Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(9):717-723. doi:10.7150/ijms.16345 This issue Cite
Research Paper
Regulatory SNPs Alter the Gene Expression of Diabetic Retinopathy Associated Secretary Factors
1. VYM Genome Research Center, National Yang-Ming University, Taipei, Taiwan;
2. Department of Ophthalmology, Taipei City Hospital, Taipei, Taiwan;
3. Department of Ophthalmology, Shin Kong Wu Ho-Su Memorial Hospital, Taipei, Taiwan;
4. Department of Ophthalmology, Taipei Medical University, Taipei, Taiwan;
5. Department of Ophthalmology, Taiwan University Hospital, Taipei, Taiwan;
6. Department of Internal Medicine, Taipei City Hospital, Taipei, Taiwan;
7. Department of Health Care Management, National Taipei University of Nursing and Health Sciences, Taipei, Taiwan;
8. Department of Medicine, National Yang-Ming University, Taipei, Taiwan;
9. Institute of Public Health, National Yang-Ming University, Taipei, Taiwan.
Received 2016-6-1; Accepted 2016-7-30; Published 2016-9-12
Abstract
Objectives: Diabetic retinopathy (DR) is a common microvascular complication in both type I and type II diabetes. Several previous reports indicated the serum centration of some secretary factors were highly associated with DR. Therefore, we hypothesis regulatory SNPs (rSNPs) genotype in secretary factors may alter these gene expression and lead to DR.
Methods: At first, pyrosequencing were applying to screen the SNPs which present allele frequency different in DR and DNR. Then individual genotyping was processed by Taqman assays in Taiwanese DR and DNR patients. To evaluate the effect of SNP allele on transcriptional activity, we measured promoter activity using luciferase reporter constructs.
Results: We found the frequencies of the CC, CG, and GG genotype of the rs2010963 polymorphism were 15.09%, 47.14%, and 37.74% in DR and 12.90%, 19.35%, and 67.74% in DNR, respectively (p = 0.0205). The prevalence of DR was higher (p = 0.00793) in patients with the CC or CG genotype (62.26% and 32.26% for DR and DNR, respectively) compared with the patients with the GG genotype. To evaluate the effect of rs2010963-C allele on transcriptional activity, we measured promoter activity using luciferase reporter constructs. The rs2010963-C reporter showed 1.6 to 2-fold higher luciferase activity than rs2010963-G in 3 cell lines.
Conclusion: Our data proposed rs2010963-C altered the expression level of VEGFA in different tissues. We suggested small increase but long term exposure to VEGFA may lead to DR finally.
Keywords: DR, SNP, VEGFA.
Introduction
Diabetic retinopathy (DR) is a common microvascular complication of diabetes and causes new cases of blindness among adults [1, 2]. It begins with mild nonproliferative abnormalities, follows by moderate and severe nonproliferative DR (NDR), and finally progresses to proliferative DR (PDR) [1, 2]. Almost all patients with type 1 diabetes and more than 60% of patients with type 2 diabetes are expected to develop retinopathy during the first decade of occurrence of diabetes [3, 4]. With the incidence of diabetes increasing in the world, the incidence of DR is expected to rise to alarming levels [5]. Diabetes duration, poor glycemic control, and hypertension are known as the primary risk factors for development of DR [5, 6]. However, some patients were found in clinical observation that had poorly controlled or long lasting duration diabetes but did not develop retinopathy, whereas others even with relatively good glycemic control developed advanced retinopathy eventually [7]. These clinical observations suggested there are other factors involved in the development of DR.
Recent studies suggest that genetic factors are important for DR. For example, a family study of Mexican-Americans with type 2 diabetes found that severe retinopathy in the proband is significantly associated with severe retinopathy in siblings [8]. A study of identical twins found a concordance for DR in 68% of type 1 and 95% of type 2 diabetes cases [9]. To explore the candidate genes associated with DR, several genome-wide associated studies were conducted to detect potential regions involved in the etiology of DR [10, 11]. By gene approach studies, several potential candidate genes in DR has been reported such as receptor for advanced glycation end products (RAGE) [12, 13], vascular endothelial growth factor (VEGF) [14-17], erythropoietin (EPO) [13, 18] and pigment epithelium derived factor (PEDF) [13]. However, these studies were not reproduced in multiple populations and molecular mechanism still remain unconfirmed.
