and thus suppress the activation of Wnt/-catenin signaling. | [30] | |
| CRC | Circ-BANP | CRC tissue and cell lines | ↑ | Regulate CRC cell proliferation | Knock-down using siRNA targeted to circ_BANP suppressed CRC cell proliferation and reduced p-Akt protein expression, suggesting that the Akt pathway might be involved in the process of circ-BANP-induced cell proliferation. | [31] |
| CircRNA-103809 | CRC tissue | ↓ | Biomarker | CircRNA-103809 level was negatively correlated with lymph node metastasis (p = 0.021) and TNM stage (p = 0.011); ROC curve was 0.699 (p < 0.0001). | [32] | |
| CircRNA-104700 | CRC tissue | ↓ | Biomarker | CircRNA-104700 level was negatively correlated with distal metastasis (p = 0.036); ROC curve was 0.616 (p < 0.0001). | [32] | |
| Hsa_circ_001988 | Colorectal tissue | ↓ | Biomarker | Hsa_circ_001988 level was negatively correlated with differentiation (p < 0.05) and perineural invasion (p < 0.05); ROC curve was 0.788, sensitivity and specificity were 0.68 and 0.73, respectively. | [33] | |
| Hsa_circ_0000069 | CRC tissue CRC cell lines | ↑ | Promote cell proliferation, invasion and migration | Knock-down of hsa_circ_0000069 notably induced G0/G1 phase arrest of cell cycle in CRC cells in vitro. | [34] | |
| ciRS-7 | CRC tissue CRC cell lines | ↑ | Over-expression of ciRS-7 resulted in a more aggressive oncogenic phenotype of CRC cell lines | Activate the EGFR/RAF1/MAPK pathway via suppression of miR-7 activity. | [35] | |
| CircRNA-ITCH | CRC tissue CRC cell lines | ↓ | Over-expressed circRNA-ITCH inhibits CRC Cell proliferation | Act as sponge of miR-7 and miR-20a to enhance ITCH expression and thereby suppression of Wnt/β-catenin signaling activation. | [14] | |
| Hsa_circ_001569 | CRC tissues CRC cell lines | ↑ | Promote cell proliferation and invasion of CRC cells | Act as a miRNA sponge to directly inhibit miR-145, and subsequently up-regulate miR-145 targets E2F5, BAG4 and FMNL2. | [36] | |
| Bladder cancer | CircRNA-MYLK | Bladder cancer tissue and cell lines | ↑ | Promote tumorigenesis, EMT and metastasis | Act as a sponge for miR-29a and activating the VEGFA/VEGFAR2 signaling pathway. | [37] |
| CircTCF25 | Bladder cancer tissue and cell lines | ↑ | Promote cancer cell proliferation and migration | Act as sponge of miR-103a-3p and miR-107, thus increasing CDK6 expression. | [38] | |
| LSCC | Hsa_circ_100855 | LSCC tissue | ↑ | Biomarker | Hsa_circ_100855 level was positively correlated with T classification (p = 0.006), lymph node metastasis (p = 0.003), primary location (p = 0.007), and clinical stage (p = 0.001). | [39] |
| Hsa_circ_104912 | LSCC tissue | ↓ | Biomarker | Hsa_circ_104912 level was negatively correlated with T classification (p = 0.010), differentiation (p = 0.039), lymph node metastasis (p = 0.020), and clinical stage (p = 0.008). | [39] | |
| ESCC | Hsa_circ_0067934 | ESCC tissue ESCC cell lines | ↑ | Promote proliferation and migration of ESCC cell lines | Promote the proliferation of ECSS cells by regulating cell cycle | [40] |
| Breast cancer | Hsa_circ_104689 | Breast cancer tissue | ↑ | Biomarker | ROC curve was 0.61, sensitivity and specificity were 0.57 and 0.55, respectively. | [41] |
| Hsa_circ_104821 | Breast cancer tissue | ↑ | Biomarker | ROC curve was 0.60, sensitivity and specificity were 0.57 and 0.57, respectively. | [41] | |
| Hsa_circ_006054 | Breast cancer tissue | ↓ | Biomarker | ROC curve was 0.71, sensitivity and specificity were 0.65 and 0.69, respectively. | [41] | |
