Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(13):1480-1485. doi:10.7150/ijms.27333 This issue Cite
Research Paper
Influence of DNA methylation on the expression of OPG/RANKL in primary osteoporosis
1. The First Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China.
2. The Second Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
3. Department of Orthopedics, People's Hospital of Sanshui, Foshan, China.
4. Departments of Diagnostics of Traditional Chinese Medicine, Guangzhou University of Traditional Chinese Medicine, Guangzhou, China.
5. Department of Orthopaedics & Traumatology, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong SAR, China.
6. The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Lingnan Medical Research Center of Guangzhou University of Chinese Medicine, Guangzhou, China.
*Peng Wang and Yanming Cao contributed equally to this work.
Received 2018-5-18; Accepted 2018-8-27; Published 2018-10-3
Abstract
Purpose: A key factor in regulating bone absorption is the proportion of RANKL/OPG. Although many reports showing diverse transcription factors or epigenetic modification could be responsible for regulating RANKL&OPG ratio, there is still little exploration on promoter methylation status of both genes in osteoporotic bone tissues. Our aim is to investigate the changes of methylation in CpG island of these genes' promoters in patients with primary osteoporosis.
Methods: The diagnosis of osteoporosis was based on the results of dual energy X-ray absorptiometry measurements. All femoral bone tissues were separated in surgeries. After extracting total RNA, we checked the relative expression levels of OPG and RANKL by quantitative real time PCR. The genomic DNA of Non-OPF (Non-osteoporotic fracture bone tissues) & OPF (osteoporotic fracture bone tissues) were treated by bisulfite modification, and methylation status of CpG sites in the CpG island of OPG/RANKL promoters were determined by DNA sequencing.
Results: RANKL expression in the OPF group was significantly higher than that in Non-OPF group, and the CpG methylation status in RANKL gene promoter was significantly lower. However, for OPG, lower gene expression level and higher methylation degree were found in the OPF group.
Conclusion: Our study demonstrated that DNA methylation influenced the transcriptional expression of OPG and RANKL, which probably take on a “main switch” role in pathogenesis of primary osteoporosis.
Keywords: DNA methylation, primary osteoporosis, bone remodeling, OPG/RANKL
Introduction
Imbalance of bone remodeling is one of important causes leading to osteoporosis. It is a continuous process, including bone matrix formation induced mainly by osteoblasts and its resorption via osteoclasts [1]. RANKL (Receptor Activator of Nuclear Factor-kappa B Ligand, the product of the TNFSF11 gene), and its receptor OPG (Osteoprotegerin, encoded by the TNFRSF11B gene), play an essential role in regulating the balance [2]. RANKL can bind up with RANK (Receptor Activator of Nuclear Factor-kappa B), then the compounds are able to promote the maturation, differentiation and activation of osteoclasts. Whereas, this binding level can be weakened down by OPG that is secreted by the osteoblasts on the level of cell envelope [3]. Summarily, OPG can inhibit bone resorption through combination with RANKL which will induce osteoporosis when it was over expressed in transgenic mice [4].
If the gene expression mechanisms of RANKL-OPG system can be identified clearly at the transcriptional level, it will benefit potential therapies for osteoporosis [5]. It is known that Fibroblast growth factor 21, P-GE2, Transforming growth factor-beta and Estrogen promote OPG expression. Also, expression of RANKL is regulated by various activity factors, like PAPSS2, IL-3 and Tumor Necrosis Factor Alpha, as well as by the Wnt ligands [6].
DNA methylation happens in the genomes of diverse organisms, and it belongs to one of the epigenetic mechanisms, including RNA interference, histone modification, chromatin remodeling. Epigenetic mechanism means the DNA sequence does not change, but the gene expression can be altered, and this transform can be transmitted steadily during development and cell proliferation [7]. In multicellular eukaryote, DNA methylation seems to be associated with a repressed chromatin state and inhibition of gene expression [8]. DNA methylation precisely acts on gene transcription to prevent gene expression, which mainly happens in gene promoter regions. There are two basic models: one is the association between DNA binding factors and their complementary DNA recognition sequences is inhibited by the methylated cytosine bases directly [9]; the other one is that MBPs (Methyl-CpG-binding proteins) recognize methylated DNA sequence and recruit co-repressors to silence gene expression [10, 11].
The role of DNA methylation of OPG-RANKL system was reported in murine models and cancer cells, even in human bone recently [5]. Whereas, there still lacks specific reports on the methylation status of RANKL-OPG gene promoter regions in osteoporosis. Therefore, we aimed to explore the changes of DNA methylation of OPG-RANKL system in patients with primary osteoporosis.