Single nucleotide polymorphism (SNP) is a DNA sequence variation occurring commonly within a population. This change in the DNA sequence may affect the process of gene expression if they occur in the putative regulatory region [19, 20]. In several previous reports, the serum centration of some DR associated genes were highly associated with DR [21-27]. Therefore, we hypothesis regulatory SNPs (rSNPs) genotype may alter gene expression of DR associated secretary factors.
Material and Methods
Sample collection
The study protocol was approved by the Research Ethics Committee of Taipei City Hospital (TCHIRB-990501, TCHIRB-1000103, and TCHIRB-1020404) and adhered to the tenets of the Declaration of Helsinki. Written informed consent was obtained from all participants before their enrollment. Eighty-four patients who had type 2 diabetes mellitus for more than 10 years (as defined by age of onset being later than 30 years of age) and exclusively used oral hypoglycemic agents were enrolled between time duration from the patient population of the Taipei City Hospital Zhongxing Branch. The criteria for diagnosis of diabetes were random blood sugars levels of over 150 mgs% and HbA1C levels of over 6%. A comprehensive ophthalmological examination included corrected visual acuity, slit-lamp biomicroscopy, and dilated fundus photography. Seven fields of 30°color fundus photographs with stereoscopic images of the optic disc and macula were taken through the dilated pupils of each patient, using a digital fundus camera (Kowa, Japan). Patients with no signs of diabetic retinopathy coupled with type 2 diabetes mellitus were assigned to the DNR study group. Patients with significant diabetic retinopathy as defined by eyes with definite neovascularization and/or vitreous/preretinal hemorrhages/microaneurysms were included in the DR study group.
Genotyping by Pyrosequencing
SNP screening analyses were performed by PSQ96MA System (Qiagen, Hilden, Germany) to determine the allele frequencies in DR and DNR population. Equal amount of genomic DNA (1ug/patient) from DR or DNR patients were equally mix as DR or DNR genomic DNA pools. Then the SNP containing DNA fragments were amplified by PCR. The internal pyrosequencing primer was designed and allele frequencies were analysis using PyroMark software (Qiagen, Hilden, Germany).
Individual Genotyping by TaqMan assays
Individual genotyping of SNPs in both DR and DNR was performed using TaqMan assays according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). The DNA samples were extracted and purified from the peripheral lymphocytes according to the manufacture's protocol. The genotyping success rate was at least 98%.
Reporter Assay
We constructed four reporter plasmids encompassing - 1272 to - 225 bp of the VEGFA promoter (+1 relative to the first nucleotide of transcription start site) which could represent the genotypes of rs2010963. The rs2010963-C was obtained by PCR from using HEK293T cell DNA. The PCR products of the VEGFA promoter were ligated into pGL4 (Promega, Madison, WI). To avoid interference from other SNPs in the VEGFA promoter region, we constructed the rs2010963-G reporter vector using the point mutation kit (Agilent, Santa Clara, CA). After incubation for 24 h, cells which transfected with reporter plasmid were collected and analyzed for luciferase activity with Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
Statistical analysis
We used the Golden Helix Package (Golden Helix, Inc., Bozeman, MT) for the statistical analysis of genotypes. The chi-square test was applied to compare discrete variables. The results were shown as hazard ratio and 95% confidence interval (CI). We considered P-values below 0.05 as statistically significant.
