Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(1):77-84. doi:10.7150/ijms.13301 This issue Cite
Research Paper
Identification of microRNAs in Throat Swab as the Biomarkers for Diagnosis of Influenza
1. Department of Health Inspection and Quarantine, School of Public Health, Sun Yat-sen University, Guangzhou, China
2. Shenzhen Entry-exit Inspection and Quarantine Bureau, Shenzhen, China
3. Biochemistry Programme, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China
4. Guangdong Medical University, Zhanjiang, China
5. Shenzhen Key Lab of Health Science and Technology, Division of Life Sciences & Health, Graduate School at Shenzhen, Tsinghua University, Shenzhen, China
6. College of Optoelectronics Engineering, Key Laboratory of Optoelectronic Devices and Systems, Ministry of Education and Guangdong Province, Shenzhen Key Laboratory of Sensor Technology, Shenzhen University, Shenzhen, China
* These authors contributed equally to this work.
Received 2015-7-21; Accepted 2015-12-15; Published 2016-1-25
Abstract
Background: Influenza is a serious worldwide disease that captures global attention in the past few years after outbreaks. The recent discoveries of microRNA (miRNA) and its unique expression profile in influenza patients have offered a new method for early influenza diagnosis. The aim of this study was to examine the utility of miRNAs for the diagnosis of influenza.
Methods: Thirteen selected miRNAs were investigated with the hosts' throat swabs (25 H1N1, 20 H3N2, 20 influenza B and 21 healthy controls) by real-time quantitative polymerase chain reaction (RT-qPCR) using U6 snRNA as endogenous control for normalization, and receiver operating characteristic (ROC) curve/Area under curve (AUC) for analysis.
Results: miR-29a-3p, miR-30c-5p, miR-34c-3p and miR-181a-5p are useful biomarkers for influenza A detection; and miR-30c-5p, miR-34b-5p, miR-205-5p and miR-449b-5p for influenza B detection. Also, use of both miR-30c-5p and miR-34c-3p (AUC=0.879); and miR-30c-5p and miR-449b-5p (AUC=0.901) are better than using one miRNA to confirm influenza A and influenza B infection, respectively.
Conclusions: Given its simplicity, non-invasiveness and specificity, we found that the throat swab-derived miRNAs miR-29a-3p, miR-30c-5p, miR-34b-5p, miR-34c-3p, miR-181a-5p, miR-205-5p and miR-449b-5p are a useful tool for influenza diagnosis on influenza A and B.
Keywords: miRNA, RT-qPCR, influenza, throat swab, biomarkers.
Background
Influenza caused by the RNA influenza virus infection has become a global public health concern because of its potential to spark pandemics and economic losses [1-2]. According to its antigenicity of nucleoprotein (NP) and M protein, it is divided into Type-A, -B and -C and the Type-A and -B are the most common seasonal influenza types [3]. Because of antigenic drift, Type-A influenza (infA) virus is the major pathogenic agent for recurrent epidemics and, more rarely, pandemics. Since 1918, several infA pandemics have been emerged and caused millions of deaths. Influenza pandemics occurred in 1918 (H1N1), 1957 (H2N2), 1968 (H3N2) [4], 2009 (H1N1) [5], and again in 2013 (H7N9) [6]. Therefore, developing a rapid, highly sensitive and specific diagnostic test for infA infection is urgently needed.
There are many influenza virus tests currently available based on the detection of immunological or genetic fingerprints. Real-time quantitative polymerase chain reaction (RT-qPCR) test on genetic fingerprints of influenza virus is considered to be the most reliable and safe method [7,8]. However, the antigenic drift from errors caused by the non-proofreading RNA polymerase and the antigenic shift from gene reassortment bar the traditional methods to report the influenza virus of unknown type. In this connection, developing a new diagnostic assay for new biomarkers may facilitate better treatments and prognosis assessment for patients with influenza virus infection.
MicroRNAs (miRNAs) are a class of small non-coding RNAs with around 22 nucleotides [9]. Studies have shown that they regulate a wide variety of biological processes including cell proliferation and apoptosis [10]. Recently, several miRNAs have been reported to be associated with influenza virus infection and replication [11]. Zhu et al. identified 153 miRNAs (146 up-regulated and 7 down-regulated) in the serum of patients with the extremely virulent and highly pathogenic H7N9 avian virus infection [12]. In 2009, He et al. found 36 pig-encoded miRNAs and 22 human-encoded miRNAs as the putative targets respectively in the swine influenza virus and Swine-Origin 2009 A/H1N1 influenza virus infected hosts [13].
