Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(9):900-906. doi:10.7150/ijms.24346 This issue Cite
Research Paper
Assessment of a combination of Serum Proteins as potential biomarkers to clinically predict Schizophrenia
1. Department of Laboratory Medicine, Hunan Provincial People's Hospital, The first affiliated hospital of Hunan Normal University, Changsha, 410005, Hunan, China
2. Department of Biochemistry and Molecular Biology, Hunan University of Chinese Medicine, Changsha 410208, Hunan, China
3. Department of Laboratory Medicine, The Second Xiangya Hospital, Central South University, Changsha 410011, Hunan, China
4. Department of Psychiatry, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China; Mental Health Institute of Central South University & Hunan Key Laboratory of Psychiatry and Mental Health, Changsha, China; China National Clinical Research Center on Mental Disorders (Xiangya) & China National Technology Institute on Mental Disorders, China.
Received 2017-12-13; Accepted 2018-4-27; Published 2018-6-4
Abstract
Schizophrenia (SZ) is a devastating psychiatric disorder. Validation of potential serum biomarkers during first-episode psychosis (FEP) is especially helpful to understand the onset and prognosis of this disorder. To address this question, we examined multiple blood biomarkers and assessed the efficacy to diagnose SZ. The expression levels of Neuregulin1 (NRG1), ErbB4, brain-derived neurotrophic factor (BDNF), DNA methyltransferases 1 (DNMT1) and ten-eleven translocation 1 (TET1) proteins in peripheral blood of 53 FEP patients and 57 healthy controls were determined by enzyme-linked immunosorbent assay (ELISA). Multivariable logistic regression including biomarker concentration as covariates was used to predict SZ. Differentiating performance of these five serum protein levels was analyzed by Receiver Operating Characteristic (ROC) curve analysis. We found that patients with SZ present a higher concentration of DNMT1, and TET1 in peripheral blood, but a lower concentration of NRG1, ErbB4 and BDNF than controls. Multivariable logistic regression showed that ErbB4, BDNF and TET1 were independent predictors of SZ, and when combined, provided high diagnostic accuracy for SZ. Together, our findings highlight that altered expression of NRG1, ErbB4, BDNF, DNMT1 and TET1 are involved in schizophrenia development and they may serve as potential biomarkers for the diagnosis of the schizophrenia. Therefore, our study provides evidence that combination of ErbB4, BDNF and TET1 biomarkers could greatly improve the diagnostic performance.
Keywords: Schizophrenia, biomarker, NRG1, ErbB4, BDNF, DNMT1, TET1
Introduction
Schizophrenia (SZ) is one of the devastating psychiatric disorders and affects more than 1% of global population. The precise pathophysiology and etiology of this disorder remains unclear and its diagnosis largely depends on interview-based subjective assessments of self-reported symptoms. Although extensive research has been carried out, no reliable biomarkers are available for the diagnosis and prognosis of SZ which make it urgent to identify biomarkers for addressing these unmet clinical needs.
Recent evidence suggests that altered intracellular signaling may contribute to the pathophysiology of schizophrenia and could be used to diagnose schizophrenia. For example, dysfunctional neuregulin1 (NRG1) and its receptor ErbB4 have been confirmed in postmortem brain tissues of SZ patients[1-3]. NRG1 is a member of the group of proteins containing epidermal growth factor (EGF)-like domains which transmit signals by activating membrane-associated tyrosine kinases[4], especially the ErbB4 receptor kinases in the central nervous system (CNS). In addition, epigenetic abnormalities, especially in DNA-methylation / demethylation network pathways, have also been identified in postmortem brains of SZ patients [5, 6]. DNA methyltransferase 1 (DNMT1) and Ten-Eleven Translocation 1 (TET1), two important component enzymes in DNA-methylation/demethylation network, were abnormally increased in SZ postmortem brains and peripheral blood lymphocytes [5-7]. Besides, reduced expression of brain-derived neurotrophic factor (BDNF), a member of nerve growth factor family, was related to the increase of 5-methyl cytosine at the BDNF promoter in the SZ patient brains, and aberrant expression of BDNF gene is implicated in several mental illness by lasting epigenetic influence[8].