Results
Pyrosequencing for rSNPs Allele Frequencies in DR and DNR DNA Pools
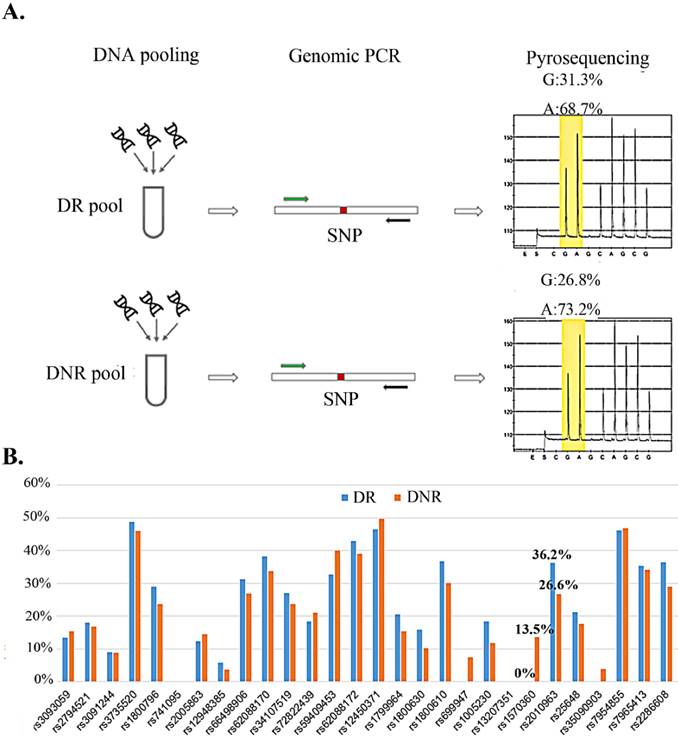
We selected nine secretary factors as our candidates for their serum centration has been reported to associate with DR in previous reports [21-27]. To explore the relation for the rSNP and the secretary factors, the SNPs which located on promoter region and minimal allele frequency > 10% in Mongoloid (information from dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) were selected as screening candidates (Table 1). Pyrosequencing technology was applying for rSNP screening for it could determine the SNP allele frequencies in DNA pools [28]. Taiwanese patients with type 2 diabetes and with or without any signs of DR were allocated to DR and DNR groups, respectively. Equal amount of genomic DNA from DR or DNR patients were equally mix as DR or DNR pools for pyrosequencing (Fig. 1A). We observed two SNPs (rs1570360 and rs2010963) presented about 10 % allele frequencies different between DR and DNR groups (Fig. 1B).
Individual genotyping for rs1570360 and rs2010963 in patients
Both rs1570360 and rs2010963 were processed individual genotyping analysis by Taqman assays. There was no genotype different for rs1570360 in DR and DNR. However, the CC, CG, and GG genotypes of the rs2010963 polymorphism were 15.09%, 47.14%, and 37.74% in DR and 12.90%, 19.35%, and 67.74%, in DNR, respectively (p=0.0205) (Table 2). The prevalence of DR was higher (p=0.00793) in subjects with the CC or CG genotype (62.26% and 32.26% for DR and DNR) compared with that in patients careering with the GG genotype (Table 3). These results suggest that the C allele of the rs2010963 is dominant relative to the G allele.
Effect of the rs2010963 polymorphism on transcriptional activity
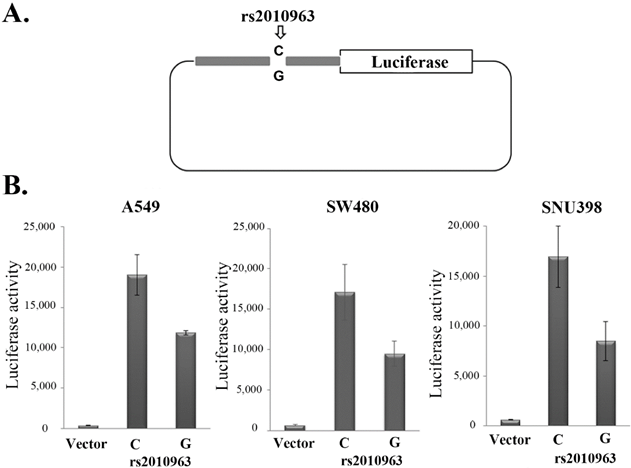
In previous studies of the VEGFA promoter, the rs2010963-G allele was suggested to generate the potential SP1 binding site [29], while the rs2010963-C generate potential GAPBα and the IRF1, 2 binding sites [30, 31]. To indicate the polymorphism of rs2010963 alter the VEGFA transcriptional activity, we generated two luciferase reporter vectors of identical sequences except for rs2010963, spanning -1272 to -225 bp of the VEGFA promoter region (Fig.2A). The rs2010963-C reporter showed 1.61, 1.8, and 2-fold higher luciferase activity compared with rs2010963-G in A549, SW480, and SNU398 (Fig.2B). Our results suggest rs2010963-C produce more VEGFA in the cell lines from different tissues.
rSNPs selection from 9 DR associated secretary factors.