To date, studies detecting the miRNAs associated with influenza virus use serum and saliva as the starting materials. Studies have also shown that the miRNAs can be stably detected in saliva samples [14,15]. Patel et al. demonstrated the possibility and feasibility of using saliva to isolate miRNAs for the detection of oral cancers [16]. Wu et al. reported that miRNA-144 is highly expressed in the saliva of patients with esophageal cancer [17]. In fact, according to the World Health Organization (WHO), saliva, because of the dilution effect, is not a good biological sample for disease marker detection. In well-equipped medical settings, serum is used commonly for detecting viral infection. Because of its invasive nature, more patient care and services are needed. Yet, results have showed that serum sample might cause more false negative results when compared to that of throat swab sample for rubella virus infection [18]. In clinics and quarantine stations, throat swab, nasal swab, nasopharyngeal swab, nasopharyngeal aspirate and nasal wash are standard protocols to obtain samples for the diagnosis of upper respiratory tract viral infections according to the WHO website [19]. Among these non-invasive sampling methods, the throat swab is the simplest because it does not need any special equipment such as vacuum for aspiration [19]. In the light of this, we investigated in this study the differential expression of potential miRNA biomarkers in the throat swabs of healthy controls and influenza patients.
The aims of this study were (i) to establish a standard method to isolate miRNAs from small volumes of throat swab sample from patients with influenza infection and (ii) to identify the miRNAs obtained from the throat swabs as biomarkers for the diagnosis of influenza.
Materials and Methods
Clinical samples
A total of 86 throat swab samples (25 patients with H1N1, 20 with H3N2, 20 with influenza B (infB) infection and 21 healthy controls) were obtained from Shenzhen Entry-exit Inspection and Quarantine Bureau, Shenzhen, China. Healthy controls were obtained randomly from individuals who had not been suffered from respiratory disease. Influenza patients recruited in this study were confirmed as being either infected with H1N1, H3N2 or infB virus by conventional RT-qPCR with standard primers. All samples were stored at -70°C after collection according to standard procedures. Participants' information is summarized in Table 1 and no statistically significant difference was found between the influenza and control group for the age and gender distribution (p > 0.05). Written informed consent was obtained from all participants before sample and data collection and this study was ethically approved by Shenzhen Entry-exit Inspection and Quarantine Bureau, Shenzhen, China.
The basic characteristics of healthy subjects and patients with influenza A (infA) H1N1, H3N2 and influenza B (infB) virus infection.
| Sample Characteristics | Healthy Control | infA-Patients | infB-Patients | |
|---|---|---|---|---|
| H1N1-Patients | H3N2-Patients | |||
| Number | 21 | 10 | 10 | 20 |
| Sex (male/female) | 14/7 | 13/12 | 13/7 | 14/6 |
| Age (mean ± SD) | 28.76±16.44 | 26.4±21.7 | 22.31±17.49 | 31.48±22.2 |
| Infectious diseases | None | H1N1 only | H3N2 only | infB only |
Sample preparation
Briefly, human throat swab samples were collected and each swab was stored in a sterile EP tube with 5 mL Hank's balanced salt solution. Then 200 μL throat swab solution was aliquoted and stored at -70°C. The whole process was carefully executed to avoid contamination. Our preliminary work confirmed that throat swab samples obtained from this procedure had contained various miRNAs (Figure S1).
miRNA isolation
The miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) was used to isolate total RNA including miRNAs from the throat swab solution according to the supplier's instructions. RNA was eluted from the RNeasy MinElute spin-column membrane with the ultrapure RNase-free water at room temperature. RNA yield and purity were determined with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington DE) through absorbance measurements at 260 and 280 nm. Only the samples with a 260/280 ratios ≥ 1.6 were used. Samples of isolated total RNA were stored at -70 °C until use. Table S1 lists the concentrations and purity of the miRNAs.
