Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(13):1547-1554. doi:10.7150/ijms.26329 This issue Cite
Research Paper
Microarray Analysis of Differentially-expressed MicroRNAs in Acquired Middle Ear Cholesteatoma
1. Department of Otolaryngology-Head and Neck Surgery, The Xiangya Hospital, Central South University, Hunan provincial key lab, Otolaryngology institute of major diseases, Changsha, Hunan 410008, China.
2. Department of Otolaryngology-Head and Neck Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China.
Received 2018-3-27; Accepted 2018-7-27; Published 2018-10-20
Abstract
Objectives: To analyze the miRNAs expression profiling between acquired middle ear cholesteatoma and normal skin, and to identify several novel miRNAs which may be involved in the etiopathogenesis of middle ear cholesteatoma.
Methods: MiRNA microarray technology was adopted to analyze the miRNA expression profiling between acquired middle ear cholesteatoma and normal skin. qRT-PCR was used to validate selected differentially expressed miRNAs.
Results: The miRNA microarray technology showed totally 44 upregulated (miRNA-21-3p, miRNA-584-5p, miRNA-16-1-3p, etc) and 175 downregulated (miRNA-10a-5p, miRNA-152-5p, miRNA-203b-5p, etc) miRNAs in cholesteatoma tissues with 2-fold change compared with normal skin. The qRT-PCR validation was in accordance with the microarray results partly: miRNA-21-3p and miRNA-16-1-3p expressed significantly higher while miRNA-10a-5p exhibited an obviously decreased expression in middle ear cholesteatoma tissues when compared with normal skin. The GO and KEGG pathway analyses provided clues that these differentially expressed miRNAs might play essential roles in the etiopathogenesis of middle ear cholesteatoma, including cell proliferation, apoptosis, cell cycle, differentiation, bone resorption and remodeling process.
Conclusions: Our study suggests possible roles of differentially expressed miRNAs in the pathogenesis of middle ear cholesteatoma. Targeting on these miRNAs may provide a new strategy for cholesteatoma therapy in the future.
Keywords: miRNA, middle ear, cholesteatoma, etiopathogenesis
Introduction
Middle ear cholesteatoma is a benign keratinizing and hyper-proliferative squamous epithelial lesion that is locally destructive and frequently recurrent. It is usually associated with chronic otitis media, which gradually expands and results in destruction of nearby bony structures, causing conductive hearing loss, facial nerve paralysis, postauricular subperiosteal abscess, labyrinthine fistulae, bacterial labyrinthitis and intracranial complications such as meningitis, sigmoid sinus thrombophlebitis, extradural abscess, and brain abscess and so on. Middle ear cholesteatoma has been estimated to affect about 9.2 per 100,000 populations a year in Europe [1], and the recurrence after surgical removal is common. Consequently, there is an urgent necessity for developing non-surgical treatment alternatives, based on the molecular mechanisms. Unfortunately, to date, the exact cellular and molecular mechanisms underlying the pathogenesis in middle ear cholesteatoma still have not been thoroughly elucidated.
MicroRNA (miRNA) is a short non-coding RNA molecule that functions in post-transcriptional regulation of gene expression. They can initiate the protein translational repression by binding to their targeted messenger RNAs (mRNAs) at the 3'-untranslated regions. Recently, more and more researchers are paying attentions to miRNA expression profiling studies since miRNAs participate in a variety of cellular pathways, including cell proliferation, differentiation, apoptosis and so on [2]. So far, the abnormal expression of miRNAs have been discovered in the etiopathogenesis of various inflammatory proliferative diseases and neoplastic diseases, such as psoriasis [3], rheumatoid arthritis [4], dermatitis [5], nasopharyngeal carcinoma [6], tongue squamous cell carcinoma [7], and so on.