| Gene | SNP | Location | Genotype | |
|---|---|---|---|---|
| CRP | C-reactive protein | rs3093059 | 5' near gene | A/G |
| rs2794521 | 5' near gene | C/T | ||
| rs3091244 | 5' near gene | A/C/T | ||
| HGF | hepatocyte growth factor | rs3735520 | 5' near gene | A/G |
| IL6 | interleukin 6 | rs1800796 | intron 1 | C/G |
| PGF | placental growth factor | rs741095 | 5' near gene | A/G |
| rs2005863 | 5' near gene | G/T | ||
| SERPINF1 | serpin peptidase inhibitor, clade F, member 1 | rs12948385 | 5' near gene | A/G |
| rs66498906 | intron 1 | A/G | ||
| rs62088170 | intron 1 | C/T | ||
| rs34107519 | intron 1 | A/G | ||
| rs72822439 | intron 1 | C/T | ||
| rs59409453 | intron 1 | A/G | ||
| rs62088172 | intron 1 | C/T | ||
| rs12450371 | intron 1 | C/T | ||
| TNF | tumor necrosis factor | rs1799964 | 5' near gene | C/T |
| rs1800630 | 5' near gene | A/C | ||
| rs1799724 | 5' near gene | C/T | ||
| rs1800610 | intron 1 | A/G | ||
| VEGFA | vascular endothelial growth factor A | rs699947 | 5' near gene | A/C |
| rs1005230 | 5' near gene | C/T | ||
| rs13207351 | 5' near gene | A/G | ||
| rs1570360 | 5' near gene | A/G | ||
| rs2010963 | exon 1 5'UTR | C/G | ||
| rs25648 | exon 1 | C/G/T | ||
| VEGFB | vascular endothelial growth factor B | rs35090903 | 5' near gene | C/T |
| VWF | von Willebrand factor | rs7954855 | 5' near gene | C/T |
| rs7965413 | 5' near gene | C/T | ||
| rs2286608 | intron 1 | A/G | ||
Genotype distribution of rs1570360 and rs2010963 in DR and DNR groups.
| Gene | SNP | Genotype | DR(n=53) | DNR(n=31) | p-value | ||
|---|---|---|---|---|---|---|---|
| VEGFA | rs1570360 | GG | 33 | 62.26% | 20 | 64.52% | |
| AG | 18 | 33.96% | 10 | 32.26% | 0.9757 | ||
| AA | 2 | 3.78% | 1 | 3.22% | |||
| VEGFA | rs2010963 | GG | 20 | 37.74% | 21 | 67.74% | 0.0205 |
| CG | 25 | 47.17% | 6 | 19.35% | |||
| CC | 8 | 15.09% | 4 | 12.90% | |||
Dominant and recessive model analysis of rs2010963 in DR and DNR groups.
| Genotype | odds ratio (95% CI) | p-value | |||||
|---|---|---|---|---|---|---|---|
| rs1570360 | AA | AG+GG | |||||
| DR (n=53) | 2 | 3.78% | 51 | 96.22% | 1.1765 (0.1023~13.5314) | 0.8962 | |
| DNR(n=31) | 1 | 3.23% | 30 | 96.77% | |||
| GG | AG+AA | ||||||
| DR (n=53) | 33 | 62.26% | 20 | 37.74% | 0.9075 (0.361~2.2814) | 0.8365 | |
| DNR(n=31) | 20 | 64.52% | 11 | 35.48% | |||
| rs2010963 | CC | CG+GG | |||||
| DR (n=53) | 8 | 15.09% | 45 | 84.91% | 1.2 (0.3298~4.3664) | 0.782 | |
| DNR(n=31) | 4 | 12.90% | 27 | 87.10% | |||
| GG | CG+CC | ||||||
| DR (n=53) | 20 | 37.73% | 33 | 62.26% | 0.2886 (0.1132~0.7357) | 0.0092 | |
| DNR(n=31) | 21 | 67.74% | 10 | 32.26% | |||
Allele frequency determine in DNA pool by pyrosequencing. A) The workflow of allele frequency determination in DNA pools by pyrosequencing. B) Minor allele frequency for 29 SNPs in DR and DNR pools.
Effects of the rs2010963 variants on luciferase reporter expression. A) The construction map of pGL4 luciferase reporter containing VEGF promoter sequence with the allele C or G allele at SNP rs2010963. B) The reporter vector with the allele C or G allele at SNP rs2010963 were transfected into A549, sw480 and SNU398 cells.