Quantitative RT-PCR
Complementary DNA (cDNA) of each miRNA was synthesized by reverse transcription with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY) according to the manufacturer's instructions by using miRNA-specific stem-loop primers (Table 2). The stem-loop at the 5'-end of the primer is designed to prevent the primers from annealing to other mature miRNA variants and the miRNA precursors (e.g. primary and pre-miRNAs), thus increasing the specificity and efficiency in the reverse transcription of a distinct form of mature miRNA to cDNA. Five nanograms of total RNA from individual throat swabs were used as the starting material. All cDNA samples were stored at -20°C until RT-qPCR analysis.
For the detection of mature miRNAs, a RT-qPCR was performed in triplicate using ABI7500 with the TaqMan MiRNA Assay (Applied Biosystems). The PCR reaction mixture of 20 µL contained: 9 µL miRNA-specific cDNA, 10 µL TaqMan 2x Universal PCR Master Mix (No AmpErase UNG), 1 µL gene-specific TaqMan MiRNA real-time PCR-Assay primer (Applied Biosystems). The thermal cycling conditions were set as follows: initial activation of Taq polymerase at 95 °C for 10 min, 40 cycles of PCR amplification at 95 °C for 15 sec, annealing/elongation at 60 °C for 1 min. Each throat swab sample for each candidate miRNA was run in triplicate. The expression level of each miRNA in an individual sample was normalized with the U6 snRNA to compensate sample variations. The relative expression of miRNA was calculated by the 2-ΔΔCt formula, in which ΔCt = Ct miRNA - Ct U6 snRNA, ΔΔCt = ΔCtpatient - ΔCtControl.
Selection and validation of throat swab miRNAs
A panel of 13 miRNAs (miR-29a-3p, miR-30c-3p, miR-34b-5p, miR-34c-3p, miR-105-3p, miR-151a-5p, miR-181a-5p, miR-200c-3p, miR-205-5p, miR-491-5p, miR-449b-5p, miR-671-5p and let-7g-5p) (Table 2) were selected based on the information in literature [3,18] and were measured in an independent cohort of 86 throat swabs with RT-qPCR. The stem-loop primers for the RT-qPCR of these miRNAs were shown in Table 2.
Statistical analysis
Statistical significance of the data was evaluated using one-way ANOVA via SPSS 13.0 and GraphPad Prism 6.01 (GraphPad Software Inc., USA). A p-value < 0.05 was considered statistically significant. Receiver operating characteristic (ROC) curve analysis was performed using the MedCalc software (version 11.4.2.0) to evaluate the diagnostic utility of the selected miRNAs. In addition, the area under the curve (AUC) values and 95% confidence intervals (CIs) were calculated to evaluate the specificity and sensitivity of the miRNA for detecting the influenza infection. The Bonferroni method was used for multiple comparison adjustment. Data were expressed as the mean ± SD.
Results
Yield of miRNAs from throat swabs
We obtained high yields of RNAs including miRNAs from as little as 200 μL out of 5 mL sample solution from throat swabs using commercial kits. This method was proven for the first time to isolate high yield of RNAs from throat swabs stored at -70 °C for up to 1 year. Table S1 shows the OD 260/280 measurements and the concentrations for the individual samples. The yield from 200 μL of throat swabs was found in the range of 45.0 to 915.0 ng.
Internal reference selection
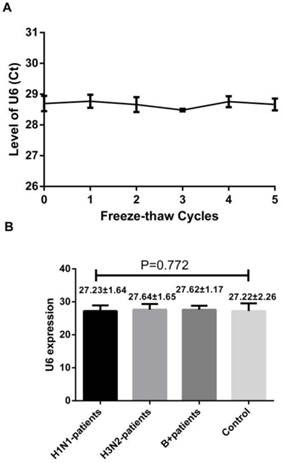
A reliable internal control should not be differentially expressed in influenza samples and control subjects and must be stable in the throat swab samples as well. U6 snRNA, a small nuclear RNA, has been selected as the normalization control for serum miRNA profiling study as it is extremely stable [9]. To confirm these, we first evaluated the stability of the U6 snRNA with repeated freeze-thaw cycles (0-5 cycles) by using RT-qPCR. We also evaluated its levels in the throat swab samples of healthy controls, H1N1, H3N2 and infB patients. Our results showed no significant difference between the samples obtained from influenza patients and healthy controls. Also, they were stable even after 5 freeze-thaw cycles testing (Figure 1, p = 0.928, one-way ANOVA). These results indicate that the U6 snRNA is a good internal control.