Because psychiatric disorders have long been considered as brain disorders, few studies focused on the resultant systemic changes, especially the changes of serum proteins which are easily accessible in clinics. Previous studies have revealed serum protein changes in SZ patients, however, the conclusions were not consistent [9-11]. Given the polygenic nature of SZ, it is widely accepted that a comprehensive multi-marker profile may have a higher predictive power in terms of sensitivity and specificity to meet the diagnostic criteria. Therefore, the aim of this study was to investigate the expression of NRG1, ErbB4, BDNF, DNMT1 and TET1 in patients' serum for the diagnosis of schizophrenia. The sensitivity, specificity and percentage of correctly classified patients were analyzed by using Receiver Operating Characteristic (ROC) curve analysis. The diagnostic efficiency of the combination of these five serum proteins was evaluated by multivariable logistic regression.
Methods
Subject selection
Patients in this study were recruited from the department of psychiatry of the Second Xiangya Hospital, Central South University, after written informed consent of participation was provided. Totally 53 drug-naïve patients with first-episode schizophrenia (26 female and 27 male) and 57 healthy controls (28 female and 29 male) were analyzed in this study. The ages of these patients ranged from 17 to 55 years old and the mean age was (28.15 ± 10.42) years old. The duration of illness was more than 1 month. All the patients were diagnosed formally according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-V) and evaluated using Positive and Negative Symptom Scale (PANSS) by a senior psychiatrist. The ages of healthy controls ranged from 18 to 54 years old and the mean age was (31.33 ± 10.69) years old. Patients were excluded from the study if they met one or more of the following criteria: other mental disorders, alcohol or substance abuse, malignant tumor, active or chronic inflammatory or autoimmune disease, diabetes mellitus, obesity (BMI > 30 kg/m2), heavy smoking (more than 18 cigarettes per day) and treatment with anti-inflammatory or immunosuppressive medication. This study was approved by the Ethics Committee of Second Xiangya Hospital, Central South University.
Enzyme-linked immunosorbent assay (ELISA)
Firstly, four milliliter venous blood was withdrawn from SZ patients and the corresponding controls in the morning into procoagulant tube prior to administration of any medication. Serum was separated by centrifugation (3500 r/min, 5 min) from coagulated blood, then was collected and stored at -80℃ until analysis. Serum NRG1, ErbB4, BDNF, DNMT1 and TET1 protein levels were measured by commercially available ELISA kits (NRG1β1/ErbB4/BDNF, R&D Systems, Minneapolis MN; DNMT1/TET1, Cusabio, Wuhan, China) following the manufacturer's instructions. The 96-well micro plates were incubated overnight with monoclonal antibody at 4℃. Samples and standard proteins were added after incubation with blocking sample buffer. Plates were then treated with enzyme-labeled polyclonal antibody. Then, H2O2 was added and the color was developed after addition of TMB solution. After adding 2 mol/L H2SO4 to stop the reaction, the absorbance at 450 nm were measured on micro plate reader. Protein concentrations were determined according to the standard curve.
Statistical Analysis
The data were statistically analyzed using SPSS version 18.0 (SPSS, Chicago, IL). Normal distribution variables were shown as mean ± standard deviation and non-normal distribution variables were shown as median and interquartile range. Categorical data were analyzed using the χ2 test. Continuous data were analyzed using Student's t-test if they displayed a standard normal distribution or Mann-Whitney U test when the variables had a skewed distribution. Kolmogorov-Smirnov test was used to assess normal distribution. Spearman correlation coefficients were calculated for associations among variables. Multivariable logistic regression including biomarker concentration as covariates was used to predict SZ. Differentiating performance of these five serum proteins for the diagnosis of Schizophrenia was tested by ROC curve analysis and the area under the curve (AUC) was calculated. The optimal cut-off point was obtained from the Youden index [maximum (sensitivity + specificity - 1)]. A p-value < 0.05 was considered as statistically significant.