It has been hypothesized that the inflammatory process and cell cytokines and mediators released in this process contribute to the etiopathogenesis of middle ear cholesteatoma. Cytokines secreted in the inflammatory response may promote the aggressive growth of cholesteatoma keratinocytes and bone resorption through activating some biochemical signaling pathways, such as phosphatidylinositol 3-kinase (PI3K)- protein kinase B (Akt) and mitogen-activated protein kinase (MAPK) signaling pathways [8]. Recently, some miRNAs have been demonstrated to act crucial biological functions in the hyper-proliferation of cholesteatoma keratinocytes, such as miRNA-21 [9] and miRNA-802 [10]. For example, miRNA-21 suppresses phosphatase and tensin homologue (PTEN) and programmed cell death 4 (PDCD4) translation and leads to cholesteatoma keratinocyte hyper-proliferation [9]. Nevertheless, there is a lack of miRNAs expression profiling analysis in acquired middle ear cholesteatoma.
In this study, we compared the miRNA expression profiling between acquired middle ear cholesteatoma and normal skin (from incision either endaural or postauricular) by miRNA microarray technology, and validated selected miRNAs using quantitative real-time polymerase chain reaction (qRT-PCR), aiming to identify several novel miRNAs which may be involved in the etiopathogenesis of middle ear cholesteatoma.
Materials and Methods
Patients and samples
Acquired middle ear cholesteatoma tissue specimens were collected from 20 patients who underwent surgical treatment for middle ear cholesteatoma from August 2016 to March 2017 at the Department of Otolaryngology Head and Neck Surgery, The Second Xiangya Hospital of Central South University. Meanwhile, 15 normal skin samples (from incision either endaural or postauricular) were obtained to be used as controls. Middle ear cholesteatoma and normal skin samples were immediately preserved in liquid nitrogen for RNA microarray or validation. The study was approved by the Ethics Committee of The Second Xiangya Hospital of Central South University on October 10, 2014 (Number: 2014114), and informed consents were obtained from all patients.
RNA extraction
Total cellular RNA was isolated from 5 middle ear cholesteatoma and 5 normal skin tissues using TRIzol reagent (Invitrogen, USA) and then purified with RNasey mini kit (QIAGEN, German) according to manufacturer's instructions. RNA concentration was measured by using nanodrop ND-1000 spectrophotometer (Thermo, USA) and RNA integrity was determined by gel electrophoresis, respectively.
MiRNA labeling and microarray hybridization
MiRNAs expression profiling was conducted through the usage of the miRCURYTM locked nucleic acid (LNA) microarray platform (Exiqon, Denmark). After quality control, miRNA labelling was performed by applying the miRCURY™ Power labeling kit (Exiqon, Denmark) according to the manufacturer's guideline. Firstly, 1μLRNA in 2.0 μL of water was combined with 1.0 μL of calf intestine phosphatase (CIP) buffer and CIP (Exiqon, Denmark). The mixture was incubated for 30 min at 37°C. Then, the reaction was terminated by incubation for 5 min at 95°C. Subseqently, the mixture was combined with 3.0 μL of labeling buffer, 1.5 μL of fluorescent label (Hy3TM), 2.0 μL of DMSO, 2.0 μL of labeling enzyme. The labeling reaction was incubated for 1 h at 16°C. Then, after incubation for 15 min at 65°C, the labeling reaction was terminated. Hybridization was followed by the labeling procedure. In detail, the Hy3™-labeled samples were hybridized on the miRCURYTM LNA Array (Exiqon, Denmark) according to manufacturer's guideline. A mixture of 25 μL Hy3™-labeled samples and 25 μL hybridization buffer were firstly denatured for 2 min at 95°C, incubated on ice for 2 min, and then hybridized to the microarray for 16-20 h at 56°C in Nimblegen Systems (Madison, USA, http://www.nimblegen.com, RRID: SCR_008571). Consequently, the hybrid chips were achieved, and were washed with Wash buffer kit (Exiqon, Denmark). Finally, the Axon GenePix 4000B microarray scanner (Axon, USA) was used to scan the hybrid chips.