Level of U6 snRNA after freezing and thawing cycles (A) and its level in the healthy subjects and patients with influenza infection (B). Results are mean ± SD (n=86), for (B), p=0.772, one-way ANOVA.
Identification of miRNAs as the influenza markers by RT-qPCR analysis
To identify the miRNAs with statistically significant differences between influenza infection and healthy controls, we tested the expression levels of 13 miRNAs (Table 2) which were selected based on the response from hosts and cultured cells with influenza infection [3,19].
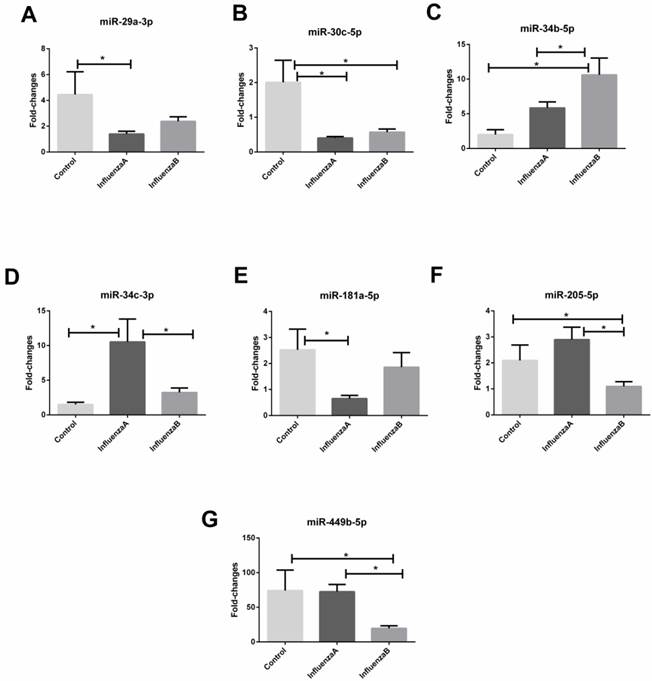
With an independent cohort of 86 throat swabs, results from the RT-qPCR show that the detection rate of miR-105-3p was <10% and the miR-491-5p had a very high Ct value (>35). Also, four other miRNAs (miR-151a-5p, miR-200c-3p, miR-671-5p and let-7g-5p) did not show any differences between the influenza samples and the control (p-value >0.05). These miRNAs were therefore excluded from further analysis. Consequently, the other seven miRNAs (miR-29a-3p, miR-30c-3p, miR-34b-5p, miR-34c-3p, miR-181a-5p, miR-205-5p, and miR-449b-5p) with a differential expression in the infA, infB and control groups were chosen as the candidate biomarkers. Figure 2 shows that miR-29a-3p, miR-30c-5p miR-34c-3p and miR-181a-5p could distinguish infA patients from healthy subjects, while miR-30c-5p, miR-34b-5p, miR-205-5p and miR-449b-5p could differentiate infB patients from healthy controls (Table 3). The levels of these 7 miRNAs in infA of different clinical subtypes (H1N1 and H3N2) were compared and results were shown in Figure S2.
The miRNA sequences and specific stem-loop primers used in this study.