Results
The demographic data of SZ patients
The demographic data of SZ patients and healthy controls were presented in Table 1. There were no significant differences in the mean age, gender, BMI and the number of cigarettes consumed per day between SZ patients and controls (p > 0.05).
Demographic data of SZ patients and controls.
| Variables | SZ (n=53) | Control (n=57) | P |
|---|---|---|---|
| Male/female | 27/26 | 29/28 | 1.000 |
| Age | 28.15 ± 10.42 | 31.33 ± 10.69 | 0.117 |
| Body mass index | 22.06 ± 3.34 | 20.96 ± 3.14 | 0.079 |
| Smokers (%) | 11 (20.75) | 14 (24.56) | 0.657 |
| No. of cigarettes smoked per day | 14.18 ± 2.93 | 12.29 ± 2.13 | 0.073 |
| Family history of psychosis | |||
| Yes (%) | 33 (62.26) | N/A | N/A |
| NO (%) | 20 (37.74) | N/A | N/A |
| Schizophrenia subtypes | |||
| Paranoid (%) | 27 (50.94) | N/A | N/A |
| Disorganized (%) | 11 (20.76) | N/A | N/A |
| Catatonic (%) | 2 (3.77) | N/A | N/A |
| Others (%) | 13 (24.53) | N/A | N/A |
| Total PANSS score | 75.81 ± 21.35 | N/A | N/A |
| Positive Symptom score | 16.58 ± 7.47 | N/A | N/A |
| Negative Symptom Score | 18.93 ± 7.83 | N/A | N/A |
Abbreviations: SZ: schizophrenia, PANSS: positive and negative symptom scores. N/A: not applicable.
NRG1
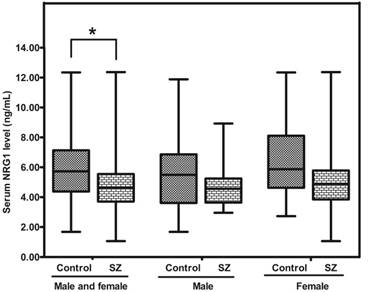
The concentration of NRG1 in SZ patients was significantly lower than that in controls [4.64 (range: 3.71-5.55) vs 5.73 (range: 4.38-7.13) ng/mL; p = 0.014], whereas there was no difference between SZ males and Control (CTR) males (p = 0.096), or between SZ females and CTR females (p = 0.089) (Figure. 1).
Representative plot showing concentrations of NRG1 protein in peripheral blood of SZ patients (n = 53) and controls (n = 57). * means P-value < 0.05, range error bars encompass the lowest and highest values. SZ: patients with first-episode schizophrenia.
ErbB4
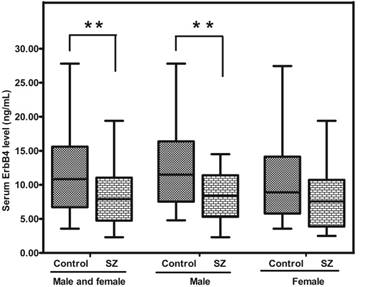
Similar with NRG1, ErbB4 expression in the peripheral blood of SZ patients was significantly lower than that in healthy controls [7.90 (range: 4.75-11.05) vs 10.83 (range: 6.72-15.60) ng/mL; p = 0.002]. Interestingly, SZ males presented a much less ErbB4 than control males (p = 0.005), while no statistical significance between SZ (female) and CTR (female) was found (p = 0.098) (Figure. 2).