Image acquisition and analysis
The GenePix Pro software, V6.0 (Axon, USA, http://www.moleculardevices.com/Products/Software/GenePix-Pro.html, RRID: SCR_010969) was applied on scanned images for grid alignment and data extraction. Replicated miRNAs were averaged and conflated. Additionally, a normalized median was obtained from miRNAs with intensities≥30 in all samples. After normalization, significant differentially expressed miRNAs between middle ear cholesteatoma and normal skin were identified through Fold change and P-value. Finally, hierarchical clustering was conducted to display differentially expressed miRNA profiling among middle ear cholesteatoma and normal skin samples.
MiRNA target prediction and network analysis
The TargetScan database (http://targetscan.org/, RRID: SCR_010845) and miRDB database (http://mirdb.org/miRDB/, RRID: SCR_010848) were adopted to predict potential target genes of the differently expressed miRNAs. Meanwhile, Gene Ontology (GO) classification (http://meme.nbcr.net/meme/cgi-bin/gomo.cgi, RRID: SCR_008864) and Kyoto Encyclopedia Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/expression, RRID: SCR_001120) were performed to acquire useful information regarding the functions of target genes underlying biological processes in the etiopathogenesis of middle ear cholesteatoma. A P<0.05 was considered to be statistically significant.
Validation for differentially expressed miRNAs using qRT-PCR
Total cellular RNA was respectively purified from 15 cholesteatoma and 10 normal skin tissues using Trizol reagent (Invitrogen, USA), precipitated with alcohol, and then stored at -70℃ until use. Extracted RNA was quantified by measuring optical density at 260 nm, and then 0.4 μg RNA was reversely transcribed to cDNA in a 20 μl reaction system according to the manufacturer's protocol. Then, quantitative PCR reactions were conducted using QuantStudioTM 5 System (Applied Biosystems, USA) with 2 μl cDNA sample adding into a 8 μl reaction system, based on the manufacturer's protocol. Amplification conditions were controlled as follows: 95℃ for 10 min for 1 cycle, followed by 40 cycles of 95℃ for 10 sec and 60℃ for 60 sec. U6 primers were applied as an internal reference for RNA template normalization. The specific amplification primer pairs were 5'-GACCCCCAGTATTAACTGTGC-3' and 5'-GTGCGTGTCGTGGAGTCG-3' for miRNA-16-1-3p forward and reverse, 5'-GGCAACCCTGTAGATCCGAA-3' and 5'-GTGCGTGTCGTGGAGTCG-3' for miRNA-10a-5p forward and reverse, 5'-TGGGTTATGGTTTGCCTGG-3' and 5'-GTGCGTGTCGTGGAGTCG-3' for miRNA-584-5p forward and reverse, 5'-GGGAACAATATCCTGGTGC-3' and 5'-CAGTGCGTGTCGTGGAGT-3' for miRNA-338-5p forward and reverse, 5'-GGCAACAGCAGTCGATG-3' and 5'-CAGTGCGTGTCGTGGAGT-3' for miRNA-21-3p forward and reverse, 5'-GCTTCGGCAGCACATATACTAAAAT-3' and 5'-CGCTTCACGAATTTGCGTGTCAT-3' for U6 forward and reverse, respectively.
Results
Clinical characteristics
Clinical characteristics for the 20 patients with middle ear cholesteatoma were as follows. Of these 20 patients with middle ear cholesteatoma, 13 (65%) were male, and 7 (35%) were female. Patients varied in age from 13 to 59 years old, with a mean age of 32.5 years old. Meanwhile, the course lasted from 1 month to 40 years, with a mean duration of 12.6 years. Totally, 15 right ears and 5 left ears were included.