| Hsa_circ_100219 | Breast cancer tissue | ↓ | Biomarker | ROC curve was 0.78, sensitivity and specificity were 0.69 and 0.71, respectively. | [41] | |
| Hsa_circ_406697 | Breast cancer tissue | ↓ | Biomarker | ROC curve was 0.64, sensitivity and specificity were 0.63 and 0.63, respectively. | [41] | |
| Circ-ABCB10 | Breast cancer tissue and cell lines | ↑ | Promote breast cancer proliferation and progression | Act as sponge of miR-1271. | [42] | |
| Hsa_circ_103110 | Breast cancer tissue | ↑ | Biomarker | ROC curve was 0.63, sensitivity and specificity were 0.63 and 0.63, respectively. | [41] |
Notes: ↑, up-regulated; ↓, down-regulated. AFP, alpha-fetoprotein; ASAP1, ArfGAP with SH3 domain, ankyrin repeat and PH domain 1; BCLC, Barcelona Clinic Liver Cancer; CDR1as, antisense to cerebellar degeneration-related protein 1 transcript, also as know CiRs-7; CRC, colorectal cancer; ESCC, esophageal squamous cell cancer; FAF1, Fas-associated factor 1; FAM120A, family with sequence similarity 120A; GC, gastric cancer; KIAA0355, encodes an uncharacterized protein; LAC, lung adenocarcinoma; LSCC, laryngeal squamous cell cancer; NRIP1, nuclear receptor interacting protein 1; NSCLC, non-small cell lung cancer; RBM22, RNA binding motif protein 22; ROC, receiver operating characteristic curve; SMYD4, SET and MYND domain containing 4; TNM, tumor-node-metastasis.
Regulatory role in gastric cancer development
Recent studies have elucidated circRNA-mediated molecular mechanisms responsible for gastric cancer. Zhang et al [21] discovered that the level of hsa_circ_100269 was down-regulated markedly while the expression of miR-630 was up-regulated in gastric cancer tissues. Furthermore, hsa_circ_100269 could directly interact with miR-630, thereby serving as a sponge to regulate the activity of miR-630 [21] (Table 1), a newly discovered miRNA that is over-expressed in a variety of tumors and involved in cell invasion and metastasis [43]. Therefore, the down-regulation of hsa_circ_100269 in gastric cancer cells will promote cell growth by releasing the activity of miR-630 (Figure 2).
Similarly, by enquiry of the circBase, Li et al [18] selected hsa_circ_104916, a 651nt circular RNA molecule generated by back splicing of exons 1, 3, 4, 5, 6 and 8 of NEK6, a serine-threonine kinase involved in mitosis progression. Analysis of gastric tissues and cell lines showed down-regulated hsa_circ_104916 that was related to deeper invasion, higher tumor stage and more frequent lymph node metastasis in patients (Table 1). Ectopic over-expression of hsa_circ_104916 was also found to effectively inhibit proliferation, migration and invasion of gastric cancer cells in vitro. The underlying mechanisms identified include hsa_circ_104916-mediated up-regulation of an epithelial molecule (E-cadherin) and down-regulation of mesenchymal molecules (N-cadherin and vimentin) and a zinc-finger transcriptional repressor (SLUG) that is active in pre-migratory neural crest cells during the epithelial to mesenchymal transition (EMT) of gastrulation[18]. Thus, hsa_circ_104916 may inhibit the migration and invasion of gastric cancer cells by inhibiting the EMT process (Figure 2), a pivotal cellular process in cancer metastasis [44]. However, the precise mechanism by which hsa_circ_104916 regulates the expression of EMT-related molecules remains unknown.