| Name | Sequence (5'- 3') |
|---|---|
| miR-29a-3p | UAG CAC CAU CUG AAA UCG GUU A |
| Stem-loop primer of miR-29a-3p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG TAA CCG AT |
| miR-30c-5p | UGU AAA CAU CCU ACA CUC UCA GC |
| Stem-loop primer of miR-30c-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG GCT GAG AG |
| miR-34b-5p | UAG GCA GUG UCA UUA GCU GAU UG |
| Stem-loop primer of miR-34b-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG CAA TCA GC |
| miR-34c-3p | AAU CAC UAA CCA CAC GGC CAG G |
| Stem-loop primer of miR-34c-3p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG CCT GGC CG |
| miR-105-3p | ACG GAU GUU UGA GCA UGU GCU A |
| Stem-loop primer of miR-105-3p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG TAG CTC AT |
| miR-151a-5p | UCG AGG AGC UCA CAG UCU AGU |
| Stem-loop primer of miR-151a-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG ACT AGA CT |
| miR-181a-5p | AAC AUU CAA CGC UGU CGG UGA GU |
| Stem-loop primer of miR-181a-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG ACT CAC CG |
| miR-200c-3p | UAA UAC UGC CGG GUA AUG AUG GA |
| Stem-loop primer of miR-200c-3p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG TCC ATC AT |
| miR-205-5p | UCC UUC AUU CCA CCG GAG UCU G |
| Stem-loop primer of miR-205-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG CAG ACT CC |
| miR-449b-5p | AGG CAG UGU AUU GUU AGC UGG C |
| Stem-loop primer of miR-449b-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG GCC AGC TA |
| miR-491-5p | AGU GGG GAA CCC UUC CAU GAG G |
| Stem-loop primer of miR-491-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG CCT CAT GG |
| miR-671-5p | AGG AAG CCC UGG AGG GGC UGG AG |
| Stem-loop primer of miR-671-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG CTC CAG CC |
| let-7g-5p | UGA GGU AGU AGU UUG UAC AGU U |
| Stem-loop primer of let-7g-5p | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG AAC TGT AC |
| U6 snRNA | TTC GTG AAG CGT TCC ATA TTT T |
| Stem-loop primer of U6 snRNA | CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG AAA ATA TG |
Comparisons of the miRNA levels in the throat swabs from the healthy controls and patients with influenza A, influenza B virus infection
| infA compared with Control | infB compared with Control | H1N1 compared with H3N2 | |||
|---|---|---|---|---|---|
| miRNAs | Relative change* | miRNAs | Relative change* | miRNAs | Relative change* |
| miR-29a-3p | ↓ | miR-30c-5p | ↓ | miR-29a-3p | ↑ |
| miR-30c-5p | ↓ | miR-34b-5p | ↑ | miR-30c-5p | ↓ |
| miR-34c-3p | ↑ | miR-205-5p | ↓ | miR-34b-5p | ↑ |
| miR-181a-5p | ↓ | miR-449b-5p | ↓ | miR-34c-3p | ↑ |
| miR-181a-5p | ↓ | ||||
| miR-449b-5p | ↓ | ||||
*↑: up-regulation; ↓: down-regulation.
Relative change in miRNAs isolated from throat swabs of healthy subjects (n=21), patients with infA (n=45) or infB infection (n=20). (A) miR-29a-3p, (B) miR-30c-5p, (C) miR-34b-5p, (D) miR-34c-3p, (E) miR-181a-5p, (F) miR-205-3p and (G) miR-449b-5p. Level of the miRNAs expression was normalized to the U6 snRNA and results are mean ± SD. Statistical analyses were determined by one-way ANOVA-test and the p-values were shown as indicated.
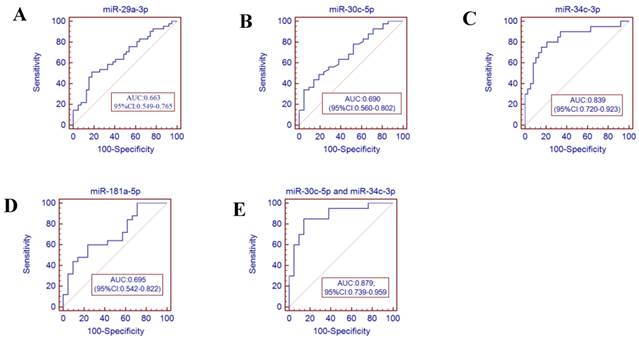
Receiver operator characteristics (ROC) curve analysis using the miRNAs isolated from the throat swabs to discriminate infA patients from healthy controls. (A) miR-29a-3p (AUC: 0.663 ,95% CI: 0.549-0.765), (B) miR-30c-5p (AUC: 0.690, 95% CI: 0.560-0.802), (C) miR-34c-3p (AUC: 0.839, 95% CI: 0.679-0.928), (D) miR-181a-5p (AUC: 0.695, 95% CI: 0.542-0.822), (E) miR-30c-5p and miR-34c-3p (AUC: 0.879, 95% CI: 0.739-0.959) , p<0.001. AUC: area under the ROC curve.