Differential expression of miRNAs in cholesteatoma and normal skin
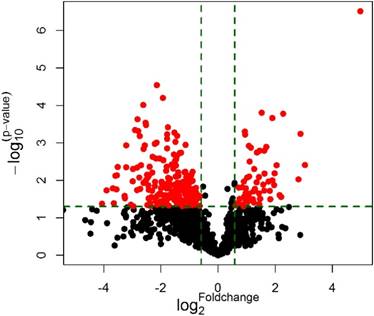
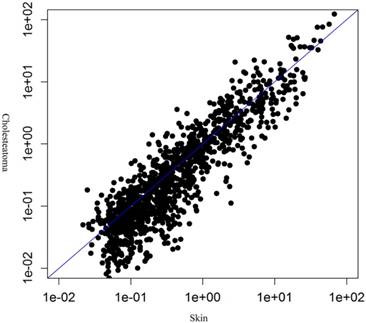
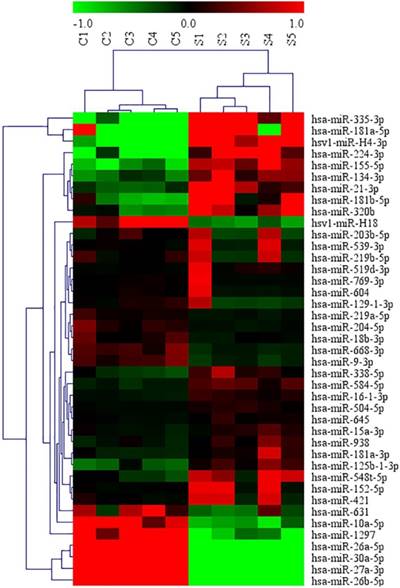
MiRNA microarray was conducted in 5 middle ear cholesteatoma tissues and 5 normal skin tissues. Compared with normal skin, totally 44 upregulated (miRNA-21-3p, miRNA-584-5p, miRNA-16-1-3p, etc) and 175 downregulated (miRNA-10a-5p, miRNA-152-5p, miRNA-203b-5p, etc) miRNAs with 2 fold change were identified in cholesteatoma tissues with statistical significance (P<0.05). The differential expression of miRNAs were presented in a volcano plot (Figure 1), and then subjected to a scatter plot analysis (Figure 2) and hierarchical clustering analysis (Figure 3). These discoveries revealed that middle ear cholesteatoma tissues were significantly different from normal skin, and these differently expressed miRNAs may play a vital role in the etiopathogenesis of middle ear cholesteatoma.
Validation for differentially expressed miRNAs using qRT-PCR
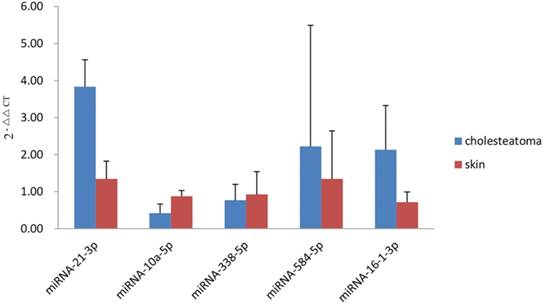
To validate the miRNAs microarray results, qRT-PCR analysis of 15 cholesteatoma samples and 10 normal skin samples was conducted for part of differentially expressed miRNAs, including 4 upregulated miRNAs (miRNA-21-3p, miRNA-16-1-3p, miRNA-584-5p, miRNA-338-5p) and 1 downregulated miRNAs (miRNA-10a-5p). The qRT-PCR validation was accordance with the microarray results partly: miRNA-21-3p and miRNA-16-1-3p expressed significantly higher (P=0.000 and P=0.000, respectively) while miRNA-10a-5p exhibited an obviously decreased expression (P=0.000) in middle ear cholesteatoma tissues when compared with normal skin. However, an increased expression of miRNA-584-5p (P=0.431) and decreased expression of miRNA-338-5p (P=0.465) were demonstrated with no statistical significance (Figure 4).
Volcano plot of microarray data. Plot showed differences between middle ear cholesteatoma and normal skin. Differentially expressed miRNAs with a fold change ≥ 2 and P<0.05 between the two groups were showed in red.