Nevertheless, current studies indicate that circRNAs, including hsa_circ_100269 [21] and hsa_circ_104916 [18], are involved in the molecular pathogenic pathway of gastric cancers. As both hsa_circ_100269 and hsa_circ_104916 are down-regulated in gastric cancer and consequently promote its pathogenesis, the over-expression of both circRNAs would have therapeutic potential for gastric cancer.
circRNAs and Hepatocellular Carcinoma (HCC)
HCC is one of the most common malignancies in the world and has been extensively researched [45]. Over the past few decades, a large number of ncRNAs, including miRNAs and lncRNAs, have been shown to be involved in human HCC [46, 47]. The more recent studies have indicated that circRNAs are also involved in the pathogenesis of HCC, ultimately revealing many more valuable clues than expected, including the therapeutic potential of these large ncRNAs for HCC.
circRNAs as potential clinical diagnostic markers of HCC
circRNAs may be expressed differentially in HCC tissues and normal hepatic tissues. Xu et al [22] found that the expression of ciRS-7 in HCC tissues was largely similar to that in matched nontumor tissues, with more than half of the HCC tissues examined showing slightly lower ciRS-7 expression. However, the instances of HCC tissue samples having high ciRS-7 expression coincided with significantly higher levels of hepatic microvascular invasion and alpha-fetoprotein, as well as younger age at diagnosis, compared with the HCC tissue samples expressing low ciRS-7. Furthermore, ciRS-7 was identified as an independent factor of hepatic microvascular invasion [22]. In another study, Shang et al [48] found that hsa_circ_0005075 was up-regulated in HCC tissues and that this differential expression was positively related to size of the HCC.
Down-regulated circRNAs have also been found in HCC tissues. For instance, an analysis of 89 paired samples of HCC and adjacent liver tissues showed that circRNA hsa_circ_0001649 was significantly down-regulated in HCC and positively related to tumor size; receiver operating characteristic curve analysis provided further support for the potential diagnostic value of circ_0001649 for HCC [15] (Table 1). In other studies, the circRNAs hsa_circ_ITCH, hsa_circ_0005986, circMTO1 and hsa_circ_0004018 have been found to be significantly down-regulated in HCC compared to para-tumorous tissue [23-26]. Furthermore, lower expression of hsa_circ_0004018 was found to be correlated with serum alpha-fetoprotein level, as well as tumor diameter, differentiation and stage. Importantly, hsa_circ_0004018 expression demonstrated a HCC stage-specific feature among diverse chronic liver diseases [26], highlighting its potential sensitivity and specificity for the diagnosis of HCC (Table 1).
Together, the set of circRNAs differentially expressed in HCC hold promise as valuable biomarkers for early diagnosis or prognosis for HCC.
Role and regulatory mechanism of circRNAs in HCC
The studies published to date demonstrate that circRNAs could be involved in the development and progression of HCC by acting as sponges of oncogenic miRNAs [24-26, 49]. Such circRNAs would be abnormally expressed in HCC tissues. Indeed, circRNA ciRS-7 possesses more than 70 conventional binding sites and has been shown to function as a so-called 'super sponge' for miR-7 [2, 50], a tumor suppressor that is often down-regulated and negatively correlated with growth and colony formation of many tumor types [51]. A recent study also showed that the ciRS-7 expression levels are significantly increased in HCC tissues and negatively related to the expression of miR-7 [27]. Moreover, knock-down of ciRS-7 was found to inhibit the proliferation and invasion of liver cancer cells through targeting of miR-7, resulting in increased expression of its target genes, namely CCNE1 and pik3cd [27].