Methods
Ethical Statement
16 primary osteoporosis patients with femoral neck/trochanter fractures (OPF, case group), and 16 patients with traumatic fractures (non-OPF, control group) were recruited in the Second Affiliated Hospital of Guangzhou Medical University. The Bone Mineral Density (BMD) of the axial bone was measured by dual-energy X-ray absorptiometry (DEXA). Bone tissue samples were obtained during internal fixation surgery. The study was approved by the local ethics board and patient gave informed written consent. Patients with secondary osteoporosis, hip osteoarthritis, and pathological fracture due to non-osteoporosis were excluded. The details of the patient's related information (Non-OPF & OPF) were exhibited in Supplementary Table 1.
RNA isolation, reverse transcription (RT) reaction and quantitative real-time PCR
We adopted the Trizol® Reagent kit (Thermo Fisher Scientific), following the manufacturer's protocol (Invitrogen, 15596-018), to purify total RNA, which was treated with DNase I to remove the DNA contamination. Reverse reaction was performed with a PrimeScriPtTM RT Master Mix (TaKaRa, RR036A). Quantitative real-time PCR was performed by SYBR® Premix Ex TaqII (TaKaRa, Code No. RR820A) with CFX96TM Real-Time PCR Detection System (Bio-Rad, Munchen, Germany). The sequences of the primers were exhibited in Supplementary Table 2. The relative level of RANKL and OPG gene mRNA were determined by calculating the values of 2-∆CT method.
DNA isolation and bisulfite treatment
Genomic DNA isolation from tissue samples (100mg/case) was performed using the phenol/chloroform method. Bisulfite modification was done as described [12], including DNA bisulfite conversion solution (6N NaOH, 4.04M NaHSO3, 10 mM Hydroquinone), which made the genomic DNA of bone tissues become bisulfite-converted DNA. Then all the samples were re-suspended in 40μl EB and stored at -80℃ until use in subsequent experiments.
Bisulfite sequencing
Bisulfite-converted genomic DNA was amplified by PCR. All PCRs were done using KAPA2G™ Fast HotStart DNA Polymerase. Genomic DNA was amplified by PCR sequences of primers used for the BSP analysis were shown in Supplementary Table 3. PCR products were authenticated through running on 1.5% agarose gels, and the product bands were dissolved using TaKaRa MiniBEST Agarose Gel DNA Extraction Kit following the manufacturer's instructions (TaKaRa). Then purified bands went to T4 ligation and cloned using pMDTM 19-T Vector Cloning Kit following the manufacturer's instructions (TaKaRa). Colonies were selected and grown overnight in LB (Luria-Bertani) medium containing ampicillin (100μg/ml) with shaking at 37℃. Plasmids were sequenced using the M13 universal reverse primer (BGI).
Statistics
Data are presented as mean ± standard deviation (SD). Comparison of two independent groups was done using Mann-Whitney U test. Statistical analysis was performed using the SPSS 20.0. A value of P < 0.05 was considered statistically significant.
Results
Differential expression of RANKL and OPG in patients with Non-OPF and OPF
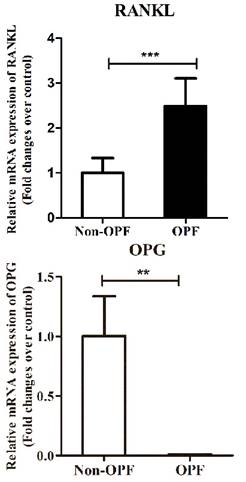
RANKL and OPG mRNA levels were detected in all the bone tissues studied. The real time PCR results showed that RNA level of RANKL in patients with osteoporosis was significantly up-regulated in comparison to the control group (Fig. 1a). As expected, the OPG mRNA expression level in Non-OPF group was significantly higher (Fig. 1b).