BDNF
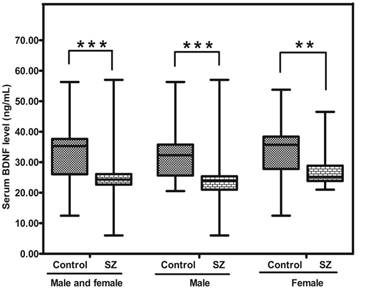
The level of BDNF protein in SZ patients was significantly lower than that in controls [24.30 (range: 22.65-26.10) vs 35.30 (range: 26.05-37.63) ng/mL; p = 0.000]. In addition, the level of BDNF protein in SZ (male) or SZ (female) was apparently lower than that in CTR (male) (p = 0.000) and CTR (female) respectively (p = 0.005) (Figure. 3).
DNMT1
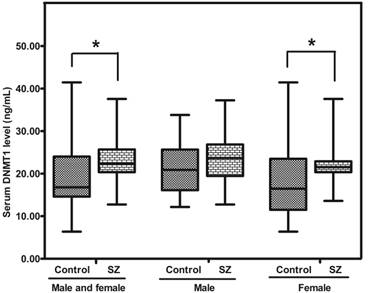
The level of DNMT1 protein in SZ was greatly higher than that in the CTR [22.35 (range: 20.36-25.67) vs 16.79 (range: 14.60-24.01) ng/mL; p = 0.011]. Meanwhile, the level of DNMT1 protein in SZ (female) was higher than that in CTR (female) (p = 0.030), while no significant difference was revealed between SZ (male) and CTR (male) (p = 0.165) (Figure. 4).
TET1
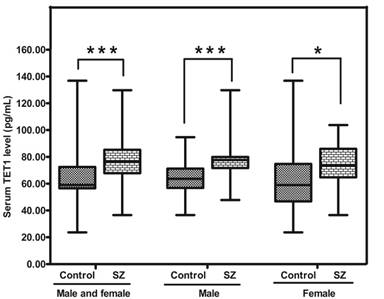
The level of TET1 protein in SZ was significantly higher (p<0.05) than that in CTR [76.50 (range: 67.85-85.25) vs 59.06 (range: 56.55-72.40) pg/mL; p= 0.000]. Moreover, the abundances of TET1 protein in SZ (male) and SZ (female) were obviously higher than those in CTR (male) (p = 0.000)and CTR (female) respectively (p= 0.049) (Figure 5).
Representative plot displaying ErbB4 protein level in peripheral blood of SZ patients (n = 53) and controls (n = 57). ** means P-value < 0.01, range error bars encompass the lowest and highest values. SZ: patients with first-episode schizophrenia.
Representative figure showing BDNF protein level in peripheral blood of SZ patients (n = 53) and controls (n = 57). ** means P-value < 0.01, *** means P-value < 0.001, range error bars encompass the lowest and highest values. SZ: patients with first-episode schizophrenia.
Representative graph showing DNMT1 protein in peripheral blood from SZ patients (n = 53) and Controls (n = 57). *means P-value<0.05, range error bars encompass the lowest and highest values. SZ, patients with first-episode schizophrenia.
Representative figure showing TET1 protein in peripheral blood from SZ patients (n = 53) and controls (n = 57). * means P-value<0.05, *** means P-value<0.001, range error bars encompass the lowest and highest values. SZ, patients with first-episode schizophrenia.
The influences of the clinical features
We also assessed the influences of the clinical features on the five serum biomarkers levels using Spearman correlations. However, no significant correlations were validated between the concentrations of the five serum proteins with age, gender, Body mass index, the rate of PANSS and smoking history among the participants with SZ (p > 0.05).