Differential expression of miRNAs between middle ear cholesteatoma and normal skin in a scatter plot analysis
Differential expression of partial miRNAs between middle ear cholesteatoma and normal skin in a hierarchical clustering analysis. The red color indicated high expression level, green indicated low expression level, and black indicated mean expression level.
qRT-PCR analysis of differentially expressed miRNAs. miRNA-21-3p, miRNA-16-1-3p, and miRNA-584-5p were upregulated in cholesteatoma, while miRNA-10a-5p and miRNA-338-5p were downregulated.
Venn diagram of target genes for candidate miRNAs. TargetScan and miRDB dababases were applied to predict target genes. The number in the intersection indicated there were 1058 target genes in total predicted by using these two databases together.
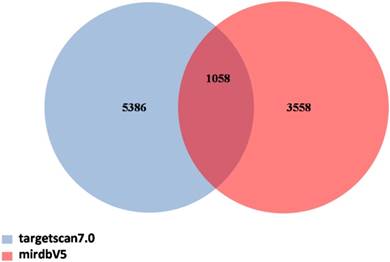
Prediction of target genes for candidate miRNAs
Target genes prediction was performed with TargetScan and miRDB databases for 19 candidate miRNAs, according to their expression levels and their previous correlations with diseases. As shown in Figure 5, a total of 1058 target genes were indentified to be regulated by 11 upregulated miRNAs (miRNA-21-3p, miRNA-584-5p, miRNA-320b, miRNA-181a-3p, miRNA-181a-5p, miRNA-16-1-3p, miRNA-181b-5p, miRNA-338-5p, miRNA-335-3p, miRNA-155-5p, and miRNA-224-3p) and 8 downregulated miRNAs (miRNA-10a-5p, miRNA-30a-5p, miRNA-203b-5p, miRNA-1297, miRNA-152-5p, miRNA-539-3p, miRNA-9-3p, and miRNA-769-3p).
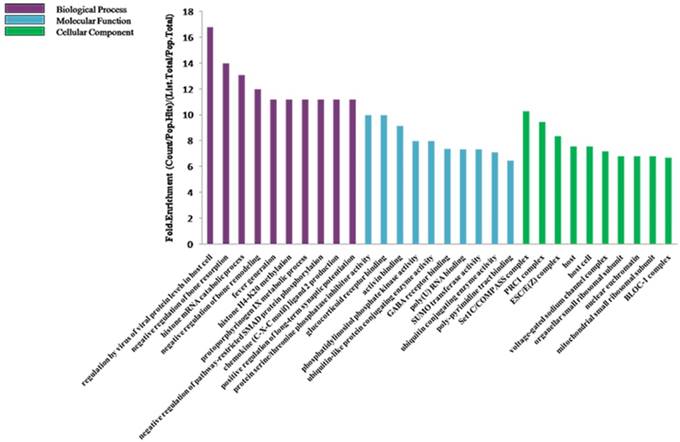
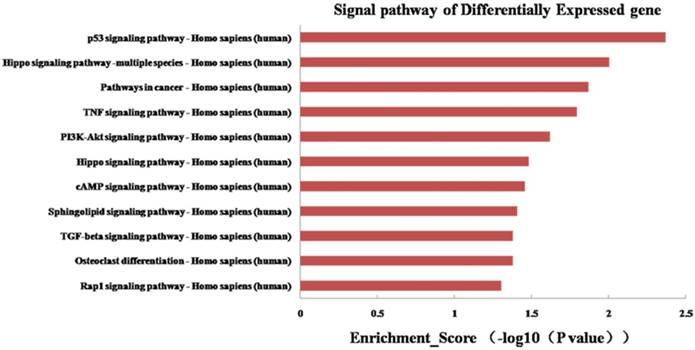
To further explore the potential functions of 19 differentially expressed miRNAs underlying biological processes in the etiopathogenesis of middle ear cholesteatoma, GO classification and pathway analysis were performed on presumptive target genes of these 19 miRNAs. The GO classification was grouped into 3 categories: molecular function, biological process and cellular component. In our study, regarding on molecular function, target genes were classified into 153 categories, playing roles in protein serine/threonine phosphatase inhibitor activity, glucocorticoid receptor binding, activin binding, and so on. While based on biological process, these target genes totally participated in 831 biological processes, including regulation by virus of viral protein levels in host cell, negative regulation of bone resorption, histone mRNA catabolic process, and so on. Moreover, categories on cellular component revealed that most target genes act as a portion of several complexes, such as Set1C/COMPASS complex, PRC1 complex, ESC/E(Z) complex, and so on (Figure 6). Additionally, the KEGG pathway analysis displayed that the putative target genes were involved in 39 signaling pathways totally, including p53 signaling pathway, PI3K-Akt signaling pathway, osteoclast differentiation, and so on (Figure 7).