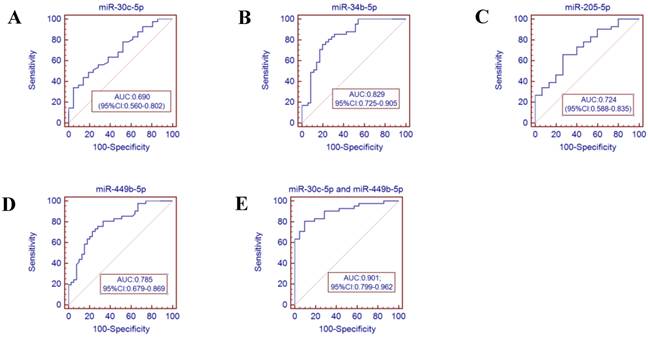
Receiver operator characteristics (ROC) curve analysis using the miRNAs isolated from the throat swabs to discriminate infB patients from healthy controls. (A) miR-30c-5p (AUC: 0.690, 95% CI: 0.560-0.802), (B) miR-34b-5p (AUC: 0.829, 95% CI 0.725-0.905), (C) miR-205-5p (AUC: 0.724, 95% CI: 0.588-0.835), (D) miR-449b-5p (AUC: 0.785, 95% CI: 0.679-0.869), (E) miR-30c-5p and miR-449b-5p (AUC: 0.901, 95% CI:0.799-0.962), p<0.001. AUC: area under the ROC curve.
Potential of miRNA biomarkers for diagnostic purpose
To further validate the diagnostic utility of the miRNA for influenza infection, we evaluated seven throat swab miRNAs (miR-29a-3p, miR-30c-5p, miR-34b-5p and miR-34c-3p, miR-181a-5p, miR-205-5p and miR-449b-5p) by ROC curve analysis. The results showed that the miR-34c-3p has the highest accuracy (AUC: 0.829, 95% CI: 0.679-0.928). The combination of miR-30c-5p and miR-34c-3p (AUC: 0.879 (p<0.001)) could discriminate infA patients from healthy controls, while the combination of miR-30c-5p and miR-449b-5p (AUC: 0.901 (p <0.0001)) could discriminate infB patients from healthy controls.
Discussion
Modulation of miRNA was firstly reported in mice during infection with the reconstructed 1918 (r1918) H1N1 virus and seasonal H1N1 virus (Tx/91) [21]. Subsequent works demonstrated that the highly pathogenic avian H5N1 influenza virus elicited distinct miRNA expression patterns in the host during infection; and the influenza virus H1N1, H7N7, H3N2, H5N1 and H7N9 induced miRNAs dysregulation in cultured cells, animal models and host [3,12,20,22-26]. In view of the importance of early diagnosis for surveillance and epidemiology studies, this non-invasive sample collecting from throat swab for miRNA study is cost effective and easy. To prove this collection method is viable and reliable, we found a highly stable internal reference U6 snRNA from throat swabs for normalization.
Our results indicate that the miR-29a-3p, miR-30c-5p, miR-34c-3p and miR-181a-5p are useful biomarkers for infA detection; and miR-30c-5p, miR-34b-5p, miR-205-5p and miR-449b-5p for infB detection. Also, using 2 miRNAs are better than using one to confirm the infA (miR-30c-5p and miR-34c-3p) and infB (miR-30c-5p and miR-449b-5p) infection.
Results from this study are consistent with the findings from literature. For example, miR-29a was found to be down-regulated in the cells and human infected with influenza H1N1 and H3N2 and mice infected with H1N1 [3, 27, 28]. miR-29a, miR-30c, miR-34b, miR-34c and miR-449b were down-regulated in A549 cells infected by influenza H1N1 strains [20]. Paresh et al. reported that the miR-29a was inversely correlated with the active HIV-1 replication in both cell models and HIV patients [29]. In addition, Zhu et al. found that this miRNA was up-regulated in the H7N9-infected serum samples but its up-regulation has yet to be confirmed by RT-PCR [12]. In addition, William et al. found that the influenza-induced expression of miR-449b interacted with the histone deacetylase HDAC1 to alter the IFN-β gene expression [30]. Loveday et al. also reported that miR-30c-5p was down-regulated 8 hours after infection in human A549 cells with swine-origin infA (S-OIV) pandemic H1N1 (2009) and the highly pathogenic avian-origin infA (A-OIV) H7N7 (2003) infection [20].
There are limitations in our study. For example, the miRNA expression patterns may be affected by other infectious agents and the immune status of the patients. For this, more samples should be used to further validate our conclusion. Second, the underlying mechanisms of the miRNAs dysregulation in the influenza patients are still elusive. This awaits further investigation.