Diagnostic efficiency of combining five proteins in serum
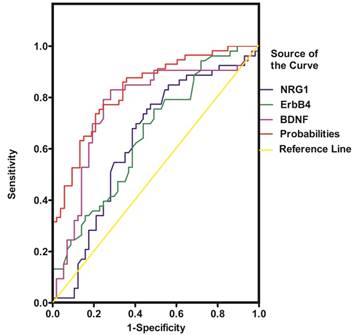
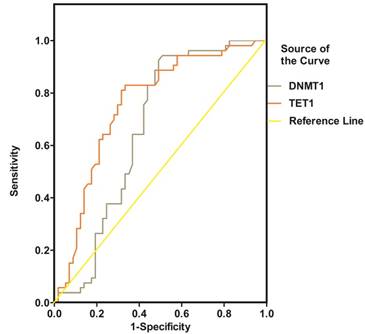
NRG1, ErbB4, BDNF, DNMT1 and TET1 were all promising predictors of SZ in the univariable logistic regression model. However, in the multivariable model, only ErbB4, BDNF and TET1 were independently associated with SZ (Table 2). The diagnostic efficiency of these three proteins was evaluated by the sensitivity, specificity, Youden index and the area under the ROC curve (AUC) (Table 3). The ROC curves for the protein concentrations were shown in Figure 6- Figure 7. A continuous combination variable model constructed from these three proteins, reported the AUC to be 0.825 (Table 3). The model provided that 41 (77.4%) of the original cases were correctly placed in SZ group, and 42 (73.7%) were correctly classified in control group. Our results have showed that the diagnostic model can accurately distinguish the SZ patients.
Predictors of schizophrenia (SZ).
| Univariable analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|
| OR (95%Cl) | p value | OR (95%Cl) | p value | ||
| NRG1 | 0.830(0.693-0.994) | 0.043 | |||
| ErbB4 | 0.884(0.814-0.960) | 0.003 | 0.883(0.803-0.972) | 0.011 | |
| BDNF | 0.904(0.857-0.954) | 0.000 | 0.915 (0.866-0.967) | 0.001 | |
| DNMT1 | 1.083(1.016-1.154) | 0.015 | |||
| TET1 | 1.050(1.022-1.080) | 0.000 | 1.052 (1.022-1.083) | 0.001 | |
OR: odds ratio, 95% CI: 95% confidence interval, NRG1: neuregulin1, BDNF: brain-derived neurotrophic factor, DNMT1: DNA methyltransferases 1, TET1: ten-eleven translocation 1.
Results of Roc Curve Analysis between the two Groups.
| Analysis | NRG1 | ErbB4 | BDNF | DNMT1 | TET1 | Probabilities |
|---|---|---|---|---|---|---|
| Sensitivity | 0.774 | 0.755 | 0.792 | 0.943 | 0.830 | 0.811 |
| Specificity | 0.526 | 0.509 | 0.754 | 0.491 | 0.667 | 0.737 |
| Youden index | 0.300 | 0.263 | 0.547 | 0.435 | 0.497 | 0.548 |
| AUC | 0.636 | 0.654 | 0.764 | 0.661 | 0.755 | 0.825 |
| Cutoff (ng/mL, pg/mL for TET1) | 5.660 | 10.765 | 26.350 | 16.675 | 65.750 | 0.461 |
| 95%CI for AUC | 0.530-0.742 | 0.553-0.756 | 0.669-0.859 | 0.555-0.767 | 0.662-0.848 | 0.747-0.903 |
ROC: receiver operating characteristic curve, AUC: area under the curve, NRG1: neuregulin1, BDNF: brain-derived neurotrophic factor, DNMT1: DNA methyltransferases 1, TET1: ten-eleven translocation 1, Probabilities: probabilities of serum ErbB4, BDNF, and TET1 levels.
ROC curve of NRG1, ErbB4 and BDNF for the diagnosis of schizophrenia. Probabilities: continuous combination variable of ErbB4, BDNF and TET1.