GO classification of genes targeted by the differentially expressed miRNAs. The predicted target genes were analysed in biological process, molecular function, and cellular component.
KEGG pathway analysis of genes targeted by the differentially expressed miRNAs.
Discussion
MiRNAs are endogenous non-coding small RNAs (containing about 20-25 nucleotides) found in animals, plants and viruses which play essential roles in post-transcriptional regulation of gene expression through targeting mRNAs [11]. In the current study, the miRNA microarray analysis was adopted to investigate the miRNA expression profiling in middle ear cholesteatoma. We totally identified 44 upregulated and 175 downregulated miRNAs in cholesteatoma tissues with statistical significance compared with normal skin. All these differentially expressed miRNAs are likely to significantly impact on gene expressions of middle ear cholesteatoma. Furthermore, to verify the microarray discoveries, qRT-PCR was conducted to detect the expression of 5 selected miRNAs, including 4 upregulated miRNAs (miRNA-21-3p, miRNA-16-1-3p, miRNA-584-5p, miRNA-338-5p) and 1 downregulated miRNAs (miRNA-10a-5p). The expression of miRNA-21-3p, miRNA-16-1-3p and miRNA-10a-5p was accordance with the microarray data, indicating that these three miRNAs might make great contribution to the etiopathogenesis of middle ear cholesteatoma. The discrepancy of expression of miRNA-584-5p and miRNA-338-5p between qRT-PCR validation and microarray analysis might result from the false positive results from microarray analysis. Moreover, a validation with a large sample size will be conducted in our further study.
In fact, researches on miRNA functions in the etiopathogenesis of middle ear cholesteatoma are rare so far, and the current researches mainly focus on miRNA-21, miRNA let-7a, and miRNA-802. Friedland et al. and Chen et al. [12, 13] detected the potential role of miRNA-21 in regulating growth and proliferation in acquired cholesteatoma. They postulated that the up-regulation of miRNA-21 could lead to suppression of PTEN and PDCD4, resulting in cholesteatoma keratinocytes proliferation and invasion. Similarly, Chen et al. [14] demonstrated that miRNA-21 was able to promote the proliferation and invasion of cholesteatoma keratinocytes. Consistent with the previous studies, the microarray data in our study showed that miRNA-21-3p was significantly upregulated with an average 7.03-fold change in cholesteatoma tissues compared with normal skin. Additionally, Chen et al. [13] found that miRNA let-7a was highly expressed in cholesteatoma tissues compared with normal skin especially in paediatric patients. However, the microarray data in our study demonstrated that the expression of miRNA-let-7a-3p and miRNA-let-7a-5p showed no statistical significance between cholesteatoma tissues and normal skin.