In contrast, Han et al [25] found that circMTO1 was significantly down-regulated in HCC tissues and the survival time of HCC patients with low expression of circMTO1 was markedly shortened. By using circMTO1 precipitation of RNA from HCC cells, the researchers identified miR-9 as the circMTO1-associated miRNA. Knock-down of circMTO1 or transfection of miR-9 mimics reduces the level of mRNA and protein expression of the tumor suppressor p21 in HCC cells. Therefore, circMTO1 suppresses HCC progression by acting as the sponge of oncogenic miR-9 to promote p21 expression, thereby regulating the expression of genes related to tumor cell proliferation and metastasis in HCC (Table 1, Figure 2).
circRNAs and Lung Cancer
Lung cancer is the leading cause of cancer-related deaths worldwide [52]. Most cases present with advanced local invasion and/or distant metastasis at diagnosis [52]. Although continuous efforts have been devoted to improving therapeutic response and the treatments for lung cancer have demonstrated survival benefits, the overall 5-year survival rate of advanced lung cancer is still below 15% [53], highlighting the need for earlier diagnosis and the related timely application of treatment. Recent studies have revealed that circRNAs may be involved in the development of lung cancer, again providing potential diagnostic, prognostic and even therapeutic clues for lung cancer.
circRNAs as potential clinical diagnostic markers of lung cancer
Recently, Yao et al [28] reported that expression of circRNA hsa_circ_100876 was higher in non-small cell lung cancer tissues than in paired adjacent tissues, and that the up-regulation of hsa_circ_100876 had a close positive relation to lymph node metastasis and tumor stage. For the 101 total lung cancer patients (representing 51 diagnosed with squamous cell carcinomas and 50 diagnosed with adenocarcinoma) assessed, the overall survival time was significantly shorter for those who showed high expression of hsa_circ-100876 than for those with low hsa_circ_100876 expression [28]. In a similar study of 87 lung cancer patients, Li et al [30] showed that the expression of circ-ITCH was significantly decreased in lung cancer tissues compared with paired noncancerous tissues.
There are likely to be many more circRNAs involved in lung cancer pathogenesis than those defined to date. By using microarrays to screen tumor-specific circRNA candidates in lung adenocarcinoma tissue, Zhu et al [29] determined that 39 circRNAs were up-regulated and 20 were down-regulated. Among these candidates, hsa_circ_0013958 was further confirmed to be highly expressed in all lung adenocarcinoma tissues as well as in plasma of these patients. The level of hsa_circ_0013958 was also found to be closely positively related to TNM stage and lymph node metastasis. Receiver operating characteristic curve analysis confirmed the high specificity (0.796) and sensitivity (0.755) of hsa_circ_0013958 for lung cancer diagnosis [29]. Thus, a set of circRNAs involved in lung cancer may be developed to serve as an early noninvasive biomarker profile that will be valuable for of diagnosis and/or prognosis of lung cancer.
circRNAs function as miRNA sponges in lung cancer
The definitive mechanism of circRNAs involvement in lung cancer pathogenesis is their regulation of miRNA sponges, by which they mediate the expression of parental genes. It has already been confirmed that circRNA hsa_circ_100876 serves as the sponge for miR-136[28]. In addition, circRNA hsa_circ_0013958, which is up-regulated in lung cancer, interacts with the tumor suppressor miR-134 but does not affect its expression [29] (Table 1). The possible effects of this demonstrated interaction include miR-134 inhibition of the development of lung cancer by down-regulation of its target gene, CCND1 [54]. Similarly, circ-ITCH has been characterized as the sponge of miR-7 and miR-214, thereby promoting the expression of their target gene (ITCH) and indirectly inhibiting activation of the Wnt/β-catenin pathway and the proliferation of lung cancer cells [30]. As such, the antitumor effects of circ-ITCH in lung cancer may involve regulation of miRNA activity, which can increase the level of ITCH and cause suppression of the canonical Wnt/β-catenin pathway (Figure 2).
circRNAs and Colorectal Cancer (CRC)
circRNAs as potential biomarkers of CRC
CRC is one of the most common cancers and the leading cause of morbidity and mortality worldwide [55]. The 5-year survival rate of patients with early CRC is 90.1% but that of CRC patients with distant metastasis is only 11.7% [55]. CRC, as a major public health problem, continues to garner much research interest and a large number of CRC-related circRNAs have been identified. Current research is focused on defining the role of these various circRNAs in the development and progression of CRC.