In summary, the objectives of this study have been fulfilled and we established a simple method to extract miRNAs from small volumes of throat swab with high yield and stability. Also, we demonstrated an association between miRNA biomarkers from throat swabs with influenza virus infection. These biomarkers are miR-29a-3p, miR-30c-5p, miR-34b-5p, miR-34c-3p, miR-181a-5p, miR-205-5p and miR-449b-5p. They could be used to differentiate infA, infB patients from healthy controls. Also, they are useful for the diagnosis of H1N1 and H3N2 infection. We expect that this non-invasive approach using the throat swab miRNAs will be an effective way for influenza diagnosis in the coming future.
Supplementary Material
Table S1, Figs.S1-S2.
Acknowledgements
This research is supported by Shenzhen Science and Technology Foundation (No. JCYJ20120618172144495, No. JCYJ20140419151618022), Shenzhen Science and Technology R&D Foundation (No. ZYA201106080020A), Shenzhen Enter-exit Inspection and Quarantine Bureau (No. SZ2014101), State Quality Inspection Administration (No. 201310087), Shenzhen/Hong Kong Innovation Circle (SGLH20121008144756945), and ITF Grant (GHX/002/12SZ), CRF grant (CUHK1/CRF/12G) and AoE (AoE/P-0/12) funding from the Hong Kong Special Administrative Region.
The authors are grateful to the patients and subjects who participated in this study.
Ethical approval
Written informed consent was obtained from all participants before sample and data collection and this study was ethically approved by Shenzhen Entry-exit Inspection and Quarantine Bureau, Shenzhen, China.
Abbreviations
AUC, area under curve; cDNA, complementary DNA; CI, confidence interval; HA, hemagglutinin; infA, influenza A; infB, influenza B; miRNA, microRNA; NA, neuraminidase; NP, nucleoprotein; ROC, receiver operating characteristic; RT-qPCR, real-time quantitative polymerase chain reaction.
Competing interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
1. Wang H, Fu C, Li K. et al. Influenza associated mortality in Southern China, 2010-2012. Vaccine. 2014;32:973-978
2. Zhao JQ, Wang X, Ragupathy V. et al. Rapid Detection and Differentiation of Swine-Origin Influenza A Virus (H1N1/2009) from Other Seasonal Influenza A Viruses. Viruses. 2012;4:3012-3019
3. Terrier JT, Carron C, Marcel V. et al. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. Journal of General Virology. 2013;94:985-995
4. Kuo RL, Yang SL, Liu YC. et al. Influenza A/B virus detection and influenza A virus subtyping with emphasis on the novel H7N9 virus by using multiplex real-time RT-PCR. J Virol Methods. 2014;208:41-46
5. Ledesma J, Pozo F, Reina G. et al. Genetic diversity of influenza A(H1N1)2009 virus circulating during the season 2010-2011 in Spain. J Clin Virol. 2012;53:16-21
6. Wang W, Peng H, Tao Q. et al. Serologic assay for avian-origin influenza A (H7N9) virus in adults of Shanghai, Guangzhou and Yunnan, China. J Clin Virol. 2014;60:305-308
7. Kis Z, Jones J, Creanga A. et al. Real-time RT-PCR assay to differentiate clades of H5N1 avian influenza viruses circulating in Vietnam. J Virol Methods. 2013;193:452-458
8. Ghebremedhin B, Engelmann I, Konig W. et al. Comparison of the performance of the rapid antigen detection actim Influenza A&B test and RT-PCR in different respiratory specimens. J Med Microbio. 2014;58:365-370
9. Zhang S, Ouyang X, Jiang X, Gu D, Lin Y, Kong SK, Xie W. Dysregulated Serum MicroRNA Expression Profile and Potential Biomarkers in Hepatitis C Virus-infected Patients. Int J Med Sci. 2015;12(7):590-598
10. Zhang WC, Liu JB, Wang GS. The role of microRNAs in human breast cancer progression. Tumor Biol. 2014;35:6235-6244
11. Song L, Liu H, Gao S. et al. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol. 2010;84:8849-8860
12. Zhu Z, Qi Y, Ge A. et al. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses. 2014;6:1525-1539
13. He T, Chen HP, Wang L. et al. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation. 2009;4:112-118