Middle ear cholesteatoma is characterized with bone resorption of the ossicular chain and bony labyrinth, which may result in hearing loss, vestibular dysfunction, facial paralysis, labyrinthine fistulae and intracranial complications. So far, the exact mechanism of bone resorption in cholesteatoma still remains unexplained. In the current study, GO classification revealed that differentially expressed miRNAs participated in negative regulation of bone resorption and remodeling biological processes, shedding light on the potential of miRNAs to be regarded as therapeutic targets in cholesteatoma. Recently, a majority of researchers hold the view that the osteoclast acts as a terminal function cell in the bone resorption process. The activation and maturity of osteoclast play a key role in the bone resorption and remodeling of middle ear cholesteatoma [15]. The KEGG pathway analysis in our study exhibited an involvement of osteoclast differentiation signaling pathway, indicating that miRNAs possessed the possibility of regulating bone resorption in cholesteatoma. Moreover, the regulatory function of miRNAs has been confirmed in some osteolytic diseases. Previous discoveries showed that osteoclasts were able to secrete miRNAs that modulated osteolytic tumor growth [16]. For instance, osteoclasts could secrete miRNA-21 that promoted tumor cell proliferation [17] and miRNA-378 that enhanced tumor growth, angiogenesis and cell survival [18]. Also, miRNA-338-3p was demonstrated to repress glucocorticoid-induced osteoclast differentiation and bone resorption through targeting on receptor activator for nuclear factor-κB ligand (RANKL) [19].
Of the miRNAs validated in this study, miRNA-16-1-3p and miRNA-10a-5p have not been investigated in cholesteatoma yet, they have already been demonstrated to play crucial roles in other diseases such as cancers. Recently, Huang et al. [20] found that miRNA-10a was markedly lower in extranodal natural killer (NK)/T-cell lymphoma (ENKTCL) tissues than in normal NK cells, indicating that miRNA-10a might be involved in the development of ENKTCL. Target genes prediction showed that PTEN is the most important target gene of miRNA-16-1-3p, and phosphoinositide-3-kinase catalytic subunit α (PIK3CA) is the most important target gene of miRNA-10a-5p. As far as we known, PTEN can dephosphorylate PIP3 at the 3' position of the inositol ring, finally leading to PI3K-Akt signaling pathway inactivation, and PIK3CA can activate the PI3K-Akt signaling pathway. In our previous study, we have demonstrated that PI3K-Akt signaling pathway is active in cholesteatoma and may play a crucial role in cholesteatoma epithelial hyper-proliferation [8]. In this study, KEGG pathway analysis also displayed that the differentially expressed miRNAs were involved in the PI3K/Akt signaling pathway, disclosing a possibility that miRNA-16-1-3p and miRNA-10a-5p might induce the hyper-proliferation of cholesteatoma tissues and regulate cholesteatoma formation through PI3K-Akt signaling pathway. The exact underlying mechanisms for these two miRNAs in cholesteatoma should be elucidated in keratinocyte cell culture model in the future study.
Conclusions
In summary, 44 upregulated and 175 downregulated miRNAs were identified in cholesteatoma tissues in the current study. The qRT-PCR validation for differentially expressed miRNAs demonstrated that miRNA-21-3p and miRNA-16-1-3p were expressed significantly higher while miRNA-10a-5p exhibited an obviously decreased expression in cholesteatoma tissues when compared with normal skin. The GO and KEGG pathway analyses provide clues that these differentially expressed miRNAs may play essential roles in the etiopathogenesis of middle ear cholesteatoma, including cell proliferation, apoptosis, cell cycle, differentiation, bone resorption and remodeling process, and so on. Further researches should be conducted to investigate the exact mechanism of these differentially expressed miRNAs in the pathogenesis of cholesteatoma. Targeting on miRNAs may provide a new strategy for cholesteatoma therapy in the future.
Abbreviations
Akt: protein kinase B; CIP: calf intestine phosphatase; ENKTCL: extranodal natural killer (NK)/T-cell lymphoma; GO: Gene Ontology; KEGG: Kyoto Encyclopedia Genes and Genomes; LNA: locked nucleic acid; MAPK: mitogen-activated protein kinase; mRNA: messenger RNA; miRNA: microRNA; NK: natural killer; PDCD: programmed cell death; PI3K: phosphatidylinositol 3-kinase; PIK3CA: phosphoinositide-3-kinase catalytic subunit α; qRT-PCR: quantitative real-time polymerase chain reaction; RANKL: receptor activator for nuclear factor-κB ligand.