Epigenetic regulation of RANKL and OPG in two groups
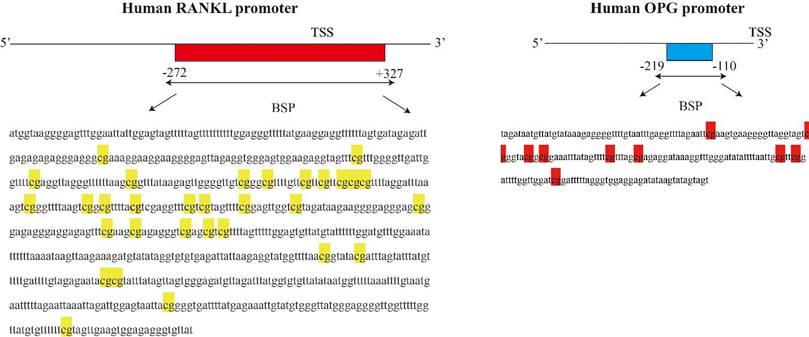
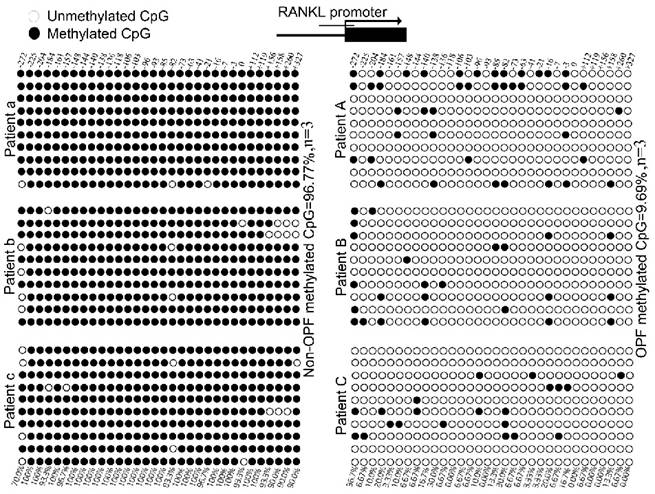
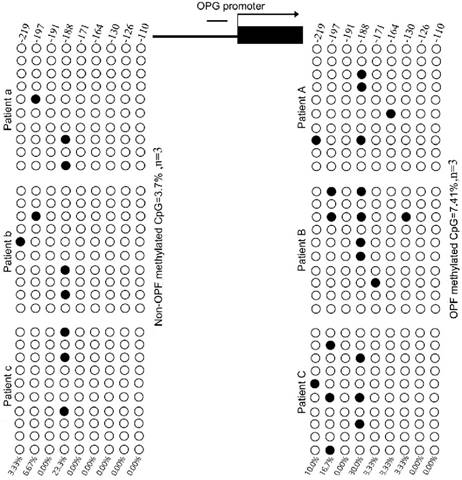
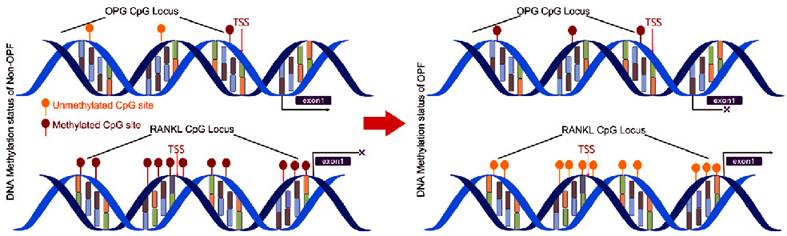
We used MethPrimer Software to analyze a length of the CpG-rich region around the transcription start site (TSS): 32 CpG sites in the RANKL gene promoter and 9 in OPG (Fig. 2). After bisulfite treatment of DNA obtained from bone tissues, we calculated the percentage of methylated CpG sites (percent CpG methylation) among the total 32 CpG sites in RANKL promoter and 9 CpG loci in OPG promoter, respectively. We found that RANKL promoter is hypermethylated whereas OPG promoter is hypomethylated in Non-OPF (96.69% and 9.69% CpG methylation). In OPF, the methylation status of RANKL and OPG promoter was opposite in comparison to Non-OPF (Fig. 3&4). In summary, at least partially, we could conclude that DNA methylation could be involved in the regulation of osteoporosis-related genes expression. Increased demethylation of CpG island in RANKL promoter and methylation in OPG promoter lead to the increase of RANKL and decrease of OPG, which contributes to the occurrence of osteoporosis and occurrence of bone fracture. The DNA methylation involved in regulating expression of RANKL/OPG are schematically illustrated in Fig. 5.
Expression level of OPG and RANKL in bone tissue samples. (a & b) Total RNA were extracted from bone tissues of patients with OPF or Non-OPF. GADPH was used as an internal control. The data are expressed as mean ± SD (n = 16).
Schematic figure indicates 32 CpG sites in CpG island of the RANKL promoter and 9 CpG sites for OPG around the transcription start site.
The methylation degree of CpG island in RANKL promoter in bone tissues of OPF and Non-OPF. DNA methylation status of RANKL promoters in three Non-OPF (patient a, b and c) and three OPF (patient A, B and C) samples using sodium bisulfite sequencing. Each PCR product was subcloned and subjected to nucleotide sequencing analysis. Sequenced clones were depicted by filled (methylated) and open (unmethylated) circles for each CpG site.