14. Salazar C, Nagadia R, Pandit P. et al. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell Oncol (Dordr). 2014;37:331-338
15. Park NJ, Zhou H, Elashoff D. et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009;15:5473-5477
16. Patel RS, Jakymiw A, Yao B. et al. High resolution of microRNA signatures in human whole saliva. Archives of Oral Biology. 2011;56:1506-1513
17. Wu WD, Hou WJ, Wu ZF. et al. miRNA-144 in the saliva is a genetic marker for early diagnosis of esophageal cancer. J South Med Univ. 2013;33:1783-1786
18. Zhu Z, Xu W, Abernathy ES. et al. Comparison of Four Methods Using Throat Swabs To Confirm Rubella Virus Infection. J Clin Microbiol. 2007;45(9):2847-2852
19. WHO guidelines for the collection of human specimens for laboratory diagnosis of avian influenza infection. http://www.who.int/influenza/human_animal_interface/virology_laboratories_and_vaccines/guidelines_collection_h5n1_humans/en/
20. Loveday EK, Diederich S, Pasick J. et al. Temporal- and Strain-Specific Host MicroRNA Molecular Signatures Associated with Swine-Origin H1N1 and Avian-Origin H7N7 Influenza A Virus Infection. J Viro. 2012;86:6109-6122
21. Li Y, Chan EY, 1 Li JN. et al. MicroRNA Expression and Virulence in Pandemic Influenza Virus-Infected Mice. J Viro. 2010;84:3023-3032
22. Rogers JV, Price JA, Wendling MQS. et al. Preliminary MicroRNA Analysis in Lung Tissue to Identify Potential Therapeutic Targets Against H5N1 Infection. Viral Immunology. 2014;25:3-11
23. Song H, Wang Q, Guo Y. et al. Microarray analysis of MicroRNA expression in peripheral blood mononuclear cells of critically ill patients with influenza A (H1N1). BMC Infectious Diseases. 2013;13:257
24. Lam WY, Yeung ACM, Ngai Karry LK. et al. Effect of avian influenza A H5N1 infection on the expression of microRNA-141 in human respiratory epithelial cells. BMC Microbiology. 2013;13:104
25. Loveday EK, Diederich S, Pasick J. et al. Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. J Gen Viro. 2015;96:30-39
26. Othumpangat S, Noti JD, Beezhold DH. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology. 2014;468-470:256-264
27. Wu ZH, Li P, Zhang CF. et al. MicroRNA Expression Profile of Mouse Lung Infected with 2009 Pandemic H1N1 Influenza Virus. PLoS One. 2013;8:e74190
28. Loo JF, Wang SS, Peng F. et al. A non-PCR SPR platform using RNase H to detect MicroRNA 29a-3p from throat swabs of human subjects with influenza A virus H1N1 infection. Analyst. 2015;140:4566-4575
29. Patel P, Ansari MY, Bapat S. et al. The microRNA miR-29a is associated with human immunodeficiency virus latency. Retrovirology. 2014;11:108
30. Buggele WA, Krause KE, Horvath CM. Small RNA profiling of influenza A virus-infected cells identifies miR-449b as a regulator of histone deacetylase 1 and interferon beta. PLoS One. 2013;8:e76560
Author contact
![]() Corresponding authors: Siu Kai Kong and Dayong Gu, Siu Kai Kong: Biochemistry Programme, School of Life Sciences, The Chinese University of Hong Kong, Room 609, Mong Man Wai Building, Shatin, NT, Hong Kong, China; Tel: +852-3943-6799; Fax: +852-2603-5123; E-mail: skkongedu.hk or Dayong Gu: Institute of Disease Control and Prevention, Shenzhen International Travel Health Care Center, Shenzhen Entry-Exit Inspection and Quarantine Bureau, Shenzhen 518055, China; Tel: +86-755-83391344; Fax: +86-755-83391344; E-mail: wanhoodcom.
Corresponding authors: Siu Kai Kong and Dayong Gu, Siu Kai Kong: Biochemistry Programme, School of Life Sciences, The Chinese University of Hong Kong, Room 609, Mong Man Wai Building, Shatin, NT, Hong Kong, China; Tel: +852-3943-6799; Fax: +852-2603-5123; E-mail: skkongedu.hk or Dayong Gu: Institute of Disease Control and Prevention, Shenzhen International Travel Health Care Center, Shenzhen Entry-Exit Inspection and Quarantine Bureau, Shenzhen 518055, China; Tel: +86-755-83391344; Fax: +86-755-83391344; E-mail: wanhoodcom.

 Global reach, higher impact
Global reach, higher impact