Discussion
In this study, we analyzed the association of 29 rSNPs which selected from 9 DR associated secretary factors in an independent cohort of Taiwanese patients with type two diabetes. At first, pyrosequencing analysis the allele frequency showed rs1570360 and rs2010963 present about 10% different allele frequency between DR and DNR pools. Individual genotyping of all our patients indicated rs2010963-C allele significant association with DR. Furthermore, our luciferase reporter assay showed the rs2010963-C allele could enhance the expression of VEGFA.
VEGF encompasses wide family of growth factors including VEGF-A,-B,-C,-D,-E, and placental growth factor (PGF) [32]. The best known and most widely used in clinical practice is VEGFA. VEGFs promote angiogenesis, cause the breakdown of the blood retinal barrier, stimulate the growth of endothelial cell, induce neovascularization, and increase vascular permeability in the ischemic retina [6, 33, 34]. Increased expression levels of VEGF and its receptors have been observed in DR patients [35, 36]. Therefore, several anti-VEGF agents are developed to treat DR such as pegaptanib, ranibizumab, and bevacizumab [37-39]. For the expression of VEGFA is important for the progression of DR, SNPs of VEGFA have been analyzed in several associated studies [14, 16, 34, 36, 40].
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is an effective method to analyze the interaction of protein and DNA. The Encyclopedia of DNA Elements (ENCODE) project (https://genome.ucsc.edu/ENCODE/) has performed more than 500 ChIP-seq experiments for hundreds transcription factors in several cell lines. Base on the CHIP-seq data from ENCODE project, 12 transcription factors binding fragments were crossed the region of rs2010963 (sup Fig.1). One of the transcription factors, TBP (TATA box binding protein), has been indicated to correlate with the expression of VEGFA in previous report [41]. In addition, our luciferase reporter assay provides direct evidence that rs2010963-C altered the expression level of VEGFA in 3 cell lines from different tissues. As we know VEGFA is a secreted factor and expressed in almost all the tissues [42]. Even though our data showed that rs2010963-C increase 1.6 to 2 folds relative to rs2010963-G, it still could be possible to lead to DR by long term exposure.
In summary, our study suggests that rs2010963 is an important genetic marker for DR in Taiwanese patients with type two diabetes. Moreover, our experiments provide a direct association between rs2010963-C and serum levels of VEGFA.
Abbreviations
DR, Diabetic retinopathy; SNP, Single nucleotide polymorphism; CRP, C-reactive protein; HGF, hepatocyte growth factor; IL6, interleukin 6; PGF, placental growth factor; SERPINF1, serpin peptidase inhibitor, clade F, member 1; TNF, tumor necrosis factor; VEGFA, vascular endothelial growth factor A; VEGFB, vascular endothelial growth factor B; VWF, von Willebrand factor.
Acknowledgements
We acknowledge the technical services provided by the High-throughput Genome Analysis Core Facility of the National Yang-Ming University VGH Genome Research Center (VYMGC). The Core Facility is supported by the National Core Facility Program for Biotechnology (NCFPB), Ministry of Science and Technology.