ROC curve of DNMT1 and TET1 for the diagnosis of schizophrenia
Discussion
Previous studies have elucidated the possibility to establish a distinct molecular network signature relevant to disease process of SZ using standard biochemical methods [12, 13]. A profile of multiple serum biomarkers associated with neuronal nutrition, neuroimmunology, and neurologic function might provide much more convincing outcome for early diagnosis of SZ compared to single marker alone[13]. In this study, we assessed the concentration of five serum proteins related to the pathophysiology of SZ to evaluate the potential function of blood markers profiling in the diagnosis of SZ patients using ROC analysis. Table 3 showed the AUC of NRG1, ErbB4, BDNF, DNMT1 and TET1 were 0.636, 0.654, 0.764, 0.661, and 0.755 respectively. Combining ErbB4, BDNF and TET1 serum markers gives the highest accuracy for separating SZ patients from healthy controls (AUC = 0.825; sensitivity = 0.811; specificity = 0.737). Furthermore, the classification with cross-validation provided that 77.4% and 73.7% of the original cases were correctly placed in SZ and control group respectively, which further suggested that a combined serum markers including ErbB4, BDNF, and TET1 may be used for the correct diagnosis of SZ.
The association between SZ and NRG1/ErbB4 signaling
In line with previous reports [14], we also detected a much lower serum concentrations of NRG1 and ErbB4 in SZ patients. The association between SZ and NRG1/ErbB4 signaling is not surprising. Individuals with decreased NRG1 mRNA in peripheral blood lymphocytes have been demonstrated to have higher risk of developing psychosis later [15]. Previous studies have shown that NRG1 promotes GABA release in mouse cortical and hippocampal slices through ErbB4 [16, 17], which was expressed specifically in interneurons [18, 19]. Intriguingly, NRG1/ErbB4 signaling remains saturated in the amygdala, which maintains high GABAergic activity and modulates the output of parvalbumin-positive interneurons [20, 21]. We proposed that decreased activity of the NRG1/ErbB4 signaling might impair the GABAergic activity, which may prompt the development of SZ. Although the serum concentrations of NRG1 and ErbB4 proteins are low, the accuracy of NRG1 and ErbB4 as diagnostic biomarkers for SZ is not high(sensitivity = 0.774 or 0.755, specificity = 0.526 or 0.509). In addition, only male patients present significantly less ErbB4 protein and thus we assume that sexual hormones especially estrogen may play a protective role in the process of schizophrenia. The gender of patients should be considered if the biomarkers are used for diagnosis of schizophrenia.
However, the results of the expression of NRG1 and ErbB4 in the schizophrenia patients still remain debatable. Petryshen et al [22] found that the expression of NRG1 and ErbB4 were up-regulated in the hippocampus and Chong et al [23] reported a protein concentration change in the prefrontal cortex. These differences may partially contribute to degrees of alteration in risk genes[24], or the duration of the modification on NRG1 signaling[25]. Besides, the specification of antibodies used in different studies against NRG1 and ErbB4 may also contribute to the differences. In addition, the complex symptoms of schizophrenia might be relevant to such inconsistencies, which need further elucidation.
The role of BDNF, DNMT1 and TET1 in SZ
It is well-documented that the concentration of BDNF is down-regulated in either serum[7] or brain[7, 26-31] of SZ patients, In the present study, we also discovered a much lower concentration of BDNF and a much higher lever of DNMT1 and TET1 in peripheral blood of SZ patients. It was previously reported that DNMT1 and TET1 were highly enriched in GABAergic neurons in the hippocampus of adult human brains and peripheral blood lymphocytes[7, 27], and BDNF in neurons and peripheral blood mononuclear cells[32]. Moreover, the decreased BDNF in the SZ patient brains was associated with the increased 5-methyl cytosine at the BDNF promoter [33]. Therefore, TET1 might interact synergistically with DNMT1 to induce transcriptional repression by directly acting at BDNF promoter[34]. As a matter of fact, these epigenetic alterations are not correlated with duration of illness, which suggests that instead of the consequence, they are the primary cause of the disease [7]. Our statistical analyses showed that among the five candidate biomarkers, the sensitivity of DNMT was the highest (sensitivity = 0.943) while BDNF was of the highest specificity (specificity = 0.754) in the context of SZ prediction, which further highlighted the diagnostic values of these markers for SZ. Furthermore, we also revealed overt gender difference in the serum BDNF protein levels between SZ patients and controls.