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant No. 81400457, Grant No.81570928), and Natural Science Foundation of Hunan Province, China (Grant No. 2017JJ3441).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
1. Kemppainen HO, Puhakka HJ, Laippala PJ. et al. Epidemiology and aetiology of middle ear cholesteatoma. Acta Otolaryngol. 1999;119:568-572
2. Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475-478
3. Xu N, Brodin P, Wei T. et al. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol. 2011;131:1521-1529
4. Long L, Yu P, Liu Y. et al. Upregulated microRNA-155 expression in peripheral blood mononuclear cells and fibroblast-like synoviocytes in rheumatoid arthritis. Clin Dev Immunol. 2013;2013:296139
5. Mannucci C, Casciaro M, Minciullo PL. et al. Involvement of microRNAs in skin disorders: a literature review. Allergy Asthma Proc. 2017;38:9-15
6. Zhang LY, Ho-Fun Lee V, Wong AM. et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454-463
7. Karatas OF, Oner M, Abay A. et al. MicroRNAs in human tongue squamous cell carcinoma: From pathogenesis to therapeutic implications. Oral Oncol. 2017;67:124-130
8. Liu W, Ren H, Ren J. et al. The role of EGFR/PI3K/Akt/cyclinD1 signaling pathway in acquired middle ear cholesteatoma. Mediators Inflamm. 2013;2013:651207
9. Chen X, Li X, Qin Z. MicroRNA-21 promotes the proliferation and invasion of cholesteatoma keratinocytes. Acta Otolaryngol. 2016;136:1261-1266
10. Li N, Qin ZB. Inflammation-induced miR-802 promotes cell proliferation in cholesteatoma. Biotechnol Lett. 2014;36:1753-1759
11. Zhang B, Wang W, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279-289
12. Friedland DR, Eernisse R, Erbe C. et al. Cholesteatoma growth and proliferation: posttranscriptional regulation by microRNA-21. Otol Neurotol. 2009;30:998-1005
13. Chen X, Qin Z. Post-transcriptional regulation by microRNA-21 and let-7a microRNA in paediatric cholesteatoma. J Int Med Res. 2011;39:2110-2118
14. Chen X, Li X, Qin Z. MicroRNA-21 promotes the proliferation and invasion of cholesteatoma keratinocytes. Acta Otolaryngol. 2016;136:1261-1266
15. Hamzei M, Ventriglia G, Hagnia M. et al. Osteoclast stimulating and differentiating factors in human cholesteatoma. Laryngoscope. 2003;113:436-442
16. Ell B, Mercatali L, Ibrahim T. et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell. 2013;24:542-556
17. O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201
18. Lee DY, Deng Z, Wang CH. et al. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350-20355
19. Zhang XH, Geng GL, Su B. et al. MicroRNA-338-3p inhibits glucocorticoid-induced osteoclast formation through RANKL targeting. Genet Mol Res. 2016;15:doi 10.4238/gmr.15037674
20. Huang H, Fan L, Zhan R. et al. Expression of microRNA-10a, microRNA-342-3p and their predicted target gene TIAM1 in extranodal NK/T-cell lymphoma, nasal type. Oncol Lett. 2016;11:345-351
Author contact
![]() Corresponding author: Wei Liu Address: Department of Otolaryngology-Head and Neck Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China. Telephone: +8613875837130 Fax number: 0731-85295235 E-mail address: liuwei007edu.cn
Corresponding author: Wei Liu Address: Department of Otolaryngology-Head and Neck Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China. Telephone: +8613875837130 Fax number: 0731-85295235 E-mail address: liuwei007edu.cn

 Global reach, higher impact
Global reach, higher impact