Funding
This study was supported by the grants from Department of Health, Taipei City Government (99001-62-031, 10001-62-019, and 10201-62-074) and Ministry of Science and Technology of Taiwan (MOST 104-2314-B-532-005).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. The New England journal of medicine. 2012;366:1227-39
2. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376:124-36
3. Klein R, Klein BE, Moss SE. Epidemiology of proliferative diabetic retinopathy. Diabetes care. 1992;15:1875-91
4. Tasman W. The natural history of active retinopathy of prematurity. Ophthalmology. 1984;91:1499-503
5. Zorena K, Raczynska D, Raczynska K. Biomarkers in diabetic retinopathy and the therapeutic implications. Mediators of inflammation. 2013;2013:193604
6. Tarr JM, Kaul K, Chopra M, Kohner EM, Chibber R. Pathophysiology of diabetic retinopathy. ISRN ophthalmology. 2013;2013:343560
7. Sobrin L, Green T, Sim X, Jensen RA, Tai ES, Tay WT. et al. Candidate gene association study for diabetic retinopathy in persons with type 2 diabetes: the Candidate gene Association Resource (CARe). Investigative ophthalmology & visual science. 2011;52:7593-602
8. Hallman DM, Huber JC Jr, Gonzalez VH, Klein BE, Klein R, Hanis CL. Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes care. 2005;28:1163-8
9. Leslie RD, Pyke DA. Diabetic retinopathy in identical twins. Diabetes. 1982;31:19-21
10. Fu YP, Hallman DM, Gonzalez VH, Klein BE, Klein R, Hayes MG. et al. Identification of Diabetic Retinopathy Genes through a Genome-Wide Association Study among Mexican-Americans from Starr County, Texas. Journal of ophthalmology. 2010. 2010
11. Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Human molecular genetics. 2011;20:2472-81
12. Ramprasad S, Radha V, Mathias RA, Majumder PP, Rao MR, Rema M. Rage gene promoter polymorphisms and diabetic retinopathy in a clinic-based population from South India. Eye. 2007;21:395-401
13. Balasubbu S, Sundaresan P, Rajendran A, Ramasamy K, Govindarajan G, Perumalsamy N. et al. Association analysis of nine candidate gene polymorphisms in Indian patients with type 2 diabetic retinopathy. BMC medical genetics. 2010;11:158
14. Churchill AJ, Carter JG, Ramsden C, Turner SJ, Yeung A, Brenchley PE. et al. VEGF polymorphisms are associated with severity of diabetic retinopathy. Investigative ophthalmology & visual science. 2008;49:3611-6
15. Bonnefond A, Saulnier PJ, Stathopoulou MG, Grarup N, Ndiaye NC, Roussel R. et al. What is the contribution of two genetic variants regulating VEGF levels to type 2 diabetes risk and to microvascular complications? PloS one. 2013;8:e55921
16. Yang X, Deng Y, Gu H, Ren X, Li N, Lim A. et al. Candidate gene association study for diabetic retinopathy in Chinese patients with type 2 diabetes. Molecular vision. 2014;20:200-14
17. Errera FI, Canani LH, Silva ME, Yeh E, Takahashi W, Santos KG. et al. Functional vascular endothelial growth factor -634G>C SNP is associated with proliferative diabetic retinopathy: a case-control study in a Brazilian population of European ancestry. Diabetes care. 2007;30:275-9
18. Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X. et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6998-7003
19. Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med (Berl). 2005;83:97-109
20. Wang X, Tomso DJ, Liu X, Bell DA. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicol Appl Pharmacol. 2005;207:84-90
21. Zhou ZW, Ju HX, Sun MZ, Fu QP, Chen HM, Ji HB. et al. Serum fetuin-A levels are independently correlated with vascular endothelial growth factor and C-reactive protein concentrations in type 2 diabetic patients with diabetic retinopathy. Clin Chim Acta. 2016;455:113-7
22. Canton A, Burgos R, Hernandez C, Mateo C, Segura RM, Mesa J. et al. Hepatocyte growth factor in vitreous and serum from patients with proliferative diabetic retinopathy. Br J Ophthalmol. 2000;84:732-5
23. Zhou J, Wang S, Xia X. Role of intravitreal inflammatory cytokines and angiogenic factors in proliferative diabetic retinopathy. Curr Eye Res. 2012;37:416-20
24. Kowalczuk L, Touchard E, Omri S, Jonet L, Klein C, Valamanes F. et al. Placental growth factor contributes to micro-vascular abnormalization and blood-retinal barrier breakdown in diabetic retinopathy. PloS one. 2011;6:e17462