Discussion
The investigation about relationship between methylation level of CpG-rich region and gene expression has been emerging constantly. There is increasing experimental evidence on the potential therapeutic effects of DNA methylation on neoplastic disorders [13], and on metabolic bone disease [14]. Nevertheless, little is known about the DNA methylation status of RANKL and OPG in osteoporosis.
Evolving evidence suggests that DNA methylation may be involved in age related diseases and bone biology [15]. Our previous studies have found that DNA methylation plays an essential role in determine fate of mesenchymal stem cells [16, 17]. In this study, we explored whether RANKL and OPG gene expression was influenced by the epigenetic modulation. As mentioned in the introduction, DNA methylation is linked with transcriptional silencing of associated genes [18]. For example, Delgado-Calle J have demonstrated increased sclerostin expression is associated with hypomethylation of the proximal promoter and first exon of SOST, whereas hypermethylation has the opposite effect [19]. Furthermore, Reppe et al found four genes (MEPE, SOST, WIF1, and DKK1) whose methylation state was highly correlated with transcription level through using the expanded Illumina Infinium 450-k platform in bone biopsies [20].
In the present study, we demonstrated that the expression trend of both genes was opposite in OPF and Non-OPF groups. We found that RANKL mRNA level was higher in OPF group. While, for OPG, its expression level was significantly lower in the OPF group. Apparently, the opposite tendency between DNA methylation and these two genes were revealed, and we concluded that cytosine methylation in CpG-rich region of RANKL and OPG promoter indeed regulated their expression. We found that 32 CpG sites in the CpG island of RANKL promoter was highly demethylated in the OPF group compared with the Non-OPF group. For OPG gene, its promoter was hypomethylated in both groups, but the level of methylation in OPF group was much higher. These results demonstrated that DNA methylation could inhibit RANKL and OPG expression, which was consistent with the quantitative real time PCR data. This strongly suggested the loss balance of OPG-RANKL system may induce osteoporosis through DNA methylation.
OPG-RANKL expression ratio always plays an important role in bone turnover status at the level of cellular and molecular biology. Many researches have defined the role of RANKL in bone remodeling and provided evidence for the therapeutic potential of RANKL inhibition in conditions of bone loss. In the clinical studies, denosumab, one RANKL inhibitor, plays an important role in post-menopausal women with osteoporosis [21]. Also, loss of OPG gene in rat and human genomic DNA can lead to severe osteoporosis [22]. Other factors, such as hormones, cytokines, histones or microRNAs may play the role of negative feedback in tissues/cells with low methylation region. Penolazzi found that the methylation of the transcription initiation site of exon F of ERα gene was closely related to the growth and differentiation activity of osteoblasts [23]. And histone methyltransferase DOT1L acts as a negative regulator in RANKL-induced osteoclasts, which may be regulated by H3K79me directly or indirectly [24].
In a word, the methylation status in CpG island of OPG and RANKL genes does have influenced their expression level in patients with OPF and Non-OPF. The lower methylation degree in the promoter region of RANKL and higher methylation rate in the promoter region of OPG indicate that pathogenesis of osteoporosis may be attributed to the degree of OPG-RANKL methylation, at least, remaining speculative at this moment.
The methylation degree of CpG island in OPG promoter in bone tissues of OPF and Non-OPF. OPG studies were performed in the same samples from Non-OPF (patient a, b and c) and three OPF (patient A, B and C) groups. 30 cloned plasmids were sequenced to locate the methylated CpG sites in all 9 loci, respectively.
The principle summary of the conversion process induced by the DNA methylation of OPG&RANKL gene promoter between Non-OPF and OPF.
Supplementary Material
Supplementary tables.
Acknowledgements
The work was partially supported by grants from National Natural Science Foundation of China (NSFC No. 81774339, 81574002) to Haibin Wang, Yanming Cao, and Guangdong Provincial Science and Technology Project (No. 2017A050506046, 2014A020221055, 2016A030313649), and We also thank Ms. Jing Zhang from Yuebin Medical Research Lab for providing technique support for this study.