It is reported that hsa_circ_BANP and hsa_circ_0000069 were significantly over-expressed in CRC tissues [31, 34], being positively correlated with patients' TNM stage[34]. However, some circRNAs are down-regulated in CRC tissues. For example, the down-regulation of hsa_circ_001988 in CRC was found to be closely related to perineural invasion and differentiation [33]. hsa_circ_103809 and hsa_circ_104700 were also found to be down-regulated in CRC tissues compared with normal tissues. The expression level of hsa_circ_103809 was significantly negatively correlated with lymph node metastasis and TNM stage, while the expression level of hsa_circ_104700 was significantly correlated with distant metastasis [32]. These findings suggest that these dysregulated circRNAs might serve as potential biomarkers for CRC (Table 1).
circRNA regulatory roles in CRC
The dysregulated CRC-related circRNAs promote tumor progression and/or metastasis by serving as sponges for miRNAs. Among the circRNAs up-regulated in CRC tissues, hsa_circ_0000069 was shown to promote proliferation, invasion and migration of CRC cells; loss-of-function analysis showed that silencing of hsa_circ_0000069 had the opposite effect, significantly inducing the cell cycle arrest of G0/G1 phase in CRC cells cultured in vitro [34]. Similarly, ciRS-7, which is up-regulated in neuroblastoma, astrocytoma, renal cell and lung carcinomas [35, 56] as well as CRC tissues [35], was shown to be a competing endogenous RNA of the tumor suppressor miR-7, capable of binding up to 73 copies of the miRNA [50]. Excessive ciRS-7 can disrupt normal miR-7 function, promoting aggressiveness in CRC cells and mediating activation of the EGFR/RAF1 pathway by competing for miR-7[35].
More recently, Xie et al [36] determined that hsa_circ_001569 was markedly up-regulated in CRC tissues and promoted the proliferation and invasiveness of CRC cells by targeting miR-145. The miR-145 has been reported to be a tumor suppressor gene implicated in prostate cancer and renal cell carcinoma [57-59]. Therefore, hsa_circ_001569 might competitively bind and inhibit the activity of miR-145, leading to increased expression of miR-145 targets (E2F5, BAG4 and FMNL2) [36] and promoting the proliferation of CRC cells (Table 1, Figure 2).
circRNAs may be down-regulated in CRC tissues to exert the same protumor activity as those up-regulated in CRC tissues. circ-ITCH spans multiple exons of the ubiquitin protein ligase (E3; ITCH) and harbors several miRNA binding sites, interacting with miR-7, miR-17, miR-214, miR-128 and miR-216b which bind to the 3'-UTR of ITCH [2, 50]. The targets of ITCH include p63, p73 and Notch1, which are often associated with tumor formation and chemosensitivity[60]. Recently, Huang et al [14] showed that circ-ITCH plays a significant role in CRC by regulating the Wnt/signaling pathway. The authors also found that circ-ITCH expression was usually down-regulated in CRC, as compared to para-cancerous tissues. Mechanism experiments further indicated that circ-ITCH acts as an antitumor agent by serving as the sponge of miR-7 and miR-20a, which inhibits the canonical Wnt/pathway and the expression of c-myc and cyclinD1, consequently inhibiting the proliferation of CRC cells (Table 1, Figure 2).