However, it should be noted that a stable or even up-regulated BDNF has also been argued in the serum of SZ patients [35, 36], therefore, the obtained results in the present study should be repeated by a further study with a large sample size. As potential diagnostic biomarkers, it will be helpful to monitor the concentrations of these 5 proteins prior to and after antipsychotics treatment. Another limitation is caused by the fact that we did not demonstrate other disorders (eg, depressive disorder) which share the common features of SZ. In addition, the concentrations of these 5 proteins before versus after antipsychotics treatment were not monitored.
In summary, this study has demonstrated the concentration changes of NRG1, ErbB4, BDNF, DNMT1 and TET1 in serum of SZ patients and suggested that they may play a critical role in the pathophysiology of SZ. The combination test of these five proteins has showed promising efficiency for the diagnosis of SZ and could be further explored for early diagnosis and clinical assessment of the schizophrenia.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81771448, No. 81503276) and the Hunan Provincial Natural Science Foundation of China (No. 2015JJ4069).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fazzari P, Paternain AV, Valiente M. et al. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464(7293):1376-1380
2. Hou XJ, Ni KM, Yang JM. et al. Neuregulin 1/ErbB4 enhances synchronized oscillations of prefrontal cortex neurons via inhibitory synapses. Neuroscience. 2014:261107-261117
3. Pitcher GM, Kalia LV, Ng D. et al. Schizophrenia susceptibility pathway neuregulin 1-ErbB4 suppresses Src upregulation of NMDA receptors. Nature medicine. 2011;17(4):470-478
4. Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Current opinion in cell biology. 2007;19(2):124-134
5. Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38(1):138-166
6. Houston I, Peter CJ, Mitchell A. et al. Epigenetics in the human brain. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38(1):183-197
7. Auta J, Smith RC, Dong E. et al. DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients. Schizophrenia research. 2013;150(1):312-318
8. Roth TL, Lubin FD, Funk AJ. et al. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological psychiatry. 2009;65(9):760-769
9. Favalli G, Li J, Belmonte-de-Abreu P. et al. The role of BDNF in the pathophysiology and treatment of schizophrenia. Journal of psychiatric research. 2012;46(1):1-11
10. Vinogradov S, Fisher M, Holland C. et al. Is serum brain-derived neurotrophic factor a biomarker for cognitive enhancement in schizophrenia? Biological psychiatry. 2009;66(6):549-553
11. Vasic N, Connemann BJ, Wolf RC. et al. Cerebrospinal fluid biomarker candidates of schizophrenia: where do we stand? European archives of psychiatry and clinical neuroscience. 2012;262(5):375-391
12. Schmidt HD, Shelton RC, Duman RS. Functional biomarkers of depression: diagnosis, treatment, and pathophysiology. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2011;36(12):2375-2394
13. Xiong P, Zeng Y, Wu Q. et al. Combining serum protein concentrations to diagnose schizophrenia: a preliminary exploration. The Journal of clinical psychiatry. 2014;75(8):e794-801
14. Zhang HX, Zhao JP, Lv LX. et al. Explorative study on the expression of neuregulin-1 gene in peripheral blood of schizophrenia. Neuroscience letters. 2008;438(1):1-5
15. Kiss I, Kelemen O, Keri S. Decreased peripheral expression of neuregulin 1 in high-risk individuals who later converted to psychosis. Schizophrenia research. 2012;135(1-3):198-199
16. Chen Y, Hancock ML, Role LW. et al. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30(27):9199-9208
17. Wen L, Lu YS, Zhu XH. et al. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(3):1211-1126