25. Patel JI, Tombran-Tink J, Hykin PG, Gregor ZJ, Cree IA. Vitreous and aqueous concentrations of proangiogenic, antiangiogenic factors and other cytokines in diabetic retinopathy patients with macular edema: Implications for structural differences in macular profiles. Exp Eye Res. 2006;82:798-806
26. Numata K, Fukuda H, Ohto M, Itou R, Nozaki A, Kondou M. et al. Evaluation of the therapeutic efficacy of high-intensity focused ultrasound ablation of hepatocellular carcinoma by three-dimensional sonography with a perflubutane-based contrast agent. Eur J Radiol. 2010;75:e67-75
27. Sun CY, Lee CC, Hsieh MF, Chen CH, Chou KM. Clinical association of circulating VEGF-B levels with hyperlipidemia and target organ damage in type 2 diabetic patients. J Biol Regul Homeost Agents. 2014;28:225-36
28. Neve B, Froguel P, Corset L, Vaillant E, Vatin V, Boutin P. Rapid SNP allele frequency determination in genomic DNA pools by pyrosequencing. Biotechniques. 2002;32:1138-42
29. Rueda B, Perez-Armengol C, Lopez-Lopez S, Garcia-Porrua C, Martin J, Gonzalez-Gay MA. Association between functional haplotypes of vascular endothelial growth factor and renal complications in Henoch-Schonlein purpura. The Journal of rheumatology. 2006;33:69-73
30. Rueda B, Gonzalez-Gay MA, Lopez-Nevot MA, Garcia A, Fernandez-Arquero M, Balsa A. et al. Analysis of vascular endothelial growth factor (VEGF) functional variants in rheumatoid arthritis. Human immunology. 2005;66:864-8
31. Rueda B, Lopez-Nevot MA, Lopez-Diaz MJ, Garcia-Porrua C, Martin J, Gonzalez-Gay MA. A functional variant of vascular endothelial growth factor is associated with severe ischemic complications in giant cell arteritis. The Journal of rheumatology. 2005;32:1737-41
32. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nature medicine. 2003;9:669-76
33. Comer GM, Ciulla TA. Pharmacotherapy for diabetic retinopathy. Current opinion in ophthalmology. 2004;15:508-18
34. Zhang XL, Wen L, Chen YJ, Zhu Y. Vascular endothelial growth factor up-regulates the expression of intracellular adhesion molecule-1 in retinal endothelial cells via reactive oxygen species, but not nitric oxide. Chinese medical journal. 2009;122:338-43
35. Vailati FB, Crispim D, Sortica DA, Souza BM, Brondani LA, Canani LH. The C allele of -634G/C polymorphism in the VEGFA gene is associated with increased VEGFA gene expression in human retinal tissue. Investigative ophthalmology & visual science. 2012;53:6411-5
36. Jain A, Saxena S, Khanna VK, Shukla RK, Meyer CH. Status of serum VEGF and ICAM-1 and its association with external limiting membrane and inner segment-outer segment junction disruption in type 2 diabetes mellitus. Molecular vision. 2013;19:1760-8
37. Adamis AP, Altaweel M, Bressler NM, Cunningham ET Jr, Davis MD, Goldbaum M. et al. Changes in retinal neovascularization after pegaptanib (Macugen) therapy in diabetic individuals. Ophthalmology. 2006;113:23-8
38. Avery RL, Pearlman J, Pieramici DJ, Rabena MD, Castellarin AA, Nasir MA. et al. Intravitreal bevacizumab (Avastin) in the treatment of proliferative diabetic retinopathy. Ophthalmology. 2006;113:1695 e1-15
39. Chun DW, Heier JS, Topping TM, Duker JS, Bankert JM. A pilot study of multiple intravitreal injections of ranibizumab in patients with center-involving clinically significant diabetic macular edema. Ophthalmology. 2006;113:1706-12
40. Han L, Zhang L, Xing W, Zhuo R, Lin X, Hao Y. et al. The associations between VEGF gene polymorphisms and diabetic retinopathy susceptibility: a meta-analysis of 11 case-control studies. Journal of diabetes research. 2014;2014:805801
41. Bieche I, Vacher S, Vallerand D, Richon S, Hatem R, De Plater L. et al. Vasculature analysis of patient derived tumor xenografts using species-specific PCR assays: evidence of tumor endothelial cells and atypical VEGFA-VEGFR1/2 signalings. BMC Cancer. 2014;14:178
42. McFee RM, Cupp AS. Vascular contributions to early ovarian development: potential roles of VEGFA isoforms. Reprod Fertil Dev. 2013;25:333-42
Author contact
![]() Corresponding author: Ching-Yao Tsai, MD & PhD, 145 Zheng Zhou Rd, Department of Ophthalmology, Taipei City Hospital, Taipei, Taiwan. E-mail: dac58gov.tw
Corresponding author: Ching-Yao Tsai, MD & PhD, 145 Zheng Zhou Rd, Department of Ophthalmology, Taipei City Hospital, Taipei, Taiwan. E-mail: dac58gov.tw

 Global reach, higher impact
Global reach, higher impact