CircRNAs in Bladder Cancer and Other Cancers
Bladder cancer is a common malignancy of the urinary system and a substantial cause of cancer deaths worldwide [31]. High recurrence rate is a typical and unresolved feature of bladder cancer [31]. Recently, Zhong et al detected the circRNA expression profiles in bladder carcinoma by microarray assay and found that circRNA-MYLK and circTCF25 were markedly up-regulated in bladder cancer, suggesting their potential as biomarkers of bladder cancer diagnosis and therapy [37, 38]. Specifically, circRNA-MYLK levels were found to be positively related to the progression of stage and grade of bladder cancer. circRNA-MYLK was also shown to accelerate the proliferation and migration of cancer cells, the tube formation of human umbilical vein epithelial cells, and the rearrangement of cytoskeleton and EMT by directly interacting with miR-29a to attenuate its inhibition of its target, VEGFA. Similarly, up-regulated circTCF25 was shown to inhibit miR-103a-3p and miR-107 activity, thereby increasing CDK6 protein level and promoting proliferation and migration of bladder tumor cell lines [38]. Collectively, these studies suggest that circRNAs, including circRNA-MYLK and circTCF25, could serve as miRNA sponges to regulate tumorigenesis and could be novel biomarkers for diagnosis of bladder cancer (Table 1, Figure 2).
circRNAs might be also involved in breast cancer. Lu et al [41] found approximately 1155 differentially expressed circRNAs in breast cancer tissues by use of human circRNA array, among which 715 were up-regulated and 440 were down-regulated. The validation study demonstrated that hsa_circ_103116, hsa_circ_104689 and hsa_circ_104821 were up-regulated, while hsa_circ_006054, hsa_circ_100219 and hsa_circ_406697 were down-regulated, suggesting that these circRNAs might be useful biomarkers of breast cancer. Liang et al [42] further demonstrated that circ-ABCB10 was significantly up-regulated in breast cancer tissue. Loss-of-function experiments showed that circ-ABCB10 knock-down suppressed proliferation and induced apoptosis of breast cancer cells; however, miR-1271 rescued the function of circ-ABCB10 on breast cancer cells, confirming the sponge effect of circ-ABCB10 on miR-1271 and suggesting a key regulatory role of circ-ABCB10 in breast cancer pathogenesis [42] (Table 1, Figure 2).
circRNAs have also been verified as functionally involved in squamous cell cancer. Xuan et al [39] found approximately 698 differentially expressed circRNAs (302 up-regulated and 396 down-regulated) in 4 paired laryngeal squamous cell cancer tissues and adjacent nontumor tissues by microarray analysis. Among this set, up-regulation of hsa_circ_100855 was shown to be associated with T3-4 stage, neck nodal metastasis and advanced clinical stage of laryngeal squamous cell cancer tissues, while the down-regulation of hsa_circ_104912 was correlated to T3-4 stage, neck nodal metastasis, poor differentiation and advanced clinical stage. In addition, Xia et al [40] showed that hsa_circ_0067934 was up-regulated in esophageal squamous cell carcinoma tumor tissues and cell lines. Up-regulation of hsa_circ_0067934 was positively related to poor differentiation, I-II T stage and I-II TNM stage, while silencing of hsa_circ_0067934 by siRNA in vitro inhibited the proliferation and migration of esophageal squamous cell carcinoma cells and blocked cell cycle progression, suggesting that hsa_circ_0067934 promotes the proliferation of esophageal squamous cell carcinoma cells by regulating the cell cycle. These data indicate the potential for circRNAs being developed as novel biomarkers or/and therapeutic targets for the diagnosis and progression of squamous cell cancers (Table 1, Figure 2).
Conclusion and Perspectives
circRNAs were previously considered splicing errors and, as such, did not receive much attention since their discovery in the 1970s. However, circRNAs are now understood to be ubiquitously expressed and to represent an abundant and stable class of RNA molecules with a range of activities, including miRNA sponging to indirectly regulate the expression of the miRNA targets involved in various physiological and pathological pathways. Recently, much evidence has been published to support the notion that a comprehensive profile of circRNAs expression is associated with tumorigenesis, with cancer type-specific distinguishments likely. In general, studies of the circRNAs characterized as related to human cancer support their development as biomarkers or therapeutic targets, and the research into such has already provided cues towards a better understanding of circRNA roles in cancer development and progression. Since the research field of circRNAs in cancers is still in its infancy, there are myriad avenues of study to elucidate the molecular and biological functions of circRNAs, including their interactions with specific miRNAs, mRNAs and proteins, which will comprise a regulatory network for cancer development and invasion, thereby providing more accurate diagnoses, prognoses and intervening targets to improve cancer outcomes.
As the field of circRNAs research is still in its infancy, the practical application of circRNAs in clinics remains a long road to walk. CircRNAs alone are not going to be sufficient as specific biomarkers for any particular cancer, as of yet. Although some studies have reported that certain circRNAs are specific for certain cancers, none of the studies have determined the level of these circRNAs in the other cancer types; this underlies the current suspect nature of the genuine specificity of these circRNAs for any singular specific cancer type. Theoretically, even if a circRNA might function as a sponge of a miRNA, that miRNA might target hundreds of genes [61], meaning that a circRNA may regulate hundreds of genes' expression, and thus making it highly unlikely that it would be absolutely specific for a single cancer type. The greatest likelihood is that circRNAs are a common driving mechanism of oncogenesis or are a common by-/end-product of oncogenesis [62].
Nevertheless, circRNAs could still be also useful as cancer markers, although not for specific types of cancer. In clinic, the diagnosis of a specific cancer relies on a combination of data, including the clinical phenotypes and many clinical parameters. In this view, circRNAs could contribute to the diagnosis of certain cancers, provided it is used in combination with other biomarkers or parameters.
It will almost certainly be a long route to the practical application of circRNAs in clinics because, as of yet, the circRNAs have not been extensively and systemically investigated for diseases; of note, there is a dearth of data obtained from preclinical studies that would validate their association with cancers. In addition, a very important aspect of future research will be to determine where circRNAs are localized in the molecular pathogenic pathway, which will provide us more knowledge on cancers and on more potential interfering targets for such.
Acknowledgements
This work was supported by grants from the National Key Research and Development Plan of China (No. 2016YFA0502203) and the National Foundation of China (No. 81670534).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74
2. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
3. Sanger HL, Klotz G, Riesner D. et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
4. Capel B, Swain A, Nicolis S. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019-30
5. Cocquerelle C, Daubersies P, Majerus MA. et al. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095-8
6. Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990;87:7628-32
7. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A. 1996;93:6536-41
8. Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. 1997;17:2985-93
9. Cocquerelle C, Mascrez B, Hetuin D. et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155-60
10. Li Y, Zheng Q, Bao C. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
11. Yu CY, Li TC, Wu YY. et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149
12. Meng S, Zhou H, Feng Z. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94
13. Li P, Chen S, Chen H. et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132-6
14. Huang G, Zhu H, Shi Y. et al. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PloS one. 2015;10:e0131225
15. Qin M, Liu G, Huo X. et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161-9
16. Venerito M, Vasapolli R, Rokkas T. et al. Gastric cancer: epidemiology, prevention, and therapy. Helicobacter. 2018;23(Suppl 1):e12518
17. Ishihara R. Infrared endoscopy in the diagnosis and treatment of early gastric cancer. Endoscopy. 2010;42:672-6
18. Li J, Zhen L, Zhang Y. et al. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521-9
19. Li WH, Song YC, Zhang H. et al. Decreased Expression of Hsa_circ_00001649 in Gastric Cancer and Its Clinical Significance. Dis Markers. 2017;2017:4587698
20. Shao Y, Li J, Lu R. et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173-80
21. Zhang Y, Liu H, Li W. et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging. 2017;9:1585-94
22. Xu L, Zhang M, Zheng X. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17-27
23. Guo W, Zhang J, Zhang D. et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169-77
24. Fu L, Chen Q, Yao T. et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878-88
25. Han D, Li J, Wang H. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
26. Fu L, Yao T, Chen Q. et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405-16
27. Yu L, Gong X, Sun L. et al. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PloS One. 2016;11:e0158347
28. Yao JT, Zhao SH, Liu QP. et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453-6
29. Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X. et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170-82
30. Wan L, Zhang L, Fan K. et al. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/beta-Catenin Pathway. Biomed Res Int. 2016;2016:1579490
31. Zhu M, Xu Y, Chen Y. et al. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138-44
32. Zhang P, Zuo Z, Shang W. et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39:1010428317694546
33. Wang X, Zhang Y, Huang L. et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020-5
34. Guo JN, Li J, Zhu CL. et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451-8
35. Weng W, Wei Q, Toden S. et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:3918-28
36. Xie H, Ren X, Xin S. et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680-91
37. Zhong Z, Huang M, Lv M. et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305-17
38. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919
39. Xuan LJ, Qu LM. et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932-9
40. Xia W, Qiu M, Chen R. et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576
41. Lu LS, Sun J, Shi PY. et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8:44096-107
42. Liang HF, Zhang XZ, Liu BG. et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566-76
43. Jin L, Yi J, Gao Y. et al. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai). 2016;48:810-9
44. Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850
45. Dhir M, Melin AA, Douaiher J. et al. A Review and Update of Treatment Options and Controversies in the Management of Hepatocellular Carcinoma. Ann Surg. 2016;263:1112-25
46. Wan Y, Cui R, Gu J. et al. Identification of Four Oxidative Stress-Responsive MicroRNAs, miR-34a-5p, miR-1915-3p, miR-638, and miR-150-3p, in Hepatocellular Carcinoma. Oxid Med Cell Longev. 2017;2017:5189138
47. Kogure T, Yan IK, Lin WL. et al. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer. 2013;4:261-72
48. Shang X, Li G, Liu H. et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine. 2016;95:e3811
49. Li Y, Dong Y, Huang Z. et al. Computational identifying and characterizing circular RNAs and their associated genes in hepatocellular carcinoma. PloS one. 2017;12:e0174436
50. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
51. Horsham JL, Ganda C, Kalinowski FC. et al. MicroRNA-7: A miRNA with expanding roles in development and disease. Int J Biochem Cell Biol. 2015;69:215-24
52. Williams CD, Gajra A, Ganti AK. et al. Use and impact of adjuvant chemotherapy in patients with resected non-small cell lung cancer. Cancer. 2014;120:1939-47
53. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5-29
54. Sun CC, Li SJ, Li DJ. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7:35960-78
55. Siegel R, DeSantis C, Virgo K. et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220-41
56. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-12
57. Xue G, Ren Z, Chen Y. et al. A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett. 2015;361:121-7
58. Lu R, Ji Z, Li X. et al. miR-145 functions as tumor suppressor and targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma. J Cancer Res Clin Oncol. 2014;140:387-97
59. Hart M, Wach S, Nolte E. et al. The proto-oncogene ERG is a target of microRNA miR-145 in prostate cancer. FEBS J. 2013;280:2105-16
60. Melino G, Gallagher E, Aqeilan RI. et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15:1103-12
61. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3-11
62. Wang Y, Mo Y, Gong Z. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25
Author contact
![]() Corresponding authors: Weiping Fan, Department of Microbiology and Immunology, Shanxi Medical University, Taiyuan 030001, China. E-mail: fanweiping26418com. Tel: +86-13934631873; Fax: +86-351-5634785. Bing Ni, Department of Pathophysiology, Third Military Medical University, 30 Gaotanyan Street, Shapingba District, Chongqing 400038, China. E-mail: nibingxicom. Tel: +86-23-68772228; Fax: +86-23-68772228
Corresponding authors: Weiping Fan, Department of Microbiology and Immunology, Shanxi Medical University, Taiyuan 030001, China. E-mail: fanweiping26418com. Tel: +86-13934631873; Fax: +86-351-5634785. Bing Ni, Department of Pathophysiology, Third Military Medical University, 30 Gaotanyan Street, Shapingba District, Chongqing 400038, China. E-mail: nibingxicom. Tel: +86-23-68772228; Fax: +86-23-68772228

 Global reach, higher impact
Global reach, higher impact