Author Contribution
LLX, HBW and PW designed the study. YMC performed in fracture surgery and got samples. PW, DXZ, YML, DW and BW conducted experiments, analyzed data. PW wrote the manuscript. LLX, HBW, WH and GL revised and edited the manuscript. All authors have read and approved the final submitted manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zebaze R M, Ghasem-Zadeh A, Bohte A. et al. Intracortical remodelling and porosity in the distal radius and post-mortem femurs of women: a cross-sectional study. LANCET. 2010;375:1729-1736
2. Marie P, Halbout P. [OPG/RANKL: role and therapeutic target in osteoporosis]. Med Sci (Paris). 2008;24:105-110
3. Piedra M, Garcia-Unzueta M T, Berja A. et al. "Single nucleotide polymorphisms of the OPG/RANKL system genes in primary hyperparathyroidism and their relationship with bone mineral density". BMC MED GENET. 2011;12:168
4. Simonet W S, Lacey D L, Dunstan C R. et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. CELL. 1997;89:309-319
5. Delgado-Calle J, Sanudo C, Fernandez A F. et al. Role of DNA methylation in the regulation of the RANKL-OPG system in human bone. EPIGENETICS-US. 2012;7:83-91
6. Glass D N, Bialek P, Ahn J D. et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. DEV CELL. 2005;8:751-764
7. Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? EPIGENOMICS-UK. 2014;6:73-88
8. Bird A P, Wolffe A P. Methylation-induced repression-belts, braces, and chromatin. CELL. 1999;99:451-454
9. Klose R J, Bird A P. Genomic DNA methylation: the mark and its mediators. TRENDS BIOCHEM SCI. 2006;31:89-97
10. Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. CELL. 1997;88:471-481
11. Nan X, Ng H H, Johnson C A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. NATURE. 1998;393:386-389
12. Zinn R L, Pruitt K, Eguchi S. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. CANCER RES. 2007;67:194-201
13. Poloni A, Maurizi G, Leoni P. Human dedifferentiated adipocytes show similar properties to bone marrow-derived mesenchymal stem cells. STEM CELLS. 2012;30:965-974
14. Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-1159
15. Reppe S, Lien T G, Hsu Y H. Distinct DNA methylation profiles in bone and blood of osteoporotic and healthy postmenopausal women. EPIGENETICS-US. 2017;12:674-687
16. Xu L, Liu Y, Sun Y. Tissue source determines the differentiation potentials of mesenchymal stem cells: a comparative study of human mesenchymal stem cells from bone marrow and adipose tissue. STEM CELL RES THER. 2017;8:275
17. Rui Y, Xu L, Chen R. Epigenetic memory gained by priming with osteogenic induction medium improves osteogenesis and other properties of mesenchymal stem cells. Sci Rep. 2015;5:11056
18. Hsieh C L. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. MOL CELL BIOL. 1997;17:5897-5904
19. Delgado-Calle J, Sanudo C, Bolado A. et al. DNA methylation contributes to the regulation of sclerostin expression in human osteocytes. J Bone Miner Res. 2012;27:926-937
20. Reppe S, Lien T G, Hsu Y H. et al. Distinct DNA methylation profiles in bone and blood of osteoporotic and healthy postmenopausal women. Epigenetics-Us. 2017;12:674-687
21. Dempster D W, Lambing C L, Kostenuik P J. Role of RANK ligand and denosumab, a targeted RANK ligand inhibitor, in bone health and osteoporosis: a review of preclinical and clinical data. CLIN THER. 2012;34:521-536
22. Whyte M P, Obrecht S E, Finnegan P M. Osteoprotegerin deficiency and juvenile Paget's disease. N Engl J Med. 2002;347:175-184
23. Penolazzi L, Lambertini E, Giordano S. Methylation analysis of the promoter F of estrogen receptor alpha gene: effects on the level of transcription on human osteoblastic cells. J Steroid Biochem Mol Biol. 2004;91:1-9
24. Gao Y, Ge W. The histone methyltransferase DOT1L inhibits osteoclastogenesis and protects against osteoporosis. CELL DEATH DIS. 2018;9:33
Author contact
![]() Corresponding authors: Liangliang Xu, Ph.D., The First Affiliated Hospital of Guangzhou University of Chinese Medicine, The First Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China. Tel: (86) 20-36585574, E-mail: xull-2016edu.cn; or Dr. Haibin Wang Guangzhou University of Chinese Medicine, Jichang Road, Baiyun District, Guangzhou, China. Tel: (86) 20-84237212, E-mail: hipkneecom.
Corresponding authors: Liangliang Xu, Ph.D., The First Affiliated Hospital of Guangzhou University of Chinese Medicine, The First Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China. Tel: (86) 20-36585574, E-mail: xull-2016edu.cn; or Dr. Haibin Wang Guangzhou University of Chinese Medicine, Jichang Road, Baiyun District, Guangzhou, China. Tel: (86) 20-84237212, E-mail: hipkneecom.

 Global reach, higher impact
Global reach, higher impact