18. Shamir A, Kwon OB, Karavanova I. et al. The importance of the NRG-1/ErbB4 pathway for synaptic plasticity and behaviors associated with psychiatric disorders. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32(9):2988-2997
19. Woo RS, Li XM, Tao Y. et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54(4):599-610
20. Bi LL, Sun XD, Zhang J. et al. Amygdala NRG1-ErbB4 is critical for the modulation of anxiety-like behaviors. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2015;40(4):974-986
21. Lu Y, Sun XD, Hou FQ. et al. Maintenance of GABAergic activity by neuregulin 1-ErbB4 in amygdala for fear memory. Neuron. 2014;84(4):835-846
22. Petryshen TL, Middleton FA, Kirby A. et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Molecular psychiatry. 2005;10(4):366-374 328
23. Chong VZ, Thompson M, Beltaifa S. et al. Elevated neuregulin-1 and ErbB4 protein in the prefrontal cortex of schizophrenic patients. Schizophrenia research. 2008;100(1-3):270-280
24. Banerjee A, Macdonald ML, Borgmann-Winter KE. et al. Neuregulin 1-erbB4 pathway in schizophrenia: From genes to an interactome. Brain research bulletin. 2010;83(3-4):132-139
25. Savonenko AV, Melnikova T, Laird FM. et al. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(14):5585-5590
26. Dong E, Ruzicka WB, Grayson DR. et al. DNA-methyltransferase1 (DNMT1) binding to CpG rich GABAergic and BDNF promoters is increased in the brain of schizophrenia and bipolar disorder patients. Schizophrenia research. 2015;167(1-3):35-41
27. Guidotti A, Auta J, Chen Y. et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60(7-8):1007-1016
28. Niitsu T, Ishima T, Yoshida T. et al. A positive correlation between serum levels of mature brain-derived neurotrophic factor and negative symptoms in schizophrenia. Psychiatry research. 2014;215(2):268-273
29. Ray MT, Shannon Weickert C, Webster MJ. Decreased BDNF and TrkB mRNA expression in multiple cortical areas of patients with schizophrenia and mood disorders. Translational psychiatry. 2014:4 e389
30. Ruzicka WB, Zhubi A, Veldic M. et al. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Molecular psychiatry. 2007;12(4):385-397
31. Sotiropoulou M, Mantas C, Bozidis P. et al. BDNF serum concentrations in first psychotic episode drug-naive schizophrenic patients: associations with personality and BDNF Val66Met polymorphism. Life sciences. 2013;92(4-5):305-310
32. Sarchielli P, Greco L, Stipa A. et al. Brain-derived neurotrophic factor in patients with multiple sclerosis. Journal of neuroimmunology. 2002;132(1-2):180-188
33. Gavin DP, Sharma RP, Chase KA. et al. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2012;37(2):531-542
34. Guidotti A, Auta J, Davis JM. et al. Toward the identification of peripheral epigenetic biomarkers of schizophrenia. Journal of neurogenetics. 2014;28(1-2):41-52
35. Huang TL, Lee CT. Associations between serum brain-derived neurotrophic factor levels and clinical phenotypes in schizophrenia patients. Journal of psychiatric research. 2006;40(7):664-668
36. Niitsu T, Shirayama Y, Matsuzawa D. et al. Associations of serum brain-derived neurotrophic factor with cognitive impairments and negative symptoms in schizophrenia. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35(8):1836-1840
Author contact
![]() Corresponding author: Yamei Tang, Department of Laboratory Medicine, The Second Xiangya Hospital, Central South University, Changsha 410011, Hunan, China Tel: +86-0731-85292037. Fax: +86-0731-85533525. E-mail address: yameitang3287edu.cn
Corresponding author: Yamei Tang, Department of Laboratory Medicine, The Second Xiangya Hospital, Central South University, Changsha 410011, Hunan, China Tel: +86-0731-85292037. Fax: +86-0731-85533525. E-mail address: yameitang3287edu.cn

 Global reach, higher impact
Global reach, higher impact