Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(1):84-92. doi:10.7150/ijms.28595 This issue Cite
Research Paper
Proteomic Study of a Parkinson's Disease Model of Undifferentiated SH-SY5Y Cells Induced by a Proteasome Inhibitor
1. Department of pediatrics, First Hospital of Jilin University, Changchun, Jilin Province, China;
2. Department of Pharmacology, College of Basic Medical Sciences, Jilin University, Chang Chun, Jilin Province, China;
3. Departments of Neurology and Neuroscience Center, First Hospital of Jilin University, Changchun, Jilin Province, China;
4. Key Laboratory of Medical Cell Biology, Institute of Translational Medicine, China Medical University, Shenyang, Liaoning Province, China.
# signify an equal contribution to the study.
* signify an equal contribution to the study.
Received 2018-7-17; Accepted 2018-11-5; Published 2019-1-1
Abstract
Parkinson's disease (PD) is one of the most common nervous system degenerative diseases. However, the etiology of this disease remains elusive. Here, a proteasome inhibitor (PSI)-induced undifferentiated SH-SY5Y PD model was established to analyze protein alterations through proteomic study. Methods: Cultured undifferentiated SH-SY5Y cells were divided into a control group and a group treated with 2.5 µM PSI (PSI-treated group). An methyl thiazolyl tetrazolium (MTT) assay was applied to detect cell viability. Acridine orange/ethidium bromide (AO/EB), α-synuclein immunofluorescence and hematoxylin and eosin (H&E) staining were applied to evaluate apoptosis and cytoplasmic inclusions, respectively. The protein spots that were significantly changed were separated, analyzed by 2D gel electrophoresis and DIGE De Cyder software, and subsequently identified by MALDI-TOF mass spectrometry and database searching. Results: The results of the MTT assay showed that there was a time and dose dependent change in cell viability following incubation with PSI. After 24 h incubation, PSI resulted in early apoptosis, and cytoplasmic inclusions were found in the PSI-treated group through H&E staining and α-synuclein immunofluorescence. Thus, undifferentiated SH-SY5Y cells could be used as PD model following PSI-induced inhibition of proteasomal function. In total, 18 proteins were differentially expressed between the groups, 7 of which were up-regulated and 11 of which were down-regulated. Among them, 5 protein spots were identified as being involved in the ubiquitin proteasome pathway-induced PD process. Conclusions: Mitochondrial heat shock protein 75 (MTHSP75), phosphoglycerate dehydrogenase (PHGDH), laminin binding protein (LBP), tyrosine 3/tryptophan 5-monooxygenase activation protein (14-3-3ε) and YWHAZ protein (14-3-3ζ) are involved in mitochondrial dysfunction, serine synthesis, amyloid clearance, apoptosis process and neuroprotection. These findings may provide new clues to deepen our understanding of PD pathogenesis.
Keywords: Parkinson's disease, ubiquitin proteasome system, SH-SY5Y cells, ptoteasomes inhibitor, proteomics
Introduction
Parkinson's disease (PD) is one of the most common nervous system degenerative diseases, which is characterized by loss of dopaminergic neurons in the substantia nigra (SN), as well as the presence of eosinophilic inclusions (Lewy bodies) in the residual neurons [1, 2]. The pathogenesis of PD involves multiple factors, including genetic mutation, mitochondrial dysfunction, oxidative stress, excitotoxicity, inflammation and ubiquitin-proteasome system damage [3]. However, the mechanisms that are currently understood cannot fully explain the pathogenesis of PD.
The ubiquitin proteasome system (UPS) is a highly efficient, selective and energy dependent protein degradation pathway with a cascade reaction of sequential E1, E2 and E3 catalysis. This pathway plays a role in protein degradation in the maintenance of a stable intracellular microenvironment [4]. Dysregulation of the UPS leads to alterations in protein homeostasis that may promote toxic protein accumulation and threaten neuronal survival [5]. The accumulation of misfolded and aggregated α-synuclein is thought to be the primary pathogenic event in familial PD [5, 6]. Therefore, it is important to deeply explore the pathogenesis of PD through a proteasome inhibitor-induced UPS dysfunction model.
SH-SY5Y cells are commonly used to establish a dopaminergic neuron model. This model is derived from SK-N-SH, a common pediatric neurogenic malignant solid tumor that originates from neuroectodermal cells [7]. Even undifferentiated SH-SY5Y cells present many characteristics of neurons, such as neurite-like cytoplasmic processes and the expressions of catecholaminergic neuron-specific tyrosine hydroxylase, dopamine β hydroxylase and dopamine transporters [7]. Protein is the material basis for the existence and movement of life. Proteins also execute cell proliferation, differentiation, senescence and apoptosis. Physiological functions and pathological changes are often accomplished by populations of proteins. Investigations in to the pathogenesis of PD through proteomics may reveal the changes that occur in PD over many levels.
In this study, we used proteomic analysis to determine the proteins that were differentially expressed in a proteasome inhibitor (PSI)-induced PD model in undifferentiated SH-SY5Y cells. The aim of the present study was to explore the pathological mechanisms involved in a proteasome deficit-induced PD model.
Results
The evaluation of PSI-treated SH-SY5Y cell viability
To evaluate the effect of PSI on SH-SY5Y cell viability, SH-SY5Y cells were treated with different concentrations of PSI (1 µM, 2.5 µM, 5 µM or 10 µM) for 24 h, 48 h and 72 h, respectively. As indicated in Figure 1, there was no change in cell viability between the control cells and 1 µM PSI treated group after 24 h. However, cell viability in the 2.5 µM PSI-treated group for 24 h was significantly lower than that of the control group (p < 0.01). Cell viability decreased further as the PSI concentration and the incubation time was increased. Thus, PSI has a dose- and time-dependent effect on cell viability.
Evaluation of proteasome inhibitor (PSI)-treated SH-SY5Y cell viability by methyl thiazolyl tetrazolium assay. Cell viability of SH-SY5Y cells was conducted following incubations of 24 h, 48 h or 72 h with different concentrations of PSI. The cell viability of the control group (0.1 % DMSO) was set to 100%. The statistical analysis method was Student's t-test. *p < 0.05 and **p < 0.01 compared to viability in the control group at the same time point; ##p < 0.01 compared to the viability in the 24 h group at the same PSI concentration; && p < 0.01 compared to the viability in the 48 h group at the same PSI concentration.
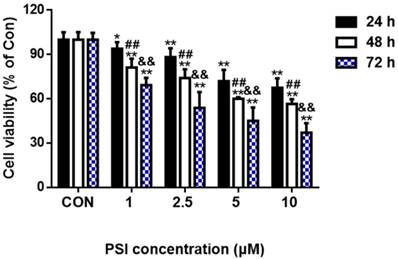
The morphological evaluation of PSI-treated SH-SY5Y cells
Cell morphology and acridine orange/ethidium bromide (AO/EB) staining tests were conducted to identify the effects of different concentrations of PSI on cell apoptosis. After treatment with PSI for 24 h, minimal morphological changes were observed between the control group and 2.5 µM PSI-treated group. As the PSI concentration increased, the morphological effects of PSI were more apparent. In the group treated with 10 μM PSI, the cell volume was lower and the neurite length was shorter than in the control group (Figure 2A). The AO/EB staining result showed early apoptotic cells in 2.5 µM PSI-treated group for 24 h (as indicated by the arrows in Figure 2B). Additionally, late apoptotic cells were observed in the group treated with 10 μM PSI for 24 h (as indicated by the arrows in Figure 2B). Excessive apoptosis may lead to intracellular protein degradation, thus, the conditions that were used in the experimental group of further experiments were 2.5 μM PSI for a 24 h incubation period.
Evaluation of proteasome inhibitor (PSI)-treated SH-SY5Y cell apoptosis by cell morphology and AO/EB staining. (A) The morphology of SH-SY5Y cells in the control and PSI-treated groups, at 200 × magnification under a light microscope. (B) The AO/EB staining of SH-SY5Y cells in the control and PSI-treated groups, at 200 × magnification under a fluorescence microscope.
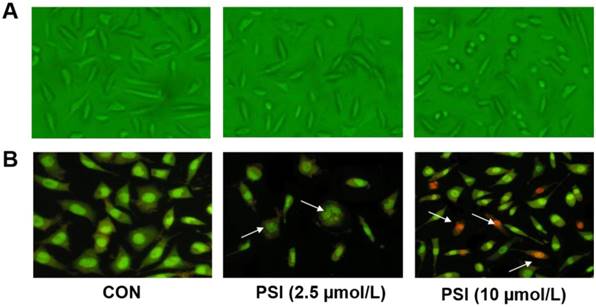
Evaluation of proteasome inhibitor (PSI)-treated SH-SY5Y cell cytoplasmic inclusions by α-synuclein immunofluorescence and hematoxylin&eosin (H&E) staining. (A) Inclusion body of α-synuclein immunofluorescence staining positive emerged in control and PSI-treated group, at 400 × magnification under a fluorescence microscope. (B) H-E staining of undifferentiated SH-SY5Y cells in the control and PSI-treated groups, at 400 × magnification under a light microscope. Lewy-like inclusion bodies in the cytoplasm are indicated by arrow.
The evaluation of cytoplasmic inclusions in PSI-treated SH-SY5Y cells
The formation of cytoplasmic inclusions is a key index through which to evaluate PD neuronal cells. Thus, we conducted α-synuclein immunofluorescence and hematoxylin and eosin (H&E) staining tests on these PSI-treated SH-SY5Y cells. In the PSI-treated group, eosinophilic inclusions, labeled with strong red fluorescence, were clearly observed in the cytoplasm of SH-SY5Y cells. Additionally, almost all of these cells showed a positive reaction for α-synuclein (Figure. 3A). In contrast, no eosinophilic inclusions were observed in the control group. Additionally, the results of the H&E staining showed no staining in the control group. Following treatment with PSI, at a concentration of 2.5 µM, clear Lewy-like inclusion bodies were observed in the cytoplasm of SH-SY5Y cells under light microscopy. However, eosinophilic inclusion bodies were not observed in the control group (Figure. 3B).
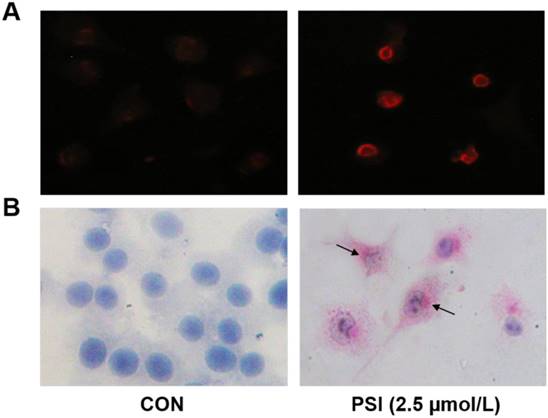
Pictures of differentially expressed proteins in proteasome inhibitor (PSI)-treated SH-SY5Y cell by 2D gel electrophoresis. (A) Gel fluorescence pictures in the control and PSI-treated groups obtained using a Typhoon 9400 laser. ①~④ display the Cy2 labeled internal standard group, Cy3 labeled control group, Cy5 labeled PSI-treated group and merge pictures, respectively. (B) Overview of the differentially expressed protein spots detected by DeCyder software.
Analysis of differentially expressed proteins in PSI-treated SH-SY5Y cells through 2D gel electrophoresis
After scanning by Typhoon 9400, three pictures were taken of each gel, corresponding to the Cy2 labeled internal standard sample (blue; Figure 4A), the Cy3 labeled samples (green; Figure 4A) and the Cy5 labeled samples (red; Figure 4A). The overlapped picture is shown in Figure 4B. DeCyder-BVA software was used to identify the proteins that were differentially expressed between the PSI-treated group and the control group (p < 0.05). In total, 18 protein spots significantly differed between the groups, including 7 proteins spots that were up-regulated and 11 protein spots that were down-regulated compared to the expression in the control group (Figure 4B and Table 1).
The identification of differential protein spots in PSI-treated SH-SY5Y cells through mass spectrometry
The peptide mass fingerprint (PMF) was analyzed using MALDI-TOF mass spectrometer analysis, and the single NCBInr database was used to identify the 18 proteins that were differentially expressed following PSI treatment. Finally, 5 protein spots were identified, and the distribution of these proteins in gel and the 2D regional amplification and abundance simulation pictures are presented in Figure 5. With functional annotation, the 5 identified proteins were classified into 5 categories (Table 2): (1) the molecular chaperone family members: mitochondrial heat shock protein 75 (MTHSP75; 127); (2) proteins related to signal transduction: tyrosine 3/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (789); (3) proteins related to energy metabolism: phosphoglycerate dehydrogenase (293); (4) proteins related to protein folding and transport function: YWHAZ protein (14-3-3 zeta protein; 837); (5) proteins related to cell proliferation, adhesion and metastasis: laminin binding protein (laminin-binding protein, LBP; 555). All of these proteins were downregulated compared to the expression observed in the control group, except for MTHSP75.
The results of differential protein spot expression between control and proteasome inhibitor (PSI)-treated SH-SY5Y cells
| protein spot | occurrence rate | ratio of PSI-treated group and control group | p value |
|---|---|---|---|
| 55 | 9 (12) | +1.50 | 0.016 |
| 101 | 9 (12) | +2.53 | 0.034 |
| 105 | 12 (12) | +2.63 | 0.026 |
| 127 | 12 (12) | +1.54 | 0.028 |
| 229 | 12 (12) | +1.33 | 0.026 |
| 293 | 12 (12) | -1.35 | 0.015 |
| 496 | 12 (12) | +1.37 | 0.018 |
| 550 | 12 (12) | -1.36 | 0.030 |
| 555 | 12 (12) | -1.36 | 0.025 |
| 590 | 12 (12) | -2.02 | 0.029 |
| 623 | 12 (12) | +1.69 | 0.027 |
| 758 | 12 (12) | -1.76 | 0.0097 |
| 789 | 12 (12) | -1.33 | 0.036 |
| 793 | 12 (12) | -1.38 | 0.046 |
| 795 | 12 (12) | -1.78 | 0.020 |
| 837 | 12 (12) | -1.43 | 0.039 |
| 902 | 12 (12) | -1.49 | 0.023 |
| 970 | 12 (12) | -1.91 | 0.0026 |
Occurrence rate refers to the number of times of a protein appeared in the total of 12 pieces of 2D gel electrophoresis map; '+' signifies an increased expression of protein in the PSI-treated group compared to the expression in the control group; '-' signifies a decreased expression of protein in the PSI-treated group compared to the expression in the control group.
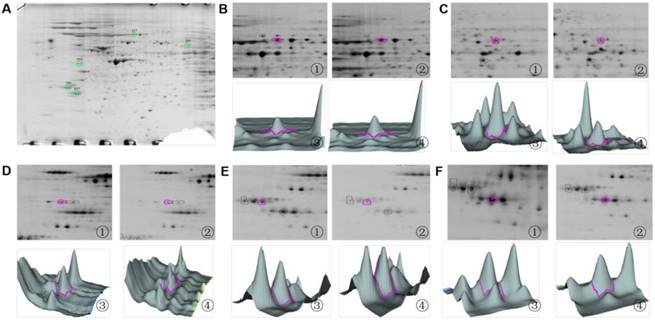
2D zooms and 3D views of differentially expressed protein spots identified by MALDI-TOF mass spectrometry. (A) Overview of differentially expressed protein spots identified in SH-SY5Y cells through MALDI-TOF mass spectrometry. (B) The 2D zoom and 3D views of spot 127: ① and ② show the 2D zooms of point 127 in the control and PSI-treated group, respectively; ③ and ④ show 3D views of 127 protein spots in the control and PSI-treated group, respectively. (C) The 2D zoom and 3D views of spot 293. ① and ② show the 2D zooms of point 293 in the control and PSI-treated group, respectively; ③ and ④ show 3D views of 293 protein spots in the control and PSI-treated group, respectively (D) The 2D zoom and 3D views of spot 555. ① and ② show the 2D zooms of point 555 in the control and PSI-treated group, respectively; ③ and ④ show 3D views of 555 protein spots in the control and PSI-treated group, respectively (E) The 2D zoom and 3D views of spot 789. (F) The 2D zoom and 3D views of spot 837. ① and ② show the 2D zooms of point 837 in the control and PSI-treated group, respectively; ③ and ④ show 3D views of 837 protein spots in the control and PSI-treated group, respectively.
The results of differential protein spot expression between control and proteasome inhibitor (PSI)-treated SH-SY5Y cells as identified through MALDI-TOF mass spectroscopy
| protein spot | NCBI database number | protein name | isoelectric point (pI) | molecular weight (kDa) | expectation | sequence coverage (%) |
|---|---|---|---|---|---|---|
| 127 | gi292059 | MTHSP75 | 6.0 | 74.05 | 0.001 | 22.2 % |
| 293 | gi23308577 | phosphoglycerate dehydrogenase | 6.3 | 57.37 | 0.000 | 20.3 % |
| 555 | gi34234 | laminin-binding protein | 4.8 | 31.89 | 0.000 | 42.1 % |
| 789 | gi5803225 | tyrosine3 / tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 4.6 | 29.33 | 0.000 | 42.7 % |
| 837 | gi4507953 | YWHAZ protein | 4.7 | 30.1 | 0.000 | 46.0 % |
Expectation refers to the error probability of the identified results; Sequence coverage refers to the amount of the identified peptide sequence that makes up the percentage of total protein sequences.
Discussion
Parkinson's disease is a lifelong and progressive neurodegenerative disease that is characterised by the loss of dopaminergic neurons and the aggregation of cytoplasmic inclusions [8]. UPS is an important protein degradation pathway in eukaryotic cells. Dysfunction of this pathway leads to the accumulation of α-synuclein and neuronal death in the substantia nigra [8, 9]. Previous studies have shown that N-benzyloxycarbonyl-Ile-Glu (O-t-butyl)-Ala-Leu-al, a proteasome inhibitor, can induce the presentation of the main features of PD in PC12 cells [10]. In the present study, we used N-benzyloxycarbonyl-Ile-Glu (O-t-butyl)-Ala-Leu-al to treat SH-SY5Y cells, a cell line that is similar to normal nerve cells in physiological and biochemical functions and is widely used in research into the pathogenesis of neurodegenerative diseases and the mechanism of drug action [11, 12]. After incubation with PSI for 24 h, the viability of SH-SY5Y cells was significantly lower than that of the untreated group. Moreover, obvious α-synuclein expression and cytoplasmic inclusions were observed in PSI-treated SH-SY5Y cells. The above results indicate that PSI may stimulate SH-SY5Y cells to express the two key pathologic characteristics of PD. Thus, our PSI-treated SH-SY5Y cells could be used as PD model in vitro.
Proteomic analysis was conducted on this PSI-treated SH-SY5Y cell PD model. 18 protein spots were significantly different to those of control cells. Of these, 5 protein spots were successfully identified, including: mitochondrial heat shock protein 75 (MTHSP75), phosphoglycerate dehydrogenase (PHGDH), laminin binding protein (LBP), tyrosine3/tryptophan5-monooxygenase activation protein (14-3-3ε) and YWHAZ protein (14-3-3ζ).
MTHSP75 is a conservative member of the human heat stress protein 70 (HSP70) family. The main function of the HSP70 family is to correct or degrade misfolded proteins in the mitochondria. Mitochondrial impairment in neuronal cells has been linked to the pathogenesis of neurological disorders [13]. Knockdown of MTHSP75 causes aggregation and dysfunction of mitochondria in yeast cells [14]. Furthermore, overexpression of MTHSP75 helps to preserve striatal bioelectrical activity under conditions of mitochondrial impairment [15]. Jochen et al. [16] found that overexpression of HSP70 could reduce the aggregation of misfolded α-synuclein, and reduce the cytotoxicity caused by the aggregation of α-synuclein. A gene expression study that profiled sporadic PD showed that decreased HSP70 expression in patients with sporadic PD lead to an impaired function of an entire repertoire of proteins [17]. Unexpectedly, our data shows a significantly higher cell number in the PSI-treated group than in the control group. This may reflect the high number of SH-SY5Y cells in the early stages of apoptosis in the PSI-treated group, and an increased MTHSP75 expression may play a protective role in defending against PSI-induced neuronal damage.
PHGDH, the first enzyme of the mammalian serine biosynthesis pathway, is widely distributed in various cell types [18]. Serine is a necessary precursor for the synthesis of proteins, lipids, glycine, cysteine, d-serine, and tetrahydrofolate metabolites. Moreover, serine can act as the intermediate in the synthesis of L-dopa [19]. A previous study reported that the PHGDH expression is significantly reduced in the aged brain. This indicates that PHGDH may be a critical factor in neurogenesis [20]. Furthermore, disorders in which the encoding of PHGDH is disrupted are characterized by severe neurological symptoms, including congenital microcephaly and psychomotor retardation [21]. Our study also shows that the PHGDH expression was significantly lower in the PSI-treated group than in the control group. This may lead to a blockage of the serine synthesis pathway, which would have a direct effect on the synthesis of a series of neurogenesis related metabolites.
LBP is a versatile molecule that serves different functions in immune regulation and lipid transfer [22]. Blood clotting is adopted by an amyloid form, and LBP has been shown to prevent the amyloid formation. Moreover, Pretorius et al. found that LPS-induced fibrin amyloid formation could be diminished by LBP [23]. A continuous report from this team showed that LBP could reverse the amyloid state of fibrin seen in PD [24]. Our study showed that the level of LBP in the PSI-treated group was lowered to 27 % of the level observed in the control group. Corresponding with the formation of α-synuclein and cytoplasmic inclusions in PSI-treated SH-SY5Y cells, this decrease in LBP expression may accelerate the PD process and the aggravation of PD related amyloid formation.
14-3-3ε and 14-3-3ζ belong to the 14-3-3 protein family. 14-3-3ε is the highest expressed member of the 14-3-3 protein family in the nigrostriatal area [25]. Many studies have illustrated that 14-3-3ε participates in cell survival [7, 26]. The loss of 14-3-3ε in drosophila resulted in a lower survival rate during the larva-to-adult transition [27]. 14-3-3ε may have an anti-apoptotic function by combining with the B-cell lymphoma 2 (Bcl-2)-associated death promoter (BAD) to release Bcl-2 [8]. In the present study, the expression of 14-3-3ε was significantly lower in the PSI-treated group than in the control group. This is consistent with the elevated cell death that was observed in the PSI-treated SH-SY5Y cells. 14-3-3ζ is a key regulator that mediates signal transduction by binding to phosphoserine-containing proteins during the hippocampal aging process [28]. Upregulation of 14-3-3ζ helps to promote hippocampal development [29]. Moreover, 14-3-3ζ is potentially a regulator with a similar function to that of β-catenin, which is a central effector of Wnt signaling in metastasis. The study of Zhang and colleagues demonstrated that 14-3-3ζ could protect dopaminergic neurons through the Wnt signaling pathway [30]. Our data shows that the expression of 14-3-3ζ was lower in the PSI-treated group than in the control group. This finding suggests that a series of phosphoserine of proteins related with 14-3-3ζ may dysfunction in the PSI-induced PD process.
Taken together, the 5 differentially expressed proteins that were identified in this study are involved in mitochondrial dysfunction, serine synthesis, amyloid clearance, the apoptotic process and neuroprotection. Based on the functional annotation, our study may provide new clues to deepen our understanding of the pathogenesis of PD.
Materials and methods
Materials and instruments
The SH-SY5Y cell line was purchased from American Type Culture Collection (ATCC; USA). PSI, N-benzyloxycarbonyl-Ile-Glu (O-t-butyl)-Ala-Leu-al, was purchased from Merck Millipore Corporation (Germany). Dulbecco's modified Eagle's medium (DMEM) and Fetal Calf Serum (FCS) were purchased from Thermo Fisher (USA). L-glutamine, dimethyl sulfoxide (DMSO) and MTT were purchased from Sigma (USA). The AO/EB kit was purchased from AMRESCO (USA). The anti-α-SYN antibody was purchased from Santa Cruz biotechnology (Santa Cruz; USA). The Cy5 labeled goat anti-mouse antibody, 2-D Quant Kit and protein clean-up kit were purchased from Biosciences-GE Healthcare Amersham (USA). Radio-Immunoprecipitation Assay (RIPA), Phenylmethanesulfonyl fluoride (PMSF), Hematoxylin, eosin and phosphatase inhibitors were purchased from Nanjing Jiancheng Bioengineering institute (China).
Cell culture and modeling
The SH-SY5Y cells were grown in DMEM containing 5 % FCS. The cells were plated at 2 × 104 cells per well in 24-well plates. The experimental group was treated with 2.5 μM PSI per well, and correspondingly, the control group was treated with the same volume of culture medium containing 0.1 % DMSO. The cells were harvested for further experiments after a 24 h incubation.
Cell viability
The SH-SY5Y cells were grown in DMEM containing 5 % FCS. The cells were plated at 5000 cells per well in 96-well plates. After treatment with different doses of PSI (1 µM, 2.5 µM, 5 µM or 10 µM) for 24 h, the culture medium was removed and 200 µL of MTT reagent, at a concentration of 1 mg/mL in phosphate buffered saline (PBS), was added to each well. After incubation at 37 °C for 4 h, MTT was removed and the blue-colored formazan product was solubilized in 150 μL DMSO. The absorbance was measured at a wave length of 570 nm after incubation and shaking for 20 min.
AO/EB Staining
After PSI treatment for 24 h, the cells were harvested and centrifuged at 1000 g × 3 min and washed twice with PBS. The cell pellets were resuspended in 500 µL of PBS. A volume of 100 µL of cell suspension was dropped on a slide with 5 uL of AO/EB dye solution. Cell apoptosis was examined and photographed using an IX-71 fluorescence microscope (Olympus; Japan).
Immunofluorescence staining of α-synuclein
After culture for 24 h, the cells were fixed by 4 % paraformaldehyde for 30 min, then washed three times with PBS and permeabilized by 0.1 % Triton X-100 for 2 min. The fixed cells were blocked by 5 % bovine serum albumin (BSA) for 30 min, then incubated with anti-α-SYN antibody (1:100 with 1 % BSA dilution) for 2 h followed by an incubation with a Cy5 labeled Goat anti-mouse (1:100 with 1 % BSA dilution) second antibody for 30 min. After three washes in PBS, the cells were then examined under an IX-71 fluorescence microscope (470/40 nm filter).
H&E staining
After being fixed by 4 % paraformaldehyde for 10 min, cells were washed in PBS and stained with Mayer's Hematoxylin for 10 min and eosin for 5 min. The samples were then passed through an ascending graded ethanol series. Morphological observations were performed using a BX40 12 J02 system biological microscope (USA).
Protein sample preparation and labeling
Four batches of undifferentiated SH-SY5Y (named: 1, 2, 3, 4) were randomly set as the experimental groups (T1~4) and the control groups (C1~4). The total proteins were extracted by RIPA lysis (1:10, w:w), and impurities were removed from the sample by a protein clean-up kit. The protein concentration was measured using a 2-D Quant Kit, a Lowry test, and was adjusted to the final concentration of 5 mg/mL. The pH of the protein solution was adjusted with 50 mM NaOH to reach 8.5~9.0. A total protein weight of 50 µg was collected from T1~4 and C1~4. These samples were labeled with 1 µL of Cy3 or Cy5 working fluid, at random. Internal marker samples were prepared from a mixture with 6.25 µg protein from each tube (T1~4 and C1~4) and labeled with 4 µL Cy2 working fluid. All samples were incubated on ice in dark conditions for 30 min. Finally, 1 µL lysine solution (10 mM) was added to terminate the reaction.
2D gel electrophoresis and image analysis
2D gel electrophoresis was performed on an Ettan DALT six electrophoresis unit (2 W per gel, 600 V, 400 mA; Amersham Biosciences; USA). The Cy2, Cy3 and Cy5 immunofluorescence was scanned using Typhoon 9400 scanner at the wavelength of 532 nm, 633 nm and 488 nm, respectively (Amersham Biosciences; USA). The digitalized images of the gels were analyzed using Image Master 2D Evolution version 2003.02 (Amersham Biosciences; USA). Spots in images were quantified and statistically evaluated using pattern analysis of computer-assisted pit assessment and the reproducibility and significance of the differences in spot sizes were determined.
Gel digestion and MALDI-TOF mass spectrometer analysis
Protein spots were automatically selected and placed in 96-well plates using an Ettan Spot Picker robotic workstation (Amersham Biosciences; USA). The gel plugs were destained twice, dehydrated, desiccated and incubated in 10 μl of 1 μg/μl modified porcine trypsin at 4°C overnight. Following this, the digested peptide fragments were extracted twice and the two combined extracts were desiccated in a new 96-well plate. A volume of 0.3 μl of the digested peptide fragments and an equal volume of 4 μg/μl a-cyano-4-hydroxy-transcinnamic acid were deposited on the surface of a sample slide (Amersham Biosciences; USA).
The PMF was analyzed using a MALDI-TOF mass spectrometer (Amersham Biosciences; USA). The most intense signals of mass spectrometry had to be characterized by at least 3 to 5 of the strongest peaks. Protein identification via PMF was initially accomplished using the ProFound search engine against the latest version of the single NCBInr database, and was further enhanced using Mowse score-based Mascot search engine against the latest version of the multiple NCBInr, SwissProt.
Statistical analysis
Differences between groups were determined using Student's t-tests. p values of less than 0.05 were considered to be statistically significant. The MTT statistical analyses were performed using GraphPad Prism version 5.01 for Windows (USA). The peptide mass fingerprint (PMF) was analyzed using an MALDI-TOF MS mass spectrometer (Amersham Biosciences; USA). Protein identification through PMF was performed using the ProFound search engine against the latest version of the single NCBInr database. The requirements of evaluation standard were as follows: sequence coverage at least 20%, expectation as much as 0.05.
Acknowledgements
This study was supported by funds from science and technology development projects of the Jilin Province (20140203011YY, 20150311013YY, 20170623062TC). Jilin Province Natural Science Foundation (Project Number 20180101154JC to Y Zhang).
Abbreviations
AO/EB: Acridine Orange/Ethidium Bromide; H&E: hematoxylin and eosin; LBP: laminin binding protein; MTHSP75: mitochondrial heat shock protein 75; MTT: methyl thiazolyl tetrazolium; PD: Parkinson's disease; PHGDH: phosphoglycerate dehydrogenase; PSI: proteasome inhibitor; 14-3-3ε: tyrosine 3/tryptophan 5-monooxygenase activation protein; 14-3-3ζ: YWHAZ protein.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zlobine I, Gopal K, Ussher JR. Lipotoxicity in obesity and diabetes-related cardiac dysfunction. Biochim Biophys Acta. 2016;1861:1555-68
2. Volpicelli-Daley LA. Effects of alpha-synuclein on axonal transport. Neurobiol Dis. 2016
3. Gautier CA, Corti O, Brice A. Mitochondrial dysfunctions in Parkinson's disease. Rev Neurol (Paris). 2014;170:339-43
4. Lip PZ, Demasi M, Bonatto D. The role of the ubiquitin proteasome system in the memory process. Neurochem Int. 2016;102:57-65
5. Lim KL, Tan JM. Role of the ubiquitin proteasome system in Parkinson's disease. BMC Biochem. 2007;8(Suppl 1):S13
6. Cook C, Petrucelli L. A critical evaluation of the ubiquitin-proteasome system in Parkinson's disease. Biochim Biophys Acta. 2009;1792:664-75
7. Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703-9
8. Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8:600-6
9. Mor DE, Tsika E, Mazzulli JR, Gould NS, Kim H, Daniels MJ. et al. Dopamine induces soluble alpha-synuclein oligomers and nigrostriatal degeneration. Nat Neurosci. 2017;20:1560-8
10. Nakashima A, Ohnuma S, Kodani Y, Kaneko YS, Nagasaki H, Nagatsu T. et al. Inhibition of deubiquitinating activity of USP14 decreases tyrosine hydroxylase phosphorylated at Ser19 in PC12D cells. Biochem Biophys Res Commun. 2016;472:598-602
11. Lebedev TD, Spirin PV, Orlova NN, Prokofjeva MM, Prassolov VS. [Comparative study of therapy targeted genes expression in neuroblastoma cell lines]. Mol Biol (Mosk). 2015;49:1048-51
12. Cheung YT, Lau WK, Yu MS, Lai CS, Yeung SC, So KF. et al. Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology. 2009;30:127-35
13. Voloboueva LA, Lee SW, Emery JF, Palmer TD, Giffard RG. Mitochondrial protection attenuates inflammation-induced impairment of neurogenesis in vitro and in vivo. J Neurosci. 2010;30:12242-51
14. Kawai A, Nishikawa S, Hirata A, Endo T. Loss of the mitochondrial Hsp70 functions causes aggregation of mitochondria in yeast cells. J Cell Sci. 2001;114:3565-74
15. Tantucci M, Mariucci G, Taha E, Spaccatini C, Tozzi A, Luchetti E. et al. Induction of heat shock protein 70 reduces the alteration of striatal electrical activity caused by mitochondrial impairment. Neuroscience. 2009;163:735-40
16. Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 Reduces alpha-Synuclein Aggregation and Toxicity. J Biol Chem. 2004;279:25497-502
17. Mandel S, Grunblatt E, Riederer P, Amariglio N, Jacob-Hirsch J, Rechavi G. et al. Gene expression profiling of sporadic Parkinson's disease substantia nigra pars compacta reveals impairment of ubiquitin-proteasome subunits, SKP1A, aldehyde dehydrogenase, and chaperone HSC-70. Ann N Y Acad Sci. 2005;1053:356-75
18. Murphy JP, Giacomantonio MA, Paulo JA, Everley RA, Kennedy BE, Pathak GP. et al. The NAD(+) Salvage Pathway Supports PHGDH-Driven Serine Biosynthesis. Cell Rep. 2018;24(e5):2381-91
19. Ghiglieri V, Mineo D, Vannelli A, Cacace F, Mancini M, Pendolino V. et al. Modulation of serotonergic transmission by eltoprazine in L-DOPA-induced dyskinesia: Behavioral, molecular, and synaptic mechanisms. Neurobiol Dis. 2016;86:140-53
20. Kinoshita MO, Shinoda Y, Sakai K, Hashikawa T, Watanabe M, Machida T. et al. Selective upregulation of 3-phosphoglycerate dehydrogenase (Phgdh) expression in adult subventricular zone neurogenic niche. Neurosci Lett. 2009;453:21-6
21. Kawakami Y, Yoshida K, Yang JH, Suzuki T, Azuma N, Sakai K. et al. Impaired neurogenesis in embryonic spinal cord of Phgdh knockout mice, a serine deficiency disorder model. Neurosci Res. 2009;63:184-93
22. Gopalakrishna R, Gundimeda U, Zhou S, Bui H, Davis A, McNeill T. et al. Laminin-1 induces endocytosis of 67KDa laminin receptor and protects Neuroscreen-1 cells against death induced by serum withdrawal. Biochem Biophys Res Commun. 2018;495:230-7
23. Pretorius E, Mbotwe S, Bester J, Robinson CJ, Kell DB. Acute induction of anomalous and amyloidogenic blood clotting by molecular amplification of highly substoichiometric levels of bacterial lipopolysaccharide. J R Soc Interface. 2016:13
24. Pretorius E, Page MJ, Mbotwe S, Kell DB. Lipopolysaccharide-binding protein (LBP) can reverse the amyloid state of fibrin seen or induced in Parkinson's disease. PLoS One. 2018;13:e0192121
25. Wang B, Underwood R, Kamath A, Britain C, McFerrin MB, McLean PJ. et al. 14-3-3 proteins reduce cell-to-cell transfer and propagation of pathogenic alpha-synuclein. J Neurosci. 2018
26. Sai Y, Peng K, Ye F, Zhao X, Zhao Y, Zou Z. et al. 14-3-3 Proteins in the regulation of rotenone-induced neurotoxicity might be via its isoform 14-3-3epsilon's involvement in autophagy. Cell Mol Neurobiol. 2013;33:1109-21
27. Ng YS, Sorvina A, Bader CA, Weiland F, Lopez AF, Hoffmann P. et al. Proteome Analysis of Drosophila Mutants Identifies a Regulatory Role for 14-3-3epsilon in Metabolic Pathways. J Proteome Res. 2017;16:1976-87
28. Conklin DS, Galaktionov K, Beach D. 14-3-3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci U S A. 1995;92:7892-6
29. Wang J, Li Q, Kong Y, Zhou F, Li J, Li W. et al. Biosystems Study of the Molecular Networks Underlying Hippocampal Aging Progression and Anti-aging Treatment in Mice. Front Aging Neurosci. 2017;9:393
30. Cao C, Chen J, Lyu C, Yu J, Zhao W, Wang Y. et al. Bioinformatics Analysis of the Effects of Tobacco Smoke on Gene Expression. PLoS One. 2015;10:e0143377
Author contact
![]() Corresponding author: Dr. Ying Zhang, Associated Professor, Tel: +86-0431-88782362, E-mail: rose19700com; Dr. Ming Zhang, Associated Professor, Tel: +86-0431-85619799, E-mail: zhangming99edu.cn.
Corresponding author: Dr. Ying Zhang, Associated Professor, Tel: +86-0431-88782362, E-mail: rose19700com; Dr. Ming Zhang, Associated Professor, Tel: +86-0431-85619799, E-mail: zhangming99edu.cn.

 Global reach, higher impact
Global reach